Abstract
The mechanisms regulating activation of monocytes remain incompletely understood. Herein we provide evidence that Kruppel-like factor 2 (KLF2) inhibits proinflammatory activation of monocytes. In vitro, KLF2 expression in monocytes is reduced by cytokine activation or differentiation. Consistent with this observation, KLF2 expression in circulating monocytes is reduced in patients with chronic inflammatory conditions such as coronary artery disease. Adenoviral overexpression of KLF2 inhibits the LPS-mediated induction of proinflammatory factors, cytokines, and chemokines and reduces phagocytosis. Conversely, short interfering RNA-mediated reduction in KLF2 increased inflammatory gene expression. Reconstitution of immunodeficient mice with KLF2-overexpressing monocytes significantly reduced carrageenan-induced acute paw edema formation. Mechanistically, KLF2 inhibits the transcriptional activity of both NF-κB and activator protein 1, in part by means of recruitment of transcriptional coactivator p300/CBP-associated factor. These observations identify KLF2 as a novel negative regulator of monocytic activation.
Keywords: inflammation, transcription, transcriptional factor, NF-κB, activator protein 1
Under physiologic conditions the peripheral blood monocyte circulates in the bloodstream in an inactive state. In response to injury or inflammatory stimuli the monocyte exits the bloodstream, enters tissues, and assumes the features of a tissue macrophage. Through elaboration of cytokines/chemokines and ingestion of noxious agents, the tissue macrophage critically regulates tissue injury and repair. However, this response must be tightly regulated, and prolonged activation can be deleterious (reviewed in ref. 1). Indeed, prolonged activation of monocytes has been implicated in the pathogenesis of many inflammatory disease states such as atherosclerosis, rheumatoid arthritis, and tumor development (reviewed in ref. 1). As such, the identification of factors that negatively regulate monocyte/macrophage activation is of considerable scientific interest.
Kruppel-like factors (KLFs) are a subclass of the zinc-finger family of transcription factors implicated in the regulation of cellular growth and differentiation (2, 3). Previous studies indicate that members of this family play a particularly important role in hematopoietic cell biology. For example, erythroid KLF (EKLF; KLF1) is a key regulator of β-globin gene synthesis and erythropoiesis (4–7). Lung KLF (LKLF; KLF2) has been shown to regulate T cell activation. KLF2 is induced during the maturation of single-positive T cells and is rapidly extinguished after single-positive T cell activation (8). Forced overexpression of KLF2 induced a quiescent T cell phenotype whereas KLF2-null T cells exhibited a spontaneously activated phenotype (9). More recently, KLF2 has also been implicated in the regulation of embryonic globin gene synthesis (10).
Although it has previously been recognized that KLF2 is also expressed in monocytes (8), its function in this cell type remains unknown. In this study we provide evidence that forced overexpression of KLF2 inhibited monocytic proinflammatory gene expression in vitro and inflammation in vivo. This inhibitory effect is mediated through inhibition of both the NF-κB and activator protein 1 (AP-1) pathways and involves recruitment of coactivators. These observations coupled with previous studies in T cell biology support a central role for KLF2 in the regulation of immune cell activation.
Results
KLF2 Expression Is Reduced with Monocyte Activation/Differentiation into Macrophages.
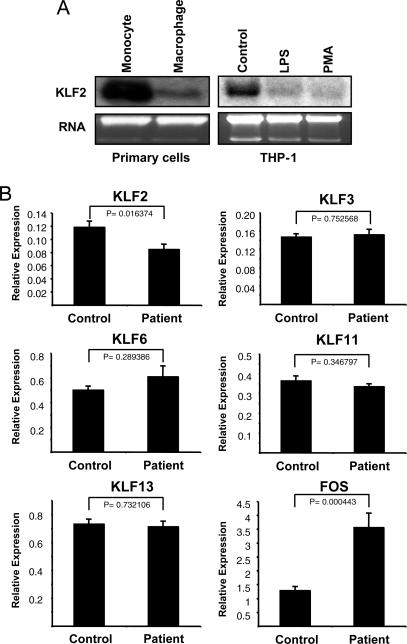
To better understand the role of KLF2 in the regulation of immune cells such as monocyte/macrophage, we first evaluated the expression of KLF2 in primary human monocytes. As shown in Fig. 1A Left, KLF2 expression is robust in primary human monocytes and strongly reduced with differentiation into macrophages after 5 days. KLF2 expression in the human monocytic cell line THP-1 was much lower than that in primary human monocytes. However, expression was further reduced upon treatment of these cells with LPS or 12-O-tetradecanoylphorbol-13-acetate, two agents well established to induce cellular activation or differentiation (Fig. 1A Right).
Fig. 1.
KLF2 expression in activated monocytes in vitro and in vivo. (A) KLF2 mRNA is reduced with monocyte differentiation and activation. Northern blot analysis was performed with 10 μg of total RNA per lane from human monocytes and macrophages (Left). A similar analysis was performed with 20 μg of total RNA from THP-1 cells cultured in FBS-free for 36 h thereafter and an additional 6-h stimulation with LPS or 12-O-tetradecanoylphorbol-13-acetate (Right). (B) KLF2 expression is reduced in monocytes from patients with atherosclerosis. Quantitative real-time RT-PCR was performed with the monocytes isolated from 14 patients (Patient) and 14 healthy age-matched controls (Control). Levels of specified genes expression were normalized with the control gene eukaryotic translation initiation factor.
We next sought to determine whether KLF2 expression is regulated in the inflammatory setting in vivo. Monocytic activation is a key pathophysiologic event in the development of atherosclerosis, a chronic low-grade inflammatory condition, so we reasoned that KLF2 expression may be reduced in these patients (11). We examined a group of 14 age-matched normal subjects and 14 patients with extensive atherosclerosis (≈50% with history of coronary artery revascularization) who are stratified by the lowest and highest levels of Finkel–Biskis–Jinkins osteosarcoma gene (FOS), respectively, which has been shown to serve as a marker of inflammation (12). As shown in Fig. 1B, KLF2 expression was significantly reduced by ≈30% in patients. In contrast, no significant effect was observed for KLF3, KLF6, KLF11, and KLF13. Consistent with our previous study, FOS expression was significantly increased in patients with coronary artery disease (12).
KLF2 Inhibits Monocyte Activation and Phagocytic Capacity.
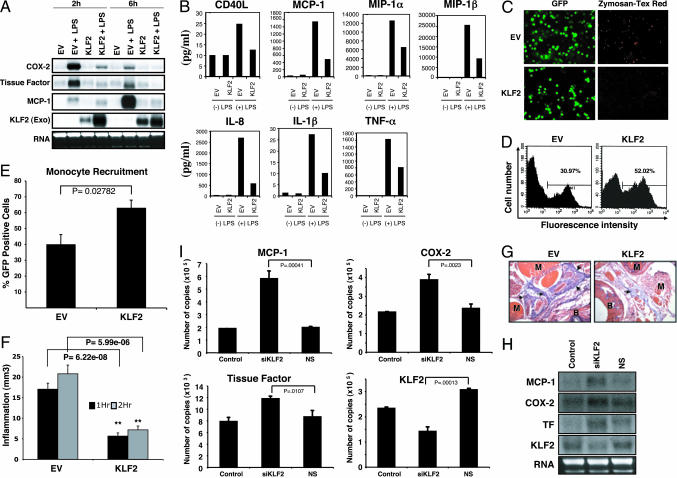
In response to inflammatory stimuli, monocytes express increased levels of proinflammatory factors and cytokines/chemokines and exhibit enhanced phagocytic capacity. To gain insight into the role of KLF2 in monocyte function, we undertook overexpression studies. THP-1 cells were infected with either control empty virus (EV) or KLF2 (Ad-KLF2) for 24–48 h and then stimulated with LPS for 2 and 6 h. As shown in Fig. 2A, treatment of EV-infected cells with LPS (25 ng/ml) resulted in a robust induction of cyclooxygenase (COX)-2, tissue factor, and monocyte chemoattractant protein (MCP)-1 mRNA. In contrast, this induction was markedly attenuated in KLF2-infected cells (Fig. 2A). Similar effects were also observed in the mouse monocyte cell line J774a (data not shown). Furthermore, overexpression of KLF2 strongly inhibited the secretion of various cytokines and chemokines. As shown in Fig. 2B, KLF2 overexpression attenuated the LPS-mediated induction of factors such as CD40L (by 49.6%), macrophage inflammatory protein 1α (48.4%), macrophage inflammatory protein 1β (64.2%), IL-1β (55.0%), IL-8 (79.1%), TNF-α (50.2%), and MCP-1 (68.8%). This effect was specific because no significant effect was observed on the levels of multiple other relevant growth factors (TGFβ1, platelet-derived growth factor, and granulocyte/macrophage colony-stimulating factor), cytokines/chemokines (IL-4, IL-10, IL-12p40, IL-12p70, IL-1α, and IFN-γ), or matrix-altering enzymes (matrix metalloproteinase types 1, 2, 3, and 9) (data not shown).
Fig. 2.
KLF2 overexpression inhibits monocyte activation. (A) KLF2 overexpression inhibits inflammatory gene expression. Northern blot analysis was performed for indicated factors with 10 μg of total RNA per lane from THP-1 overexpressed with Ad-KLF2 (KLF2) or EV as a control after stimulation with LPS for various time points as indicated. (B) KLF2 overexpression inhibits cytokine/chemokine elaboration. One million THP-1 cells per milliliter were infected with EV or KLF2 adenovirus for 24 h followed by LPS stimulation for an additional 2 h or 6 h. After stimulation, culture supernatants were harvested and assessed by SearchLight Proteome Arrays/multiplex sandwich ELISA. Each sample was evaluated in duplicate, and two independent experiments were evaluated for a panel of biological markers. A similar result was observed with different experiments. (C) KLF2 inhibits phagocytosis. EV- and KLF2-infected THP-1 cells were stimulated with LPS (25 ng/ml), and a phagocytosis assay was performed as described in Materials and Methods. (D and E) KLF2 does not inhibit monocytic recruitment. In D, a representative flow cytometric analysis of GFP-positive cells recruited to the peritoneal cavity after i.p. thioglycolate injection is shown. The gated bar indicates the percentage of GFP-positive cells in the analysis. In E the summary results from n = 5 mice per group are provided. (F) Reconstitution of immunodeficient mice as described in Materials and Methods with KLF2-overexpressed J774a cells reveals reduced edema in both 1-h and 2-h periods of carrageenan-induced inflammation. Data are expressed in ± SEM (n = 4). (G) KLF2 reduces tissue edema. Cross sections of the carrageenan-injected paws (×100 magnification) were stained with hematoxylin and eosin. Paws from mice reconstituted with KLF2-overexpressed monocytes show reduced extravasated fluid marked with arrows. Landmarks such as muscles (M) and bones (B) are indicated. (H and I) Knockdown of KLF2 increases proinflammatory gene expression. J774a cells were transfected with nonspecific (NS) or siRNA to KLF2 (siKLF2) for 48-h target genes assessed by Northern blot analysis (H) and quantitative real-time PCR analysis (I). Graphs in I represent combined data from three experiments.
A classic characteristic of the activated monocyte/macrophage is phagocytosis. To determine whether KLF2 overexpression affects the phagocytic ability of monocytes, we overexpressed KLF2 or control (EV) in THP-1 cells, stimulated with LPS, and assessed the ability of these cells to take up Texas red-labeled zymosan A bioparticles. As shown in Fig. 2C, control virus-infected cells ingested the zymosan bioparticles. In contrast, uptake by KLF2-expressing monocytes was markedly reduced (Fig. 2C). Taken together, these data demonstrate that KLF2 can inhibit monocyte secretion of cytokines/chemokines and phagocytosis.
KLF2 Does Not Inhibit Monocyte Recruitment.
We next sought to assess the effect of KLF2 on monocyte function in vivo. In response to an injury or an inflammatory stimulus, monocytes are recruited from the bloodstream to tissues. We first undertook studies to assess whether monocyte recruitment was altered by KLF2 expression. To minimize the contribution of other cell types we used C.B-17-Scid-beige mice that are deficient in T, B, and natural killer cells. These animals (n = 5 per group) were then subjected to total-body irradiation and reconstituted with J774a cells adenovirally infected with either control virus (EV) or KLF2 by means of tail vein injection (described in Materials and Methods). Animals were then subjected to i.p. injection of thioglycolate (a potent chemical irritant that induces monocyte recruitment), and the number of GFP-positive cells was assessed 48 h later by flow cytometric analysis. As shown in Fig. 2 C and D, KLF2 did not inhibit the recruitment of GFP+ J774a monocytes to the peritoneal cavity. Indeed, we reproducibly observed that KLF2-transduced animals exhibited a significant increase in GFP+ cells. These data suggest that KLF2 does not inhibit but rather augments monocytic recruitment to an inflammatory site.
KLF2 Inhibits Carrageenan-Induced Inflammation.
After recruitment and migration into tissues monocytes secrete cytokines, chemokines, and growth factors to perpetuate the inflammatory response. As shown in Fig. 2B, we noted that KLF2 overexpression attenuated the elaboration of several cytokines and chemokines. To determine whether KLF2 affects monocytic proinflammatory function, we used a well established model for inflammation: carrageenan-induced footpad injection. Injection of this chemical has been previously shown to reproducibly induce an inflammatory response characterized by paw edema (13–15). C.B-17-Scid-beige mice (n = 4 per group) were irradiated as described above, reconstituted with control or KLF2-expressing J774a cells, and then challenged with carrageenan injection in the footpad (described in Materials and Methods). Paw volume was determined by using a hydroplethismometer modified for small volumes (Ugo Basile, Milan). As shown in Fig. 2F, EV-infected animals exhibited a 17 ± 1.08-mm3 and 23.3 ± 3.05-mm3 increase in paw volume after 1 and 2 h of carrageenan injection, respectively. In contrast, animals reconstituted with KLF2-expressing J774a cells exhibited only a 6.25 ± 0.85-mm3 and 7.5 ± 0.95-mm3 increase in paw volume at 1 and 2 h after carrageenan injection, respectively (Fig. 2F). Consistent with this gross effect, histologic analysis of the paws revealed reduced fluid extravasation, which leads to edema formation (Fig. 2G, arrows).
Effect of KLF2 Knockdown on Inflammatory Gene Expression.
To determine the importance of KLF2 in monocytic inflammatory gene expression, small interfering RNA (siRNA)-mediated knockdown studies were also undertaken. As shown in Fig. 2H, by comparison with untreated (control) and nonspecific siRNA-treated cells, knockdown of KLF2 (≈60% knockdown) in J774a cells resulted in an induction of inflammatory genes such as MCP-1 and a more modest effect on COX-2 and tissue factor expression levels. These results were also verified with real-time PCR analysis (Fig. 2I).
KLF2 Inhibits NF-κB and AP-1 Promoter Activities.
KLF2 can potently inhibit the LPS-mediated induction of MCP-1, COX-2, and tissue factor (Fig. 2A). The induction of these targets by proinflammatory stimuli is known to be regulated by transcriptional pathways such as NF-κB and AP-1. We reasoned that, in principle, KLF2 may inhibit these pathways at multiple levels such as expression, DNA binding, or induction of transcriptional activity.
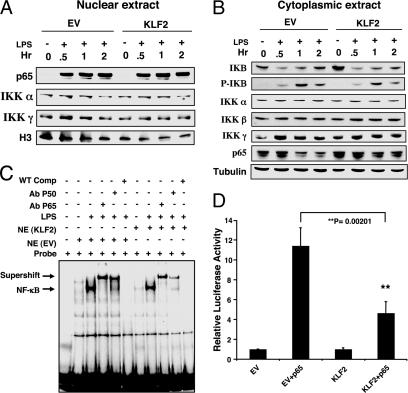
We first focused on NF-κB given its central role in inflammation. Overexpression of KLF2 did not alter nuclear accumulation of p65 or nuclear levels of IκB kinase (IKK) α and IKKγ (Fig. 3A). Furthermore, KLF2 overexpression did not alter the kinetics of IκB phosphorylation or degradation or cytoplasmic levels of IKKα, IKKβ, and IKKγ (Fig. 3B). Consistent with these observations, KLF2 did not affect NF-κB binding to DNA. As shown in Fig. 3C, treatment of control virus infected cells (EV) with LPS induced a single major band. Competition and supershift studies verified that this band represents NF-κΒ. A nearly identical pattern of NF-κB binding was observed in KLF2-overexpressing cells. To assess whether KLF2 can affect NF-κB-mediated transcriptional activation, gene reporter assays were undertaken. As shown in Fig. 3D, transfection of p65 with an NF-κB luciferase reporter plasmid induced transcriptional activity by ≈11 fold. This induction was strongly reduced in the presence of KLF2, indicating that KLF2 inhibits NF-κB transcriptional activity. Similar effects of KLF2 were observed for the AP-1 pathway (Fig. 5 A–C, which is published as supporting information on the PNAS web site).
Fig. 3.
KLF2 inhibits NF-κB transcriptional activity. (A and B) KLF2 does not alter expression of components of the NF-κB pathway. THP-1 cells were infected with the adenovirus (EV or KLF2), stimulated with LPS for 30 min, 1 h, or 2 h, and assessed for expression of the indicated factors by Western blot analysis using nuclear and cytoplasmic extracts. (C) KLF2 does not affect NF-κB DNA binding. THP-1 cells were infected with the adenovirus (EV or KLF2) and stimulated with LPS for 1 h, and nuclear extracts were used for gel-shift assays. The NF-κB band is designated by an arrow. Specificity was verified by competition and supershift studies. (D) KLF2 inhibits p65-mediated transactivation of the NF-κB concatemer. Transient transfection studies were performed in RAW264.7 cells with the indicated constructs. These experiments were performed in triplicate and repeated at least three times.
Recruitment of the Coactivator CBP-Associated Factor (PCAF) by KLF2 as a Unifying Mechanism.
Taken together, the results in Fig. 3 indicate that KLF2 can inhibit NF-κB-mediated transactivation independent of effects on the DNA binding of these factors. One possible mechanism is recruitment of critical coactivators. For example, optimal NF-κB binding requires interaction with several key coactivators such as steroid receptor coactivator (SRC)-1, PCAF, and p300/CBP. KLF2 may interact with one or more of these factors, recruit them away from NF-κB, and, as consequence, reduce NF-κB-mediated transcriptional activity.
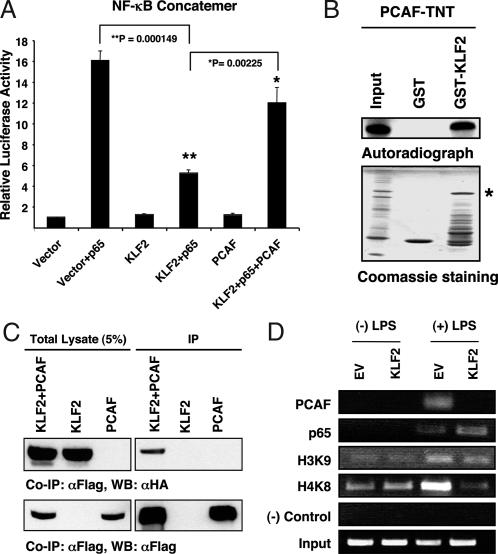
To determine the role of PCAF in KLF2-mediated inhibition of NF-κB transcriptional activity we performed cotransfection studies with exogenous PCAF. Transfection of PCAF (but not p300; data not shown) significantly rescued the KLF2-mediated repression of the NF-κB concatemer (Fig. 4A). A partial rescue was also observed on KLF2's ability to inhibit AP-1 transcriptional activity (Fig. 5D). To determine whether KLF2 and PCAF interact with each other, we performed GST pull-down and coimmunoprecipitation assays. Using in vitro transcribed and translated products, we found that KLF2 and PCAF can interact directly in a cell-free system (Fig. 4B). To know whether PCAF binds to KLF2 in vivo, we overexpressed KLF2 (hemagglutinin-tagged) and PCAF (Flag-tagged) in COS-7 cells. Immunoprecipitation with an anti-Flag antibody followed by Western blotting with an anti-hemagglutinin antibody showed that KLF2 binds with PCAF in cells (Fig. 4C).
Fig. 4.
KLF2 interacts directly with PCAF. (A) PCAF overexpression rescues KLF2-mediated inhibition of NF-κB transcriptional activity. Transient transfection studies with the indicated plasmids were performed in RAW264.7 cells as previously described (n = 9–12 per group). (B) KLF2 interacts with PCAF in a cell-free system. Binding experiments were performed by using in vitro transcribed and translated products (TNT) of PCAF and GST-KLF2. Autoradiograph data (Upper) and Coomassie staining of the gel (Lower) indicate loading amount and PCAF protein (∗). Input is 2% of the PCAF-TNT. (C) KLF2 and PCAF interact in cells. COS-7 cells were transfected with the indicated constructs, and immunoprecipitation studies were performed as described in Materials and Methods. (D) KLF2 reduces PCAF recruitment. THP-1 cells were infected with Ad-KLF2 or EV containing virus and stimulated with LPS, and ChIP assays were performed for the indicated factors on the COX-2 promoter (see Materials and Methods for details).
To determine whether the mechanisms underlying the observed effects are operative in vivo, we undertook chromatin immunoprecipitation (ChIP) studies. As expected, LPS stimulation of THP-1 cells led to the recruitment of p65 and PCAF to the COX-2 promoter (Fig. 4D). In addition, concomitant acetylation of HH3 and HH4 were observed. However, in the context of KLF2 overexpression, PCAF recruitment and histone acetylation are both strongly attenuated (Fig. 4D). Consistent with our gel-shift assays, no significant effect of KLF2 was observed on p65 recruitment.
Discussion
In response to an inflammatory challenge, the monocyte migrates into tissues, expresses proinflammatory factors, elaborates cytokines and chemokines, and exhibits enhanced phagocytic capacity. Although the activated monocyte plays an important physiologic role in the body's response to tissue injury and repair, mechanisms must exist to regulate this response. In this study we identify KLF2 as a potent negative regulator of monocytic activation.
Our observations provide insights regarding how KLF2 affects many of the cardinal features of the activated monocyte. The first key point is that KLF2's antiinflammatory effects are not secondary to a perturbation in monocyte recruitment to an inflamed site. As shown in Fig. 2 D and E, reconstitution experiments using irradiated SCID-beige mice challenged with i.p. thioglycolate revealed that accumulation of KLF2-infected cells is, in fact, higher than control virus-infected cells. However, despite this observation, paw edema in the carrageenan model of inflammation is markedly attenuated (Fig. 2 F and G). As such, the antiinflammatory effect of KLF2 is likely due to inhibition of monocyte proinflammatory gene expression and not recruitment. Consistent with this hypothesis, we found that KLF2 overexpression significantly inhibited expression of several cytokines/chemokines (IL-1β, IL-8, TNFα, MCP-1, macrophage inflammatory proteins, and CD40L; Fig. 2B) and inflammatory factors (e.g., tissue factor and COX-2; Fig. 2A). Furthermore, the net effect of inhibiting these factors was to reduce autocrine/paracrine activation of other immune and nonimmune cells in the local environment, leading to a reduction in inflammation and paw edema (Fig. 2 F and G).
Our studies also provide insights regarding the molecular basis for KLF2's antiinflammatory effects. Both the NF-κB and AP-1 pathways are well established as key regulators of cellular gene expression in response to a broad range of inflammatory stimuli (16, 17). In response to cytokines, both NF-κB and AP-1 are activated and subsequently accumulate in the nucleus, bind DNA, and induce target genes. The induction of target gene transcription is a complex process that requires interaction with coactivator molecules. Our data suggest that KLF2 does not affect protein expression, nuclear accumulation, or DNA binding of either NF-κB or AP-1 (Figs. 3 and 5). However, KLF2 does potently inhibit the transcriptional activity of both pathways (Figs. 3D and 5C). To further understand the basis for these observations we considered the possibility that KLF2 may bind to coactivators that are necessary for optimal NF-κB activity. The recruitment of rate-limiting amounts of coactivators has previously been implicated as the basis for transcriptional repression observed in other systems such as the mutual antagonism between nuclear hormone receptors and AP-1 proteins (18). Previous studies suggest that NF-κB requires coactivators such as p300/CBP or PCAF for transcriptional activation (19, 20). As such, we reasoned that KLF2 may bind to one or more of these coactivators and, as a consequence, render them incapable of assisting NF-κB to induce transcriptional activity. It follows that reconstitution of these factors in excess should ameliorate KLF2's inhibitory effect. Indeed, under the experimental conditions used here, we observed by transient transfection that PCAF overexpression rescued KLF2-mediated inhibition of NF-κB transcriptional activity (Fig. 4A). The importance of PCAF is further highlighted by the fact that KLF2 and PCAF can directly interact in cells or in a cell-free system (Fig. 4 B and C) and that recruitment of PCAF away from the NF-κB complex was observed in cells by ChIP assays (Fig. 4D). Finally, we note that, in contrast to PCAF, overexpression of p300/CBP was unable to rescue KLF2's inhibitory effect in macrophage cell lines. This observation is noteworthy because previous studies demonstrate that p300 can rescue KLF2's inhibitory effects in COS-7 cells (21). The basis for this difference is unclear but may due to use of different cell types.
Our observations regarding KLF2 in monocyte biology provide interesting parallels with those observed previously in T cell biology. For example, the reduction in KLF2 levels with monocyte differentiation/activation (Fig. 1A) is akin to the reduction in KLF2 observed by Kuo et al. (8) after T cell activation. Importantly, we have extended this observation in vivo and provided evidence that patients with chronic inflammatory conditions such as atherosclerosis also exhibit reduced expression of KLF2 (Fig. 1B). This reduction may constitute an important mechanism by which inflammatory stimuli carry out their function, i.e., by decreasing the expression of antiinflammatory factor such as KLF2 while activating proinflammatory factors such as NF-κB and AP-1. Although the molecular basis for the reduction in KLF2 remains unknown in monocytes and T cells, recent studies by Kumar et al. (22) implicate p65 and histone deacetylases in repressing expression of this factor. Whether similar mechanisms are operative in immune cells is unclear, but it raises the interesting possibility that KLF2 and p65 may function in a mutually antagonistic manner and that the balance of these two factors may dictate the degree of cellular activation.
Another interesting parallel between KLF2's role in T cell and monocyte biology is derived from our overexpression studies. Buckley et al. (9) provided evidence that forced expression of KLF2 in Jurkat cells induces T cell quiescence as evidenced by a reduction in cell size, synthetic capacity, and expression of activation markers. Similarly, we find that KLF2 overexpression can strongly inhibit characteristic features of an activated monocyte (Figs. 2 and 3). Taken together, the observations in this study coupled with previous observations in T cell biology strongly support an important regulatory role for KLF2 in immune cell activation.
Materials and Methods
Cell Culture and Reagents.
Monocytic cell lines (THP-1, J774a, and RAW264.7) as well as COS-7 cells were obtained from American Type Culture Collection and cultured following the instructions as described (23). LPS (Salmonella typhimurium) and 12-O-tetradecanoylphorbol-13-acetate and anti-tubulin antibody were obtained from Sigma-Aldrich. Antibodies for Western blots such as p50, p65, inhibitory KB (IKB), phospho-IKB, IKKα, IKKβ, IKKγ, c-fos, c-jun, and protein A/G plus agarose beads were purchased from Santa Cruz Biotechnology. Histone H3 antibody was purchased from Cell Signaling Technology (Beverly, MA). For ChIP assays, we used the ChIP kit from Upstate Biotechnology (Lake Placid, NY). The ChIP antibodies for PCAF and p65 were from Santa Cruz Biotechnology, and those for HH3 and HH4 were from Upstate Biotechnology. Adenoviral constructs were generated by the Harvard Gene Therapy Initiative (Boston). Polybrene was purchased from Specialty Media (Northampton, MA). The c-jun expression constructs were kindly provided by A. Mauviel (University of Paris, Paris), and the c-fos expression construct was obtained from M. Karin (University of California at San Diego, La Jolla). Human peripheral blood mononuclear cells were isolated by using the Ficoll–Paque method from blood obtained from Brigham and Women's Hospital as previously described (24). Procedures were reviewed and approved by the Brigham and Women's Hospital and Harvard Medical School Standing Committee.
KLF2 Expression in Human Subjects with Atherosclerosis and Normal Age-Matched Controls.
The recruitment and selection criteria for patients and controls have been described previously (12). Monocytes were purified from peripheral blood by using antibodies against CD14 bound to magnetic beads (Dynal Biotech, Brown Deer, WI) as described (12). Purification of mRNA from monocyte lysates was performed by binding to poly (dT) magnetic beads (Dynal Biotech) and reverse-transcribed by using SuperScript II (Invitrogen). Primer sequences for all of the assessed KLFs and FOS are provided in Table 1, which is published as supporting information on the PNAS web site. Standard quantitative RT-PCR was performed in duplicate at least two to three times by using SYBR Green (Molecular Probes) and TaqMan protocols on the 7900HT (Applied Biosystems, Foster City, CA). RT-PCR data were normalized by measuring average cycle threshold (Ct) ratios between candidate genes and the control gene, eukaryotic translation initiation factor (EIF3S5 or translation initiation factor). The formula 2Ct(Candidate)/2Ct(Control) was used to calculate normalized ratios (25).
Adenoviral Infection of THP-1 Cells.
Five million THP-1 cells were plated in a 100-mm dish and infected with control (Ad-GFP) or KLF2 (Ad-KLF2) virus at a multiplicity of infection of 50 in the presence of polybrene (final concentration, 8 ng/ml). In general, ≈80–90% infection was achieved within 24–48 h of incubation with adenovirus, at which time cells were used for experimentation.
Northern/Western Blot Analyses and Transient Transfection Assays.
See Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
GST Pull-Down and Coimmunoprecipitation Assays.
Recombinant GST-KLF2 protein was synthesized by the GST Gene Fusion System (Amersham Pharmacia Biotech) and purified by the bulk GST Purification Module (Amersham Pharmacia Biotech) following the manufacturer's protocol. A GST pull-down assay was performed as described previously (21).
For immunoprecipitation assays, COS-7 cells were transfected with the indicated expression plasmids (Flag-PCAF and hemagglutinin-KLF2) and harvested in RIPA buffer 48 h later. Cell lysates were subjected to immunoprecipitation with 5 μg of αFlag M2 monoclonal antibody (Sigma) at 4°C for 2 h followed by incubation with protein A/G Sepharose beads overnight at 4°C. The beads were washed, and the proteins were separated by SDS/PAGE as previously described (26).
EMSA and ChIP Assays.
Nuclear extracts (10 μg per lane) from THP-1 cells were prepared, and mobility-shift analyses were performed as previously described (23). DNA probes were generated to the AP-1 and NF-κB consensus sequences (AP-1, 5′-CGC TTG ATG ACT CAG CCG GAA-3′; NF-κB, 5′-AGT TGA GGG GAC TTT CCC AGG C-3′). Antibodies against c-jun, c-fos, p50, p65, or IgG1 (Sigma) were incubated with nuclear extracts for 2 h at 4°C before adding the radiolabeled oligonucleotide. DNA binding complexes were run in a 6% polyacrylamide gel. Autoradiography was performed after drying the gel.
ChIP assays were performed according to Miao et al. (27) by using the ChIP assay kit (Upstate Biotechnology) following the manufacturer's protocol. THP-1 cells were infected with Ad-KLF2 or EV control virus and stimulated with LPS, followed by crosslinking with 1% formaldehyde. PCR primers were used as described for the COX-2 promoter region (27), and amplification was performed following the same conditions.
Phagocytosis Assays.
Infected THP-1 cells (1 × 106 per ml) were stimulated with LPS and cultured in the presence or absence of zymosan A (Saccharomyces cerevisiae) bioparticles, Texas red conjugate (Molecular Probes) for 1 h at 37°C. Cells were viewed for internalization of particles under immunofluorescence after blocking the external signal with Trypan blue (GIBCO). Assays were performed in triplicate for at least three independent experiments.
Multiplex Sandwich ELISA.
THP-1 cells were infected with Ad-GFP (EV) and Ad-KLF2 (KLF2) for 24 h and treated with LPS (25 ng/ml) for 2 h and 6 h, and supernatants were collected after spinning. Search Light Proteome Arrays/multiplex sandwich ELISA was performed from these supernatants by Pierce. Each sample was evaluated in duplicate, and two independent experiments were evaluated.
Induction of Edema in the Mouse Paw.
Six- to 8-week-old male C.B-17-Scid-beige mice (from Taconic Farms) weighing 24–26 g were separated in groups (n = 4). Each group of animals received 3 Gy of total-body irradiation 24 h before tail vein injection of 1 × 107 macrophage (J774a) per mouse infected for 48 h with Ad-GFP-KLF2 (KLF2) or Ad-GFP (EV) as a control. Twenty-four hours after tail vein injection each group of animal received subplanter injection of 100 μl of carrageenan (1% wt/vol, Sigma) into the hind footpads (15). Animal care and procedures were reviewed and approved by the Harvard Medical School Standing Committee on Animals and performed in accordance with the guidelines of the American Association for Assessment and Accreditation of Laboratory Animal Care and the National Institutes of Health. Paw volume was measured by using a hydroplethismometer specially modified for small volumes (Ugo Basile) immediately before the subplanter injection and 1 and 2 h thereafter. The increase in paw volume was evaluated as the difference between paw volume at each time point and the basal paw volume. In some experiments paws were fixed in 10% buffered formalin (neutral pH), and frozen sections were stained with hematoxylin and eosin by using Dana–Farber/Harvard Cancer Center Pathology Core Facilities.
Thioglycolate-Induced Recruitment Studies.
Six- to 8-week-old male C.B-17-Scid-beige mice weighing 24–26 g were separated in groups (n = 5). Each group of animals received 3 Gy of total-body irradiation 24 h before tail vein injection of 1 × 107 macrophage (J774a) per mouse infected for 48 h with Ad-GFP-KLF2 or Ad-GFP as a control. Twenty-four hours after macrophage injection, 1 ml of thioglycolate (3% wt/vol, Sigma) was injected i.p (28), and recruitment of GFP-positive macrophages was assessed 48 h later. Flow cytometric analysis was performed for the GFP-positive cells as previously described (29).
siRNA-Mediated Knockdown of KLF2.
Mouse KLF2-directed siRNA (5′-GCACGGAUGAGGACCUAAAUU) and a nonspecific control siRNA (5′-CCUGGCGCCUUCGGUCUUUUU) were purchased from Dharmacon (Chicago). J774a cells were plated 1 day before transfection in antibiotic and serum-free DMEM. On the day of transfection, 60 nmol/liter specific siRNA or nonspecific siRNA was incubated with Lipofectamine 2000 (Invitrogen) at room temperature for 30 min before adding to the J774a cells. Four hours later medium was replaced with antibiotic containing serum-free DMEM for 48 h. Cells were harvested for RNA to assess for KLF2 knockdown and target gene expressions.
Statistics.
Data are expressed as mean ± SE. For comparison between two groups, an unpaired Student t test was used. For multiple comparisons, ANOVA followed by an unpaired Student t test was used. A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants HL69477, HL72952, HL75427, HL76754, and P01 HL48743 (to M.K.J.); HL67755 (to M.W.F.); and F32 HL78183 (to A.K.) and an American Heart Association Postdoctoral Fellowship (to Z.L.).
Abbreviations
- MCP
monocyte chemoattractant protein
- COX
cyclooxygenase
- ChIP
chromatin immunoprecipitation
- AP
activator protein
- PCAF
CBP-associated factor
- KLF
Kruppel-like factor
- EV
empty virus
- siRNA
short interfering RNA
- IKK
IκB kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. R.M. is a guest editor invited by the Editorial Board.
References
- 1.Lawrence T., Willoughby D. A., Gilroy D. W. Nat. Rev. Immunol. 2002;2:787–795. doi: 10.1038/nri915. [DOI] [PubMed] [Google Scholar]
- 2.Bieker J. J. DNA Cell Biol. 1996;15:347–352. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg M. W., Lin Z., Fisch S., Jain M. K. Trends Cardiovasc. Med. 2004;14:241–246. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Miller I. J., Bieker J. J. Mol. Cell. Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng W. C., Southwood C. M., Bieker J. J. J. Biol. Chem. 1994;269:1493–1500. [PubMed] [Google Scholar]
- 6.Perkins A. C., Sharpe A. H., Orkin S. H. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 7.Nuez B., Michalovich D., Bygrave A., Ploemacher R., Grosveld F. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 8.Kuo C. T., Veselits M. L., Leiden J. M. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 9.Buckley A. F., Kuo C. T., Leiden J. M. Nat. Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 10.Basu P., Morris P. E., Haar J. L., Wani M. A., Lingrel J. B., Gaensler K. M., Lloyd J. A. Blood. 2005;106:2566–2571. doi: 10.1182/blood-2005-02-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libby P. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 12.Patino W. D., Mian O. Y., Kang J. G., Matoba S., Bartlett L. D., Holbrook B., Trout H. H., III, Kozloff L., Hwang P. M. Proc. Natl. Acad. Sci. USA. 2005;102:3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucci M., Gratton J. P., Rudic R. D., Acevedo L., Roviezzo F., Cirino G., Sessa W. C. Nat. Med. 2000;6:1362–1367. doi: 10.1038/82176. [DOI] [PubMed] [Google Scholar]
- 14.Winter C. A., Risley E. A., Nuss G. W. Proc. Soc. Exp. Biol. Med.; 1962. pp. 544–547. [DOI] [PubMed] [Google Scholar]
- 15.Bucci M., Roviezzo F., Posadas I., Yu J., Parente L., Sessa W. C., Ignarro L. J., Cirino G. Proc. Natl. Acad. Sci. USA. 2005;102:904–908. doi: 10.1073/pnas.0408906102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayden M. S., Ghosh S. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 17.Karin M., Liu Z., Zandi E. Curr. Opin. Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 18.Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 19.Sheppard K. A., Rose D. W., Haque Z. K., Kurokawa R., McInerney E., Westin S., Thanos D., Rosenfeld M. G., Glass C. K., Collins T. Mol. Cell. Biol. 1999;19:6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wardell S. E., Boonyaratanakornkit V., Adelman J. S., Aronheim A., Edwards D. P. Mol. Cell. Biol. 2002;22:5451–5466. doi: 10.1128/MCB.22.15.5451-5466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SenBanerjee S., Lin Z., Atkins G. B., Greif D. M., Rao R. M., Kumar A., Feinberg M. W., Chen Z., Simon D. I., Luscinskas F. W., et al. J. Exp. Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A., Lin Z., Senbanerjee S., Jain M. K. Mol. Cell. Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg M. W., Jain M. K., Werner F., Sibinga N. E., Wiesel P., Wang H., Topper J. N., Perrella M. A., Lee M. E. J. Biol. Chem. 2000;275:25766–25773. doi: 10.1074/jbc.M002664200. [DOI] [PubMed] [Google Scholar]
- 24.Das H., Groh V., Kuijl C., Sugita M., Morita C. T., Spies T., Bukowski J. F. Immunity. 2001;15:83–93. doi: 10.1016/s1074-7613(01)00168-6. [DOI] [PubMed] [Google Scholar]
- 25.Cerutti J. M., Delcelo R., Amadei M. J., Nakabashi C., Maciel R. M., Peterson B., Shoemaker J., Riggins G. J. J. Clin. Invest. 2004;113:1234–1242. doi: 10.1172/JCI19617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray S., Feinberg M. W., Hull S., Kuo C. T., Watanabe M., Sen-Banerjee S., DePina A., Haspel R., Jain M. K. J. Biol. Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 27.Miao F., Gonzalo I. G., Lanting L., Natarajan R. J. Biol. Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 28.Shi C., Zhang X., Chen Z., Sulaiman K., Feinberg M. W., Ballantyne C. M., Jain M. K., Simon D. I. J. Clin. Invest. 2004;114:408–418. doi: 10.1172/JCI21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das H., Sugita M., Brenner M. B. J. Immunol. 2004;172:6578–6586. doi: 10.4049/jimmunol.172.11.6578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.