Abstract
Despite being scarce in the human genome, active L1 retrotransposons continue to play a significant role in its evolution. Because of their recent expansion, many L1s are not fixed in humans, and, when present, their mobilization potential can vary among individuals. Previously, we showed that the great majority of retrotransposition events in humans are caused by highly active, or hot, L1s. Here, in four populations of diverse geographic origins (160 haploid genomes), we investigated the degree of sequence polymorphism of three hot L1s and the extent of individual variation in mobilization capability of their allelic variants. For each locus, we found one previously uncharacterized allele in every three to five genomes, including some with nonsense and insertion/deletion mutations. Single or multiple nucleotide substitutions drastically affected the retrotransposition efficiency of some alleles. One-third of elements were no longer hot, and these so-called cool alleles substantially increased the range of individual susceptibility to retrotransposition events. Adding the activity of the three elements in each individual resulted in a surprising degree of variation in mobilization capability, ranging from 0% to 390% of a reference L1. These data suggest that individual variation in retrotransposition potential makes an important contribution to human genetic diversity.
Keywords: human variation, population genetics, retrotransposon
Several types of DNA polymorphisms contribute to human genetic diversity. Among them are SNPs, microsatellite polymorphisms, variable number tandem repeats, copy number variation of large deletions and duplications, and presence/absence of young retrotransposons (1–3). Here, we describe another type of polymorphism, variation in retrotransposition capability, due to SNPs within hot L1s.
L1 retrotransposons have populated eukaryote genomes for >150 million years and account for ≈17% of the human genome (1). L1s are scattered throughout the genome and include 5′-truncated, rearranged, and mutated elements along with intact, full-length (FL) 6-kb copies (1, 4). Presently, L1s are the only autonomous mobile elements in the human genome (4). Whether active or inactive, L1-derived sequences contribute to genome variability by promoting ectopic recombination or by altering the regulatory properties and expression patterns of genes (5–8). However, only the youngest, active L1s can generate genomic variability through insertional mutagenesis (8–11), deletion (12–14), exon-shuffling (15), or transmobilization of processed pseudogenes (16) and nonautonomous sequences, like SVAs and Alus (17–19). Importantly, only mobile L1s can ensure their own survival in the human genome.
Mobile L1s are bicistronic entities flanked by a 5′ UTR carrying internal sense (20) and antisense (5) promoters and a 3′ UTR with a weak polyA signal. The 5′ cistron ORF1 encodes a 40-kDa RNA-binding protein (21) with chaperone activity (22). The 3′ cistron ORF2 encodes a 150-kDa protein with conserved endonuclease (23), reverse transcriptase (24), and zinc knuckle (25) domains. Both proteins are required for retrotransposition (23, 26), which occurs through a FL, polyadenylated RNA intermediate by target-primed reverse transcription that generates target site duplications flanking the retrotransposed copy (27, 28).
In order for an active L1 to affect an individual genome or a population, the L1 must first be present in the genome. This obvious requirement is not necessarily fulfilled by active L1s, because they belong to the youngest subfamily (L1-Ta). Indeed, their recent mobilization is reflected by presence/absence polymorphism in individuals and populations (29, 30), which represents a significant source of diversity in the contemporary human genome (2, 29–33). From activity analyses of L1 elements with functional ORFs in the human genome working draft (HGWD), we estimated that the average individual contains 80–100 potentially mobile L1s, and 6 L1s per haploid genome (12 per individual) are highly active or hot (34). Significantly, hot L1s account for the bulk of L1s known to retrotranspose in present-day humans (34).
The aggregate of active L1s carried by an individual determines his/her overall retrotransposition capability. A second individual, however, is likely to have a different set of active L1s and a different retrotransposition capability. Indeed, in addition to presence/absence polymorphism, large differences in retrotransposition activity of two alleles of LRE1, the first active human L1 isolated, demonstrated the potential contribution of alleles of active L1s to individual variability (35–37).
Based on these preliminary data, we analyzed sequence polymorphisms in three testable hot L1s (34) and the individual variation produced by the mobilization capacity of allelic variants. Each hot L1 was analyzed in 161–206 haploid genomes from different geographic origins. We found one previously uncharacterized allele in every three to five L1s sequenced and three to four wide-ranging activity levels per locus. Many alleles had activity levels <25% of a reference L1, and we call them cool. When the allelic activity potential at all three loci was combined, hot and cool L1s, along with presence/absence polymorphism, suggested that there is substantial individual variation in retrotransposition capability.
Results
Presence/Absence Polymorphism.
Three of six hot L1s described in ref. 34 were excluded from our study because two are very rare (Al356438 and Ac004200), and one is inserted within older L1 sequences (Al137845). We determined presence/absence frequencies for the remaining three hot elements in 161–206 haploid genomes of different geographic origins (Table 1). The frequency of each L1 varied from population to population (Table 1), but none of the elements departed significantly from Hardy–Weinberg equilibrium in any population. Overall insertion frequencies of L1A (Al512428), L1B (Ac02980), and L1C (Ac021017) were consistent with previous analyses (2, 29–31, 34), indicating that the Ta-1 subfamily is expanding in humans, and most of its members are polymorphic (29).
Table 1.
Insertion polymorphism frequencies and unbiased heterozygosity per population
| African | Asian | European | South American | Indo/Pak | Pacific | Total freq. | Het* | |
|---|---|---|---|---|---|---|---|---|
| L1A (A1512428) | n = 48 | n = 50 | n = 50 | n = 40 | n = 8 | n = 10 | n = 206 | |
| Freq. present | 0.35 | 0.12 | 0.16 | 0.1 | 0 | 0.5 | 0.19 | 0.31 |
| L1B (Ac02980) | n = 33 | n = 41 | n = 36 | n = 31 | n = 12 | n = 8 | n = 161 | |
| Freq. present | 0.67 | 0.12 | 0.42 | 0.68 | 0.33 | 0.63 | 0.46 | 0.5 |
| L1C (Ac021017) | n = 48 | n = 50 | n = 50 | n = 40 | n = 8 | n = 8 | n = 206 | |
| Freq. present | 0.1 | 0.86 | 0.44 | 0.45 | 0.3 | 0.63 | 0.46 | 0.5 |
n, number of genomes analyzed; Het*, unbiased heterozygosity; Pak, Pakistani; Freq., frequency.
Sequence Polymorphism.
We obtained complete sequence data for 35 of 40 genomes with L1A, 59 of 67 genomes with L1B, and 78 of 96 genomes with L1C (Fig. 1 A, B, and C). We found a previously uncharacterized allele in every 5 genomes for L1A and in every 3 genomes for L1B and L1C. L1A had 17 polymorphic sites among 8 alleles, whereas L1B and L1C had 19 and 26 polymorphic sites among 18 and 26 alleles, respectively. Unexpected changes included a 1-nt insertion in an allele of L1C, a 3-nt deletion shared by three alleles of L1B, and a total of four different nonsense mutations, two in a relatively common allele of L1A and two in separate alleles of L1C. Finding nonsense-containing alleles of active L1s led us to analyze nonsense-containing L1s in HGWD for active alleles, see Supporting Discussion in Supporting Text, which is published as supporting information on the PNAS web site. Nucleotide diversity levels fell within ranges described for humans across coding and noncoding regions (Table 2) (38–40).
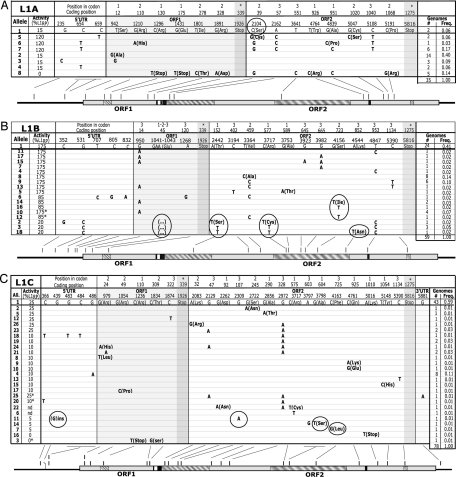
Fig. 1.
Alleles and activity variants of L1A, L1B, and L1C. (A–C Top) Nucleotide changes relative to HGWD sequence (allele 1). Amino acid changes are in parentheses. The retrotransposition activity (%L1rp) of each allele is shown at left. (A–C Bottom) A scaled L1 sequence. Lines indicate the location of each change. ORF1 and ORF2 (gray boxes) appear separated by the inter ORF (white box). Hatched boxes represent (left to right) leucine zipper, endonuclease, reverse transcriptase, and zinc knuckle. Black boxes in ORF2 represent sites A and B, putative ORF1p-binding sites to L1 RNA (57). (A) L1A. 17 polymorphic sites distributed in 8 alleles (35 genomes). A circle denotes the change responsible for an 87% reduction in activity. (B) L1B. Nineteen polymorphic sites distributed in 18 alleles (59 genomes). Circles indicate potential changes that reduce activity by 50–88%. (C) L1C. Twenty-six polymorphic sites distributed in 26 alleles (72 genomes). Circles denote changes potentially responsible for an 80% reduction in activity. For alleles marked with an asterisk or denoted nd, the activity was not tested because the alleles could not be cloned. ∗, the activity value was predicted from sequence similarities to closely related, tested alleles; nd, the activity value could not be predicted because the amino acid changes were not present in other alleles.
Table 2.
Population summary statistics, neutrality tests, and coalescence calculations
| Locus name | L1A (A1512428) | L1B (Ac02980) | L1C (Ac021017) |
|---|---|---|---|
| Sample size | 35 | 59 | 78 |
| Number of alleles | 8 | 18 | 26 |
| Number of polymorphic sites | 17 | 18† | 25‡ |
| Nucleotide diversity (π) (×10−3) | 0.81 | 0.35 | 0.21 |
| Tajima's D | 0.59 (NS) | −1.38 (NS) | −2.39(P < 0.01) |
| Fu and Li's D* | 1.22 (NS) | −2.13 (NS) | −5.18(P < 0.05) |
| Fu's Fs | 2.31 (NS) | −9.72 (P < 0.01) | −28.65(P < 0.01) |
| Coalescence time (×103 years) | 590 ± 160 | 230 ± 70 | 120± 35 |
D and D* were calculated with dnasp and Fs with arlequin (1,000 simulations). Coalescence time, assuming a nucleotide substitution rate 2.3 × 10−8 per site per generation and a generation time of 25 years. NS, not significant.
†In addition, one codon is deleted in alleles 2, 3, and 18.
‡In addition, allele 11 has a G insertion.
We used three tests to assess whether the three L1s were evolving neutrally (Table 2). No test for L1A was significant. For L1C, all three tests, as well as the Fu's FS test for L1B, generated significant negative values. These results were more significant (P < 0.01) for both elements in the case of Fu's FS, a test known to be sensitive to an excess of low-frequency alleles (41). Indeed, L1B and L1C have a high proportion of singleton alleles, 20% (12 of 59) and 27% (21 of 78), respectively (Fig. 1). Formally, these results indicate that L1B and L1C depart from neutral evolution, as discussed below.
Network Phylogenies of L1A, L1B, and L1C.
We reconstructed phylogenies of the three L1s by using Median Joining (MJ) networks (42, 43) (Fig. 5 A, C, and D). Because the ancestral allele is usually the most frequent (38), and MJ networks integrate both allele frequencies (area of the nodes, Fig. 5 and Table 3, which are published as supporting information on the PNAS web site) and evolutionary relationships (network position), the ancestral allele can usually be inferred by its size and position within the network. For both L1B and L1C, the most likely ancestral allele was easily inferred as allele 1, because it was central in the network and had the highest allele frequency. For L1A, allele 1, the most central allele (Fig. 5A), was not the most frequent, whereas allele 2, although not central, had the highest frequency in the sample. The consensus sequence for the element is ambiguous at nucleotide 2,104, where either C (Ser-39, allele 1) or G (Cys-39) is possible. Consensus Cys-39 was not among the alleles, either because it was not sampled or because it became extinct (43). When reconstructed as sequence φ, Cys-39 was central in the network (Fig. 5B). Moreover, Cys-39 is found in 54% of L1 clade non-LTR retrotransposons, whereas Ser-39 is found in only 9% (44). To test the retrotransposition activity of consensus Cys-39, we recreated it in a chimeric element, which had high activity (see Supporting Methods and Supporting Results in Supporting Text), whereas Ser-39 (allele 1) had low activity (Fig. 1). Thus, although consensus Cys-39 is less parsimonious, its network features, evolutionary conservation, and activity level all suggest that it is the most likely ancestor of L1A.
Allelic Variation in Retrotransposition Capability: Hot and Cool Alleles.
We tested the mobilization capability of 46 of 52 (88%) alleles of L1A, L1B, and L1C in a retrotransposition assay in human 143B cells (34, 45). Supported by a statistical algorithm, we clustered the alleles into percent activity categories ([x]) according to their mean activity and variance (Fig. 1 A, B, and C; and see Materials and Methods and Supporting Methods in Supporting Text). The activity categories of L1A were [0], [15], and [120]; activity categories of L1B were [20], [85], and [175]; and activity categories of L1C were [0], [5], [10], and [25] (Fig. 1 A, B, and C). The presumed ancestor of L1B and L1C, as determined by the network phylogenies, along with the hypothetical ancestor of L1A, reconstructed sequence φ (Supporting Text), were in the highest activity categories. We found nucleotide changes that significantly altered the retrotransposition activity (see Discussion). For each element, an analysis of the changes and their effect on L1 structure is presented in Supporting Results in Supporting Text.
For the three elements, 22 of 46 tested alleles had 25% or greater activity compared with the reference L1 (L1rp) and were hot (34). The remaining 24 alleles (5 of 8 L1A, 3 of 16 L1B, and 16 of 22 L1C) had activity <25% of L1rp and were cool. Of all elements at loci containing hot L1s, 33% (57 of 170) were cool (Fig. 1).
L1 Retrotransposition Potential in Individuals and Populations.
The number of possible genotypes in an individual for a locus with n alleles is n(n + 1)/2 or 36 for L1A, 171 for L1B, and 351 for L1C, with 8, 18, and 26 alleles, respectively. Because three or four activity categories were identified per locus, the many possible genotypes reduce to only 6, 10, and 9 possible phenotypes, respectively (Table 4, which is published as supporting information on the PNAS web site). Of those possible phenotypes, 66%, 80%, and 44% per locus correspond to hot L1 phenotypes, defined as having a biallelic activity ≥25% that of L1rp (Table 4). The remaining phenotypes with biallelic activity <25% of L1rp were defined as cool L1 phenotypes. After assigning the activity value to the allelic variants that each individual carried at L1A, L1B, and L1C loci, we observed that only 11% (9 of 80), 44% (35 of 79), and 45% (36 of 80) of phenotypes per locus were hot (Table 4).
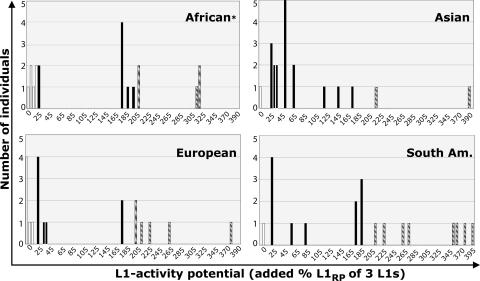
For each individual, we added the activity values per locus to obtain the total L1 activity potential (3 L1s combined; see Table 5, which is published as supporting information on the PNAS web site). Fig. 2 shows the wide distribution of L1 activity potentials per individual in every population (from 0% to >300%). Of the 80 individuals, one was excluded because his L1B element could not be isolated (Table 5). Among the remaining 79 individuals, 18% did not have a total hot L1 phenotype, 56% had a hot phenotype between 25% and 200%, and 26% had a very hot phenotype ≥200% (Fig. 2). This latter group is likely at higher risk than the others of undergoing a retrotransposition event.
Fig. 2.
Combined retrotransposition potential of three hot L1s per individual in four populations. From 26% (African) to 55% (South American) of individuals per population have a unique L1 activity potential. White, black, and hatched bars represent individuals lacking a hot L1 phenotype (<25%), having an intermediate L1 activity, and having a high L1 activity (>200%), respectively. ∗, The African distribution is based on 19 individuals (Table 5).
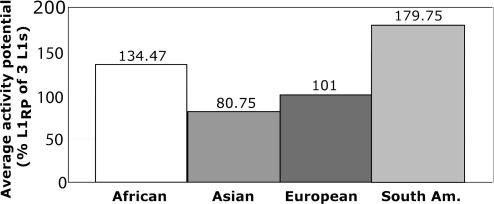
To obtain an overall L1 activity potential per population, we added the value from all individuals in a population and divided by the number of individuals (Table 5 and Fig. 3). We tested whether the different populations were statistically different in their overall L1 activity potential. There was a >2-fold difference between the relative activity potential of the highest (South Americans, 180%) and the lowest group (Asians, 81%). The hypothesis that all population means are equal was marginally rejected by an ANOVA test (P = 0.036) with South American and African means differing from those of Asians and Europeans. Note that the variation in L1 activity potential among individuals within populations is much larger (0–390%) than that among individuals between different populations (81–180%), a result consistent with other human population studies (46).
Fig. 3.
Average retrotransposition potential of three hot L1s in four populations. The total retrotransposition potential of L1A, L1B, and L1C for each individual was divided by the number of individuals in the population to determine the average retrotransposition potential in each population. The means of the four populations are not equal by ANOVA (P = 0.036).
Discussion
Thanks to our natural mutagenesis system combining genotype with functional assays, our data contribute to the complex structure/function map of human L1s (36, 47–49), identifying essential, preferred, and dispensable amino acids. In ORF2p, C39, A604, and F725 are essential because their nonconservative, nonsynonymous substitutions produced 85–95% reductions in retrotransposition activity. C39 is at the center of endonuclease subdomain II, four amino acids upstream of D43, a characterized active-site residue of exonuclease III activity (50). A604 is in reverse transcriptase (RT) subdomain III and, along with F605, is conserved in all non-LTR retrotransposons of various species (51). F725 in RT subdomain VI (9) is slightly less conserved than F605 but also appears to be essential for retrotransposition. Substitutions in preferred amino acids resulted in 30–50% reductions in activity. These reductions were found within ORF1 (R24 and R49) and ORF2 (S723, Q925, and Y1054). Finally, dispensable amino acids, substitution of which does not affect L1 mobility, were found in ORF1 (S12) and in ORF2 (S245, A290, V459, A951, and C1020). Consistent with phylogenetic analyses, these amino acids are located outside conserved domains (44, 50). Of 8 L1A, 16 L1B, and 22 L1C alleles tested, 62%, 18%, and 22%, respectively, had dramatic reductions (80–100%) in retrotransposition activity, not surprising, given that ORF2p is a multifunctional modular protein and that an intact ORF1p is also required for retrotransposition (26).
A major conclusion of this work, that hot and cool alleles of highly active L1s produce extensive variation in individual retrotransposition capability, rests on the proposition that L1 activity in cell culture mirrors L1 activity in vivo. But L1 expression in vivo depends on a number of factors not evaluated in the cell culture assay, including chromatin status, presence of appropriate transcription factors in the appropriate cell type, and DNA methylation, among others (see ref. 11 for review). As a first approximation, we asked whether the genomic region into which the element is inserted allows its expression. L1A is located within intron 21 of gene C6orf32–001 on chromosome 6 (see Supporting Discussion in Supporting Text for possible effects of L1A on expression of this gene). According to the University of California Santa Cruz genome browser, this gene is highly expressed in blood cells, indicating a transcriptionally active environment for the expression of L1A.
L1B and L1C do not reside within or close to known or predicted genes, and their immediate surrounding chromatin status is unknown, although they are in regions of low guanosine plus cytosine (GC) content (39.2%) (52). For L1B, our presence/absence PCRs in three male genomes analyzed suggest that it may have retrotransposed, carrying a 3′ transduction. These preliminary data, combined with our activity data indicating that L1B is the most active natural L1 described to date, suggest that L1B may be a “master self-propagating” element actively expanding in present-day human genomes.
Although our studies suggest that L1A and L1B are expressed in vivo, proof of that assertion is lacking. However, four of four hot L1s analyzed to date, three in this study and the disease-causing hot L1 (LRE1) (36, 37), had common alleles that demonstrated highly variable retrotransposition activity in cell culture. These data suggest that the great bulk of hot L1s responsible for most in vivo retrotransposition have both hot and cool alleles.
Allelism data allowed us to address whether the three Ta-1 subfamily members are evolving neutrally. Because of an excess of rare alleles, L1B and L1C showed departure from neutrality that can be explained by either natural selection or demographic processes (38, 39, 41). The excess of singletons could be due to recent positive or negative selection on neighboring loci to which L1B and L1C are linked (selective hitchhiking) (39). On the other hand, because human populations have been subject to extreme demographic expansion in the last 50,000–100,000 years (38, 39, 41), a conservative explanation for the excess of singleton alleles of L1B and L1C is the drastic changes in human population size since their insertion.
Assuming a neutral substitution rate of 2.3 × 10−8 per site per generation and an average human generation time of 25 years (53), we estimate the time to the most recent common ancestor as ≈590,000 years for L1A, ≈230,000 years for L1B, and ≈120,000 years for L1C (Table 2). These estimates are consistent with the estimated age of the L1 subfamilies to which the elements belong. L1A is a Ta-1nd member, an older subfamily than Ta-1d, to which L1B and L1C belong (34, 29).
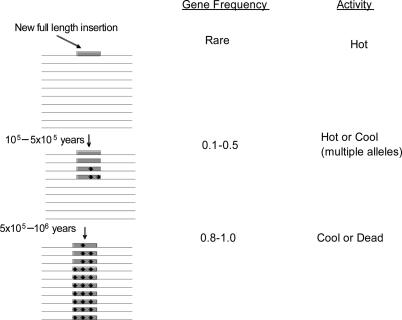
From this large-scale study of alleles of young L1s currently expanding in the human genome, we suggest a model for how L1 insertions evolve in a population (Fig. 4). The present work and the fact that nearly all present-day, disease-causing L1 insertions are hot (34, 54) suggest that new insertions in a population are derived from hot L1s. Later, as a new insertion increases in gene frequency through genetic drift, it also acquires random mutations, some of which reduce its retrotransposition potential from hot to cool, this being the status of the three elements studied here. As the L1's gene frequency increases toward fixation, alleles continue to accumulate mutations that render them either cool or dead for subsequent retrotransposition.
Fig. 4.
Model of the evolution of an L1 insertion in a population. Data presented here and evidence that hot L1s account for most new insertions (34) suggest that new insertions are derived from hot L1s. Data on alleles of L1A, L1B, L1C, and LRE1 (36, 37) indicate that, after a hot L1 reaches an intermediate gene frequency in the population, it has a significant proportion of cool alleles. As an L1 approaches fixation, mutations produce cool alleles and dead alleles. Shaded box, L1 insertion in chromosomes (lines); black dots, mutations.
Combining the retrotransposition potential of the three hot L1s studied, we found that 18% (14 of 79) of individuals lacked a hot phenotype (<25%, Fig. 2), whereas another 26% (21 of 79) have a very high retrotransposition capability (>200%, Fig. 2). Thus, nearly half of individuals fall at the extremes of the distribution of retrotransposition capability of these elements, suggesting that individuals vary significantly in their risk of a new insertion during meiosis or during development of their offspring.
But would this degree of individual variation still stand if other hot L1s in the population were included in the analysis? In other words, do L1A, L1B, and L1C combined constitute a significant fraction of the hot L1 activity in world populations? In HGWD, L1A, L1B, and L1C were among six elements with gene frequencies ≤0.47 that were hot (L1A, L1B, L1C, and Al137845 were common, and Ac004200 and Al356438 were rare) (34). Using the gene frequencies of the four common hot L1s in our 80 individuals, we estimate that L1A, L1B, and L1C account for 2/3 of these L1s in the population. Boissinot et al. (55) isolated Ta-1 elements from four other humans of diverse origin and, by extrapolation, found that Ta-1 elements in HGWD account for ≈1/2 of common Ta-1 elements in the population. Although these estimates are fraught with potential error, the data suggest that L1A, L1B, and L1C account for at least 1/3 of the common hot L1 activity (2/3 × 1/2). After other hot L1s are studied in these and other individuals, we predict that the proportion of individuals at the extremes of the distribution of retrotransposition capability will decrease somewhat, but the difference in retrotransposition potential of individuals at those extremes will increase. Thus, we conclude that individual variation in retrotransposition capability is an important contributor to human genetic diversity.
Materials and Methods
L1 Elements Analyzed.
Sequence polymorphisms in three FL L1 elements were analyzed. Al512428, Ac02980, and Ac021017, which we call L1A, L1B, and L1C, respectively, belong to the replicatively dominant Ta-1 subfamily (29) that is subdivided into two groups depending on the presence (Ta-1d, L1B, and L1C) or absence (Ta-1nd and L1A) of nucleotide G74. For details of the genomic locations and insertion signatures of the elements, see Table 6, which is published as supporting information on the PNAS web site.
DNA Samples.
One hundred sixty-one to 206 gender-typed haploid genomes from human variation panels of subSaharan African, African American, Asian, European, South American, Indo/Pakistani, and Pacific origins were obtained from Coriell Cell Repositories and the laboratory of M.A.B. Because of sample size heterogeneity, some populations were pooled to increase statistical power. For specific details see Supporting Methods in Supporting Text.
Presence/Absence Polymorphism, PCR, and Sequencing.
Presence/absence polymorphism status was determined by using a three-primer–two-PCR assay (30). Sequence polymorphisms of FL L1s were obtained by direct sequencing of the PCR products of heterozygous and homozygous genomes. The final assignment of nucleotide changes in homozygous genomes was made after sequencing of cloned products. Procedures to perform presence/absence polymorphism assays to obtain high-quality DNA and the primers used are described in Supporting Methods in Supporting Text; and see Table 7, which is published as supporting information on the PNAS web site. To minimize the amplification of potential PCR errors, we used Phusion Hi-fi DNA Polymerase (MJ Research, Cambridge, MA), which has the lowest error rate of currently available proofreading enzymes. For both sequencing and cloning, we systematically pooled products of at least three independent PCRs.
Sequence and Polymorphism Analyses.
Sets of 11 overlapping sequences covering each FL L1 were imported into sequencher 4.5 and aligned to HGWD sequence. Nucleotides different from HGWD sequence were verified manually in the chromatograms and considered new alleles. In cases of unclear nucleotide readings, DNA was repurified from new PCR products and resequenced. Allele files were generated with macvector 8.02, and consensus sequences were obtained with clustalw. The evolutionary history of mutations was reconstructed with Median Joining (MJ) networks (42). Neutrality departure tests (Tajima's D, Fu and Li's D*, and Fu's Fs) were performed with dnasp and arlequin software suites.
Cloning of L1 Alleles, Transfection, Retrotransposition Assay, and Definition of Activity Categories.
FL L1s were isolated from heterozygous and homozygous genomes, cloned into a vector carrying the EGFP retrotransposition cassette, sequenced to verify allelic changes, and tested for activity in human 143B thymidine kinase (TK)− cells (45). Details of cloning, transfection, and activity categories definition by using cart software can be found in SupportingMethods in Supporting Text. Under our PCR conditions, very few cloned elements had PCR errors (estimated error rate ≈1.9 × 10−5), and those clones were discarded. Thus, activity differences observed for different clones of an L1 were relatively minor and due to biological variation intrinsic to the assay. Activity differences among L1s cloned here and in previous analyses (34, 56) are discussed in Supporting Discussion in Supporting Text.
Supplementary Material
Acknowledgments
We thank D. Babushok for helpful suggestions and C. N. Talchai for experimental work. This work was supported by National Institutes of Health grants (to H.H.K. and M.A.B.) and National Science Foundation grants (to M.A.B.).
Abbreviations
- FL
full-length
- HGWD
human genome working draft.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Myers J. S., Vincent B. J., Udall H., Watkins W. S., Morrish T. A., Kilroy G. E., Swergold G. D., Henke J., Henke L., Moran J. V., et al. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 4.Kazazian H. H., Jr Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 5.Speek M. Mol. Cell. Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J. S., Szak S. T., Boeke J. D. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 7.Perepelitsa-Belancio V., Deininger P. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 8.Furano A. V. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- Moran J. V., Gilbert N. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 836–869. [Google Scholar]
- 10.Chen J. M., Stenson P. D., Cooper D. N., Ferec C. Hum. Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 11.Ostertag E. M., Kazazian H. H., Jr. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert N., Lutz-Prigge S., Moran J. V. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 13.Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., Parmigiani G., Boeke J. D. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert N., Lutz S., Morrish T. A., Moran J. V. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran J. V., DeBerardinis R. J., Kazazian H. H., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 16.Esnault C., Maestre J., Heidmann T. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 17.Dewannieux M., Esnault C., Heidmann T. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 18.Ostertag E. M., Goodier J. L., Zhang Y., Kazazian H. H., Jr Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Xing J., Grover D., Hedges Kyudong Han D. J., Walker J. A., Batzer M. A. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 20.Swergold G. D. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohjoh H., Singer M. F. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 22.Martin S. L., Bushman F. D. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Q., Moran J. V., Kazazian H. H., Jr, Boeke J. D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 24.Mathias S. L., Scott A. F., Kazazian H. H., Jr, Boeke J. D., Gabriel A. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 25.Fanning T., Singer M. Nucleic Acids Res. 1987;15:2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 27.Cost G. J., Feng Q., Jacquier A., Boeke J. D. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 29.Boissinot S., Chevret P., Furano A. V. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 30.Sheen F. M., Sherry S. T., Risch G. M., Robichaux M., Nasidze I., Stoneking M., Batzer M. A., Swergold G. D. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badge R. M., Alisch R. S., Moran J. V. Am. J. Hum. Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett E. A., Coleman L. E., Tsui C., Pittard W. S., Devine S. E. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent B. J., Myers J. S., Ho H. J., Kilroy G. E., Walker J. A., Watkins W. S., Jorde L. B., Batzer M. A. Mol. Biol. Evol. 2003;20:1338–1348. doi: 10.1093/molbev/msg146. [DOI] [PubMed] [Google Scholar]
- 34.Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H., Jr Proc. Natl. Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dombroski B. A., Mathias S. L., Nanthakumar E., Scott A. F., Kazazian H. H., Jr Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 36.Farley A. H., Luning Prak E. T., Kazazian H. H., Jr Nucleic Acids Res. 2004;32:502–510. doi: 10.1093/nar/gkh202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz S. M., Vincent B. J., Kazazian H. H., Jr, Batzer M. A., Moran J. V. Am. J. Hum. Genet. 2003;73:1431–1437. doi: 10.1086/379744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bamshad M., Wooding S. P. Nat. Rev. Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 39.Przeworski M., Hudson R. R., Di Rienzo A. Trends Genet. 2000;16:296–302. doi: 10.1016/s0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- 40.Li W. H., Sadler L. A. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreitman M. Annu. Rev. Genomics Hum. Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- 42.Bandelt H. J., Forster P., Rohl A. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 43.Cordaux R., Hedges D. J., Batzer M. A. Trends Genet. 2004;20:464–467. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 44.Malik H. S., Burke W. D., Eickbush T. H. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 45.Ostertag E. M., Prak E. T., DeBerardinis R. J., Moran J. V., Kazazian H. H., Jr Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jorde L. B., Wooding S. P. Nat. Genet. 2004;36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 47.Dhellin O., Maestre J., Heidmann T. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulpa D. A., Moran J. V. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 49.Yang N., Zhang L., Zhang Y., Kazazian H. H., Jr Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin F., Olivares M., Lopez M. C., Alonso C. Trends Biochem. Sci. 1996;21:283–285. [PubMed] [Google Scholar]
- 51.Xiong Y., Eickbush T. H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliver J. L., Carpena P., Roman-Roldan R., Mata-Balaguer T., Mejias-Romero A., Hackenberg M., Bernaola-Galvan P. Gene. 2002;300:117–127. doi: 10.1016/s0378-1119(02)01034-x. [DOI] [PubMed] [Google Scholar]
- 53.Nachman M. W., Crowell S. L. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brouha B., Meischl C., Ostertag E., de Boer M., Zhang Y., Neijens H., Roos D., Kazazian H. H., Jr Am. J. Hum. Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boissinot S., Entezam A., Young L., Munson P. J., Furano A. V. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lavie L., Maldener E., Brouha B., Meese E. U., Mayer J. Genome Res. 2004;14:2253–2260. doi: 10.1101/gr.2745804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hohjoh H., Singer M. F. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.