Abstract
Here we define a function of metastasis-associated protein 1 (MTA1), a presumed corepressor of estrogen receptor α (ERα), as a transcriptional activator of Breast Cancer Amplified Sequence 3 (BCAS3), a gene amplified and overexpressed in breast cancers. We identified BCAS3 as a MTA1 chromatin target in a functional genomic screen. MTA1 stimulation of BCAS3 transcription required ERα and involved a functional ERE half-site in BCAS3. Furthermore, we discovered that MTA1 is acetylated on lysine 626, and that this acetylation is necessary for a productive transcriptional recruitment of RNA polymerase II complex to the BCAS3 enhancer sequence. BCAS3 expression was elevated in mammary tumors from MTA1 transgenic mice and 60% of the human breast tumors, and correlated with the coexpression of MTA1 as well as with tumor grade and proliferation of primary breast tumor samples. These findings reveal a previously unrecognized function of MTA1 in stimulating BCAS3 expression and suggest an important role for MTA1-BCAS3 pathway in promoting cancerous phenotypes in breast tumor cells.
Keywords: BCAS3, coactivator, estrogen receptor
In recent years, many coregulator proteins that modulate nuclear receptor activity have been identified, raising questions about the complexity of nuclear receptor action (1–3). Coregulators alter chromatin by numerous strategies including ATP-dependent remodeling and histone modifications (1–3). For example, HDAC activity-containing NuRD (for nucleosome remodeling and histone deacetylation) complex also possesses ATP-dependent nucleosome remodeling activity due to associated Mi-2/CHD family of proteins. Another distinguishing feature of NuRD is the inclusion of MTA1 or MTA2. MTA1 was originally identified as being overexpressed in metastatic carcinomas (4) and contains several defined domains, including a zinc finger and a SANT domain (5). The dynamics of histone acetylation provide an attractive mechanistic foundation for the reversible activation and repression of transcription. In addition to histones, many transcriptional regulators, including components of the basal transcriptional machinery, are also acetylated (6). Thus acetylation, acetyltransferases, and deacetylases have diverse consequences apart from their effects on chromatin (7).
Genetic or epigenetic events alter gene expression and are involved in carcinogenesis. DNA amplification is an important mechanism that allows cancer cells to increase expression of oncogenes. The chromosomal region 17q23, which contains several oncogenes, is amplified in ≈20% of primary breast tumors and associated with cancer progression and poor prognosis (8). A gene of unknown function, FLJ20128 (later named Breast Carcinoma Amplified Sequence 3, BCAS3), also localized to this area. BCAS3 gene was amplified and overexpressed in breast cancer cell lines. Furthermore, the last two exons of BCAS3 were translocated to 20q13, another commonly amplified region in breast cancers, resulting in a fusion mRNA that was highly overexpressed (9). Thus, BCAS3 gene may be important in the process of breast tumorigenesis. However, a lack of information about the function of the protein, the absence of homology with any other known protein, and the absence of upstream regulator have impeded insights into its potential regulators and targets in breast cancer cells.
Expression of MTA1 has been shown to be closely correlated with aggressiveness in several types of cancers, including breast cancer (10). Overexpression of MTA1 results in increased anchorage-independent growth and growth of tumor xenografts in breast and pancreatic cancer cells (11, 12). Although MTA1 is a part of the NuRD complex and associated with HDACs, its precise function as a corepressor remained speculative until recently, when MTA1 was found to act as a repressor for ligand-induced estrogen receptor (ER) transactivation in breast cancer cells (12). Surprisingly, however, recent studies have implicated two coactivators of ER, MICoA and NRIF3, as MTA1-binding partners, giving support to the notion that coactivators and corepressors may coexist in the same complex (13, 14). Furthermore, in a transgenic mouse model of MTA1 (15), up-regulation of cyclin D1 was observed, prompting the authors to speculate that MTA1 may not be a universal corepressor. To explore the possibility of MTA1 having a role outside of its corepressor abilities, we undertook the current investigation and identified BCAS3 as a target of MTA1. Here, we investigated acetylation of MTA1 in a physiological setting and the consequences of such a modification upon MTA1's ability to control BCAS3 expression in breast cancer.
Results
For further details, see Supporting Text, Tables 2–4, and Figs. 7–9, which are published as supporting information on the PNAS web site.
Identification of a Chromatin Target of MTA1.
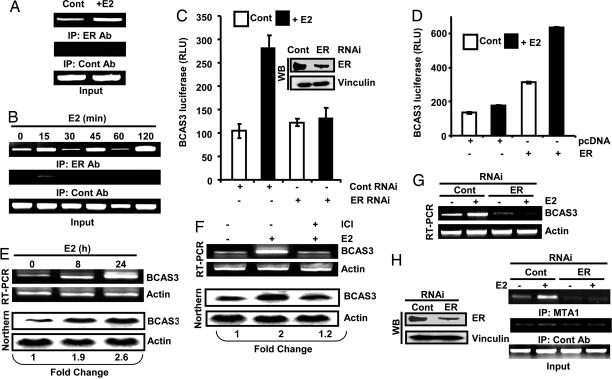
To identify targets of MTA1, we used a chromatin immunoprecipitation (ChIP)-based screening method and identified several targets (Table 2) including a 600-bp fragment that mapped to the second intron of the BCAS3, ≈12 kb away from the transcriptional start site of the gene. Subsequently, studies were undertaken to confirm that BCAS3 was indeed a bona fide in vivo target of MTA1. PCR using specific primers (BCAS3 F and BCAS3 R of Table 4) indicated that MTA1 could be recruited to the ChIP-pull down fragment in the BCAS3 intron under basal conditions and estrogen (E2) treatment increased MTA1 occupancy on the BCAS3 intron (Fig. 1A). ChIP analysis for 2-kb upstream (using primers UBCAS3 F and UBCAS3 R of Table 4) and downstream (using primers DBCAS3 F and DBCAS3 R of Table 4) of the pull down fragment indicated that this fragment appears to be the major MTA1 binding site in the BCAS3 gene (data not shown). Because MTA1 had been reported to be a corepressor for ER (12), we evaluated the status of BCAS3 under conditions of MTA1 up-regulation and depletion. To detect BCAS3 protein, anti-BCAS3 antibody was generated and characterized (Fig. 7). We found that BCAS3 mRNA and protein was higher in MTA1-overexpressing cells (T11 cells, ref. 12) (Fig. 1B). Likewise, knockdown of endogenous MTA1 by MTA1-specific RNA interference (RNAi) (16) in MCF-7 cells decreased the amounts of BCAS3 mRNA and protein levels (Fig. 1C).
Fig. 1.
Recruitment to the regulatory region and modulation of BCAS3 by MTA1. (A) MTA1-associated chromatin from MCF-7 cells associates with regulatory region of BCAS3. (B) Levels of BCAS3 and MTA1 in MCF-7/MTA1 (T11 clone) and MCF-7/pcDNA cells. (C) Effect of MTA1 knockdown by RNAi on the levels of BCAS3. (D) Effect of T7-MTA1 or pCDNA on the BCAS3-luc activity in MCF-7 cells. Inset, expression of transfected T7-MTA1. (E) Status of BCAS3-luc activity in MCF-7/MTA1 and HC11/MTA1 clones with respective controls. (F) Recruitment of MTA1 onto BCAS3 regulatory region upon E2 stimulation. (G) BCAS3-luc reporter activity in MCF-7 cells transiently transfected with T7-MTA1 and treated with E2. (H and I) Effect of E2 treatment on BCAS3-luc activity and BCAS3 protein in MCF-7/pcDNA and MCF-7/MTA1 cells. Luciferase activity is represented as fold induction by E2 as compared to control (n = 3). (J) Effect of MTA1 depletion on BCAS3 expression in MCF-7 cells treated with or without E2 (n = 3).
Because of the unexpected observation of MTA1 as a stimulator of BCAS3 gene expression and to validate the functionality of the regulatory sequence, a luciferase vector containing BCAS3 regulatory region and MTA1 expression vector was cotransfected. The results indicated a dose-dependent increase in the activity of the reporter gene in response to MTA1 expression in MCF-7 cells (Fig. 1D), indicating that MTA1 behaves as a coactivator in the context of BCAS3 gene in breast cancer cells. There was also elevated BCAS3–luciferase reporter gene activity in the MTA1 overexpressing stable clones than in the pcDNA clone (Fig. 1E). Other MTA family members had no significant effect on BCAS3 regulatory region in response to E2 stimulation as seen by cotransfection studies of BCAS3 luciferase activity with MTA2, MTA3 and MTA1s (Fig. 8), implying that the coactivator functions seem to be restricted to MTA1 in the context of BCAS3 gene.
Estrogen Regulation of BCAS3 Expression.
Next we determined the kinetics of MTA1 recruitment onto the regulatory region of BCAS3 in response to E2 signaling. In MCF-7 cells, MTA1 was recruited onto the enhancer region gradually with a maximum occupancy seen at 45 min after treatment followed by a gradual decrease in occupancy (Fig. 1F). Transient cotransfection of MTA1 expression plasmid with BCAS3-luciferase reporter vector resulted in increased reporter activity. Estrogen stimulation further augmented the activity of the reporter gene (Fig. 1G), indicating that MTA1 has a specific role in E2 induction of the BCAS3 gene. The role of MTA1 in E2 stimulation of BCAS3 was confirmed by the higher activity of the reporter gene and elevated BCAS3 protein in response to E2 stimulation in MTA1-overexpressing stable clones (Fig. 1 H and I). As expected, E2 stimulation of cells further enhanced the levels of BCAS3. To validate a role of MTA1 in E2 regulation of BCAS3, we next depleted MTA1 in MCF-7 cells and found a significant down-regulation of protein level upon stimulation by E2 (Fig. 1J), indicating that MTA1 is indispensable for induction of BCAS3 expression by E2.
Because MTA1 was recruited to the BCAS3 enhancer sequence in response to E2 stimulation, we examined a potential role for ER signaling in the regulation of BCAS3 expression. By ChIP analysis, we found that E2 stimulation of MCF-7 cells induced the recruitment of ERα onto the regulatory region (Fig. 2A) with a cyclic pattern involving ERα recruitment occurring at 15-min intervals (Fig. 2B). Knockdown of ERα and cotransfection of BCAS3-luciferase reporter vector in MCF-7 cells abrogated E2-stimulated luciferase activity relative to that in con-RNAi-transfected cells (Fig. 2C). Transient transfection of ERα expression vector along with BCAS3-luciferase reporter vector in ERα negative HeLa cells also enhanced reporter gene activity in the presence of ER; the activity was further stimulated with E2 treatment (Fig. 2D). MCF-7 cells stimulated by E2 for either 8 or 24 h showed significant up-regulation of BCAS3 mRNA levels (Fig. 2E) that could be blocked by the pure anti-estrogen ICI-182780 (Fig. 2F). Furthermore, the E2-induced increase in BCAS3 expression was abolished when ERα was depleted in MCF-7 cells by using ERα-specific RNAi (Fig. 2G), suggesting that the E2-stimulated up-regulation of BCAS3 expression is specific and mediated by ERα.
Fig. 2.
Occupancy of BCAS3 enhancer module by ERα and regulation of gene expression. (A) E2 stimulation promotes ERα recruitment onto BCAS3 regulatory region in MCF-7 cells. (B) ERα occupancy of BCAS3 regulatory sequence upon E2 treatment. (C) Effect of ERα depletion upon E2 stimulation of BCAS3-luc activity in MCF-7 cells. (D) BCAS3-luc activity in HeLa cells transfected with ERα or vector and treated with E2. (E) Effect of E2 stimulation on BCAS3 mRNA in MCF-7 cells. (F) Inhibition of E2-induced BCAS3 expression by ICI-182780 in MCF-7 cells. (G) Effect of ERα knockdown on BCAS3 expression in MCF-7 cells treated with or without E2. (H) (Right) Recruitment of MTA1 onto BCAS3 regulatory region in response to E2 stimulation in ER-depleted MCF-7 cells. (Left) Status of ERα knockdown by RNAi (n = 3).
Molecular Basis of Estrogen Responsiveness of BCAS3.
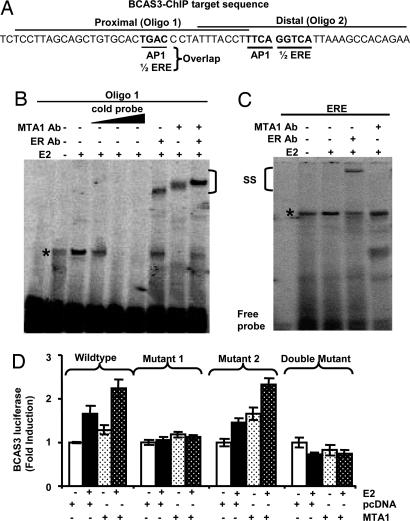
To determine whether ERα was essential for MTA1 occupancy of the BCAS3 regulatory region, ChIP analysis was carried out after ERα depletion and E2 stimulation using MTA1 antibody. Results showed that MTA1 could not be optimally recruited onto the BCAS3 enhancer sequence in the absence of ERα upon E2 stimulation (Fig. 2H). Analysis of the BCAS3 enhancer sequence suggested the presence of two potential ERE half-sites (TGACC) in the vicinity of AP1-binding sites (Fig. 3A). We next examined the potential involvement of these ERE half-sites in the regulation of BCAS3 expression by ERα and MTA1. Specific32P-labeled oligonucleotides encompassing either proximal or distal ERE half-sites were incubated with either control or E2-treated MCF-7 nuclear extract. Results showed the formation of higher order protein–DNA complexes, which could be competed out with 100-fold excess cold probe for the proximal ERE half-site (Fig. 3B, lanes 2–6); no specific protein–DNA complexes could be detected in the oligo designed against the distal site (data not shown). These complexes could be supershifted with anti-ERα or MTA1-specific antibodies (Fig. 3B, lanes 7–9), indicating that both ERα and MTA1 are directly recruited to the region containing the proximal ERE half-site. Nuclear extracts from the above experiments were also subjected to gel-shift and supershift analysis with oligo containing an ERE-full site. Anti-ERα, but not MTA1, antibody could supershift the complex formed (Fig. 3C). Mutation of the proximal ERE half-site (TGAC to TGTA) in the BCAS3–luciferase reporter construct substantially reduced E2's ability to induce reporter gene activity (Fig. 3D). However, mutation of the distal site (GTCA to TACA) did not change E2-induced reporter activity, whereas a double mutant with both sites mutated also compromised the E2-stimulated gene activity (Fig. 3E). These results suggest that proximal ERE half-sites play an important role in the E2-mediated up-regulation of BCAS3 enhancer activity.
Fig. 3.
MTA1 and ERα regulate BCAS3 gene expression via an ERE half-site. (A) Schematic representation of the two ERE half-sites in BCAS3 regulatory sequence. (B) Gel-shift and supershift assay using labeled oligo in nuclear extracts from MCF-7 cells treated with E2. SS, supershift with ERα or MTA1 Abs. (C) Gel shift assay with same nuclear extracts using ERE probe. SS, supershift with MTA1 or ERα Abs. (D) Effect of MTA1 upon luciferase activity driven by WT or mutated BCAS3 on either or both ERE half-sites. Luciferase activity is represented as fold induction by E2 as compared to control (n = 3).
Identification of Acetylation of MTA1 in Vivo.
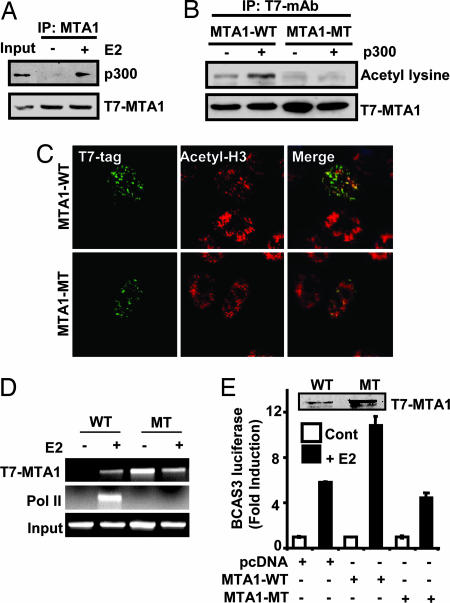
Previous studies have shown that MTA1 can be found in complexes containing coactivators with associated HAT (13). Therefore, we explored whether MTA1 interacts with the components of the HAT complex, and found that MTA1 associates with p300 in MCF-7 cells stably expressing T7-MTA1 (Fig. 4A). Because p300 has HAT activity, physical interaction of MTA1 with p300 raised the possibility that MTA1 could be a substrate for p300. Inspection of the primary amino acid sequence of MTA1 revealed a potential acetylation motif encompassing Lys-626 in the context of the GKSYP motif (data not shown). This acetylation motif was selectively present in MTA1 protein and not in other members of the MTA family. To determine whether MTA1 could be specifically acetylated by p300, T7-MTA1 expression vector was transfected with either control or HA-p300-expression vector. Immunoprecipitation of the transfected MTA1 was done by using antibody specific to T7-tag followed by Western blotting using acetyl-lysine antibody. MTA1 was indeed acetylated in vivo, and p300 cotransfection increased the acetylation level (Fig. 4B). To identify the acetylation site, we mutated the potential lysine residue into alanine, generating T7-MTA1-K626A mutant, and found its inability to be acetylated by exogenous p300 (Fig. 4B).
Fig. 4.
Identification of MTA1 as an acetylated protein. (A) CoIP and Western blot analysis showing MTA1 association with p300 in vivo. (B) MCF-7 cells transfected with T7-MTA1 or -MTA1-K626A mutant with lysine to alanine substitutions in the GKSYP motif, and p300 were IP with anti-T7 and analyzed with antibodies to acetylated lysine or T7. (C) Localization of T7- MTA1-WT or T7-MTA1-K626A (green) and acetyl-H3 (red) in MCF-7 cells. (D) MCF-7 cells transfected with MTA1-WT or MTA1-K626A mutant were treated with E2 and ChIP assay was performed with T7- MAb. Double ChIP was carried out with Pol II antibody. (E) Effect of MTA1-WT and MTA1-K626A mutant on PGL2-BCAS3-luc activity in MCF7 cells. Luciferase activity is represented as fold induction by E2 as compared to control (n = 3).
Having established that MTA1 is indeed acetylated on K626 in vivo, we sought to determine the functional consequences of MTA1 acetylation. Analysis of the effect of acetylation on MTA1 localization was evaluated by transfection of wild-type MTA1 or MTA1-K626A mutant followed by staining for the T7-tag and acetylated H3. MTA1-WT colocalized with acetyl-H3, a marker of active chromatin in the cell, whereas the K626A mutant showed no such colocalization (Fig. 4C), suggesting that acetylation may be important for MTA1 activity as a coactivator.
We next examined the effect of acetylation status of MTA1 on its occupancy of the BCAS3 enhancer sequence by transfection of T7-MTA1-WT or MTA1-K626A expression vector followed by ChIP analysis using anti-T7 mAb. Elutes from the first ChIP were reimmunoprecipitated with an antibody against RNA polymerase II (Pol II), a marker of functionally active transcriptional complex. Pol II was recruited to the BCAS3 enhancer region in E2-stimulated cells expressing MTA1-WT but not those expressing MTA1-K626A (Fig. 4D), suggesting in vivo existence of an MTA1/Pol II active complex.
To determine the functional consequence of MTA1 acetylation on MTA1-mediated BCAS3 gene expression, vectors expressing T7-MTA1-WT or MTA1-K626A were cotransfected into MCF-7 cells along with BCAS3-reporter vector. We found that MTA1-K626A was incapable of inducing transcriptional activation in response to E2, whereas MTA1-WT activated BCAS3-luciferase (Fig. 4E). These findings indicated that the acetylation of MTA1 is important for the ability of MTA1 to associate with HAT complex proteins, presumably by dissociating MTA1 from HDAC2, and that such changes might influence the coactivator function of MTA1 upon the BCAS3 regulatory region.
Potential Role of BCAS3 in Breast Cancer.
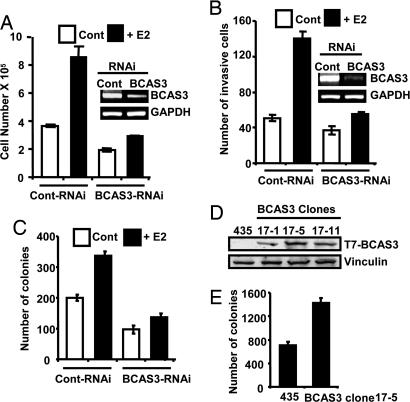
Having established that BCAS3 is regulated by MTA1 and ERα, we next asked whether endogenous BCAS3 was important for the tumorigenic properties manifested by breast cancer cells. MCF-7 cell growth was effectively inhibited by down-regulation of BCAS3 by specific small interfering RNA (Fig. 5A). Similarly, highly motile ZR-75 cells that could be induced to move across a membrane in response to the ligand in the Boyden chamber assay were unable to do so in the absence of BCAS3 (Fig. 5B). Also, ligand-stimulated anchorage-independent growth was compromised when the amount of BCAS3 protein was decreased (Fig. 5C), indicating that BCAS3 is essential for typical tumorigenic properties of cancerous cells. To delineate the potential effects of BCAS3 on the biology of breast cancer cells, we also established stable MDA-MB-435 clones expressing T7-tagged BCAS3 or control vector (Fig. 5D). Deregulated BCAS3 expression increased anchorage independent growth of the MDA-MB-435 cells on soft agar (Fig. 5E).
Fig. 5.
BCAS3 deregulation impacts pathophysiological effects of E2. (A) Effect of BCAS3 knockdown on MCF-7 cell proliferation in response to E2. A total of 10,000 cells per well were plated in six-well dishes, and BCAS3 knockdown was performed followed by E2 treatment. Cell number was counted 4 days later by using a Coulter Counter (n = 3). (B) BCAS3 knockdown reduces E2-induced invasion of ZR-75 cells. Cells were transfected with BCAS3 RNAi and, 48 h after transfection, 20,000 cells were loaded on the upper well of a Boyden chamber coated with matrigel. Cells that invaded the membrane were counted after 24 h (n = 9). (C) BCAS3 knockdown inhibits anchorage-independent growth of ZR-75 cells. Cells were transfected with BCAS3 RNAi and, 48 h after transfection, 10,000 cells were used for soft agar assay. The colonies were counted after 2 weeks of growth (n = 3). (D) Expression of T7-BCAS3 in stable clones. (E) BCAS3 overexpression promotes anchorage-independent growth of MDA-MB-435 cells. Conditions used were the same as described for C (n = 3).
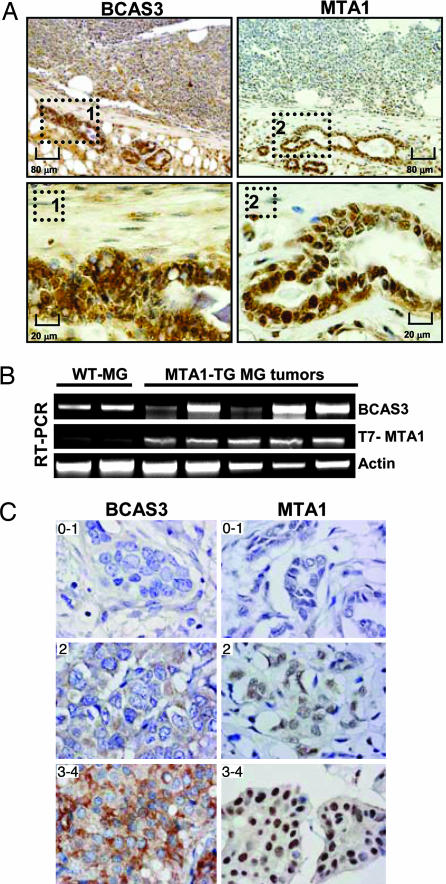
Because BCAS3 resides in a locus that is frequently amplified in breast cancers (17) and given the fact that it is essential in cancerous phenotypes and can promote anchorage-independent growth of breast cancer cells (this study), we sought to investigate the expression levels of BCAS3 in breast cancers. In MTA1 transgenic mice that developed breast tumors (15), BCAS3 was up-regulated in the tumors, as evidenced by immunohistochemistry (Fig. 6A) using an anti-peptide BCAS3 antibody (Fig. 9) and RT-PCR (Fig. 6B). Immunohistochemical examination of BCAS3 and MTA1 expression in the paired tumor and normal tissues showed a positive correlation between two proteins (Fig. 9).
Fig. 6.
BCAS3 deregulation in breast tumors in mice and humans. (A) Immunohistochemical analysis of BCAS3 and MTA1 expression in breast tumors from MTA1-TG mice. (Upper) Lower magnification (×100) pictures. (Lower) Higher magnifications (×400) of the same samples. (B) RT-PCR analysis of BCAS3 and T7-MTA1 in mammary tumors from MTA1-TG and WT-mouse mammary tissue. (C) BCAS3 and MTA1 staining in primary breast cancer samples. Breast cancer tissue microarrays were stained with BCAS3 and MTA1 antibodies and the staining was scored from 0 (negative) to 5 (strongest staining).
Based on above studies, BCAS3 and MTA1 status was assessed in a patient cohort consisting of 380 premenopausal breast cancer patients (18). The expression of BCAS3 and MTA1 was first evaluated from 0 (negative) to 4 (strongest intensity) but were later divided into three groups, 0–1, 2, and 3–4, as illustrated in Fig. 6C. BCAS3 stained mostly in the cytoplasm. In addition to cytoplasmic staining, we consistently found easily detectable nuclear staining in the samples. MTA1 stained with varying intensity in the nuclei of tumor cells, and also some cytoplasmic staining could be detected that seemed to correlate with the intensity in the nucleus. There was a strong correlation between BCAS3 and MTA1 expression in the tumor samples (Table 1). Also, both BCAS3 and MTA1 correlated to tumor type where high expression of BCAS3 was more common in medullary types and high expression of MTA1 was more common in lobular types (Table 3). BCAS3 also correlated to tumor grade and proliferation, whereas MTA1 only correlated to proliferation (Table 3). Because the levels of cytoplasmic BCAS3 staining graded with progression, it is possible that BCAS3 may have functions in the cytoplasm.
Table 1.
Analysis for correlation between BCAS3 and MTA1 expression in primary breast cancer samples
| MTA1 nuclear staining | BCAS3 cytoplasmic staining |
|||
|---|---|---|---|---|
| 0–1 (n = 144) | 2 (n = 147) | 3–4 (n = 89) | P value* | |
| 0–1 (n = 126) | 66 | 33 | 13 | <0.001 |
| 2 (n = 138) | 33 | 60 | 38 | |
| 3–4 (n = 100) | 19 | 39 | 32 | |
*Correlation was calculated using Spearman's ρ.
Discussion
Increasing numbers of studies have implicated MTA1 as a metastasis-associated gene associated with the degree of invasion and metastasis. The mechanism by which MTA1 might aid metastasis has not yet been elucidated. Considering that MTA1 has been identified as part of the NuRD complex, it is to be expected that chromatin targets of MTA1 would provide explanations for the metastasis-promoting abilities of the protein. BCAS3, an MTA1 target, would fit into the mold of a gene that is important in tumorigenesis because BCAS3 gene is localized to a very frequently amplified region in breast cancer cells and is overexpressed in breast cancer (8, 9). During the course of our investigation into regulation of BCAS3 by MTA1, we found BCAS3 to be an E2-inducible gene involving an ERE half-site preceded by an AP1 site, as in many other ER-responsive genes (19). Furthermore, the acetylation status of MTA1 may also be a determinant of its coactivator activity in a target-specific manner.
Another notable finding from the current study is the presence of a regulatory module in the intronic region of the BCAS3 gene, ≈12 kb from the transcriptional start site. Although the presence of a regulatory region in the intron is unusual, the observation is not without precedent (20). Also, the presence of the ERE half-site was found to be essential for significant activation of the BCAS3 enhancer sequence by estradiol and was found to recruit Pol II and other coactivator molecules (this study), demonstrating that the intronic site is transcriptionally operational. Because the target gene promoter chromatin is a highly dynamic structure, the transactivating functions of coactivators are likely to be influenced by corepressors and any deregulation of one component will have functional implications for the action of other components. In this context, we found that a presumed corepressor, MTA1, can influence the gene expression of BCAS3 as a coactivator and thus behaves as a bifunctional coregulator.
MTA1 has been recently shown to be essential for transformation by MYC and acts downstream of the MYC oncogene (21). Although an upstream regulator of MTA1 has been identified, a downstream effector of MTA1 that could be involved in the cancer progression is lacking. Identification of BCAS3 as a transcriptional target of MTA1 highlights an important pathway through which MTA1 may participate in the progression of cancerous phenotypes.
Materials and Methods
Cellular and Biochemical Assays.
Cell extracts, immunoprecipitation, Western blot analysis, confocal microscopy, EMSA, cell proliferation, invasion, and soft agar assays was performed as described (22). ER gel-shift oligonucleotides were purchased from Santa Cruz Biotechnology.
Cell Treatment.
For E2 treatments, regular medium was replaced by minimum essential medium without phenol red containing 5% charcoal stripped serum. Estrogen concentration used for all experiments was 10−9 M. For all ChIP analyses, E2 treatment was for a duration of 45 min, whereas for all remaining experiments, treatment was for 24 h unless otherwise specified. ICI-182780 treatment was done 1 h before E2 treatment.
ChIP Assays.
ChIP assay was conducted following the same procedure as reported in ref. 12. Antibodies for MTA1 (Santa Cruz Biotechnology), ERα (Chemicon International) and T7-tag (Novagen) were used, and the precipitated DNA was either used for cloning or amplified by using specific primers (Table 4). For identification of MTA1 in vivo chromatin targets, the ChIP DNA fragments were cloned into pBluescript and sequenced. A blast search was performed by using the obtained DNA sequence against the human genome sequence to identify the genes with which MTA1 might be associated (Table 2).
Plasmid Construction and Mutagenesis.
To clone the BCAS3 regulatory region, we amplified the regulatory region from DNA isolated from MCF-7 cells by using the ChIP-PCR primers, BCAS3F and BCAS3R (Table 4) and cloned into pGL2 luciferase reporter vector as described in Supporting Text. QuikChange kit (Stratagene) was used for mutagenesis.
Generation of BCAS3 Antiserum.
A 14-mer peptide encoding amino acids 578–592 (ANNAGLKREKDQSKQ) of BCAS3 was synthesized, and the conjugated peptide was used to produce rabbit polyclonal antiserum (Invitrogen).
Patient Material.
From 1986 to 1991, a total of 564 premenopausal breast cancer patients with invasive stage II disease were enrolled in a Swedish clinical trial from where the samples were obtained and analyzed by G.L. as per Malmö University Hospital policy. A detailed description of the study design has been reported (18).
Tissue Microarray and Immunohistochemistry.
Immunohistochemistry was performed as reported (23) using antibodies against BCAS3 (1:500 dilution) and MTA1 (1:25 dilution). Details are provided in Supporting Text.
Statistical Methods.
Spearman's ρ and Pearson χ2 were used for comparison between BCAS3 and other categorized variable, whereas Kruskal–Wallis test was used for comparison of medians for continuous variables. All P values corresponded to two-sided tests, and values <0.05 were considered significant. Statistical analyses were performed by using spss 11.0 software.
Supplementary Material
Acknowledgments
We thank Rozita Yarmand-Bagheri and Zhibo Yang for help in transgenic and confocal microscopy studies. This study was supported by National Institutes of Health Grants CA65746, CA098823, and CA109379 (to R.K.).
Abbreviations
- ER
estrogen receptor
- ChIP
chromatin immunoprecipitation
- E2
estrogen
- RNAi
RNA interference
- Pol II
RNA polymerase II.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Fowler A. M., Alarid E. T. Sci. STKE. 2004;2004:e51. [Google Scholar]
- 2.McKenna N. J., O'Malley B. W. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 3.McKenna N. J., Lanz R. B., O'Malley B. W. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 4.Toh Y., Pencil S. D., Nicolson G. L. J. Biol. Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 5.Bowen N. J., Fujita N., Kajita M., Wade P. A. Biochim. Biophys. Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Huo X., Zhang J. J. Cell. Mol. Med. 2005;9:103–112. doi: 10.1111/j.1582-4934.2005.tb00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar R., Gururaj A. E., Vadlamudi R. K., Rayala S. K. Clin. Cancer Res. 2005;11:2822–2831. doi: 10.1158/1078-0432.CCR-04-1276. [DOI] [PubMed] [Google Scholar]
- 8.Monni O., Barlund M., Mousses S., Kononen J., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M. L., et al. Proc. Natl. Acad. Sci. USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barlund M., Monni O., Weaver J. D., Kauraniemi P., Sauter G., Heiskanen M., Kallioniemi O. P., Kallioniemi A. Genes Chromosomes Cancer. 2002;35:311–317. doi: 10.1002/gcc.10121. [DOI] [PubMed] [Google Scholar]
- 10.Kumar R., Wang R. A., Bagheri-Yarmand R. Semin. Oncol. 2003;30:30–37. doi: 10.1053/j.seminoncol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Hofer M. D., Menke A., Genze F., Gierschik P., Giehl K. Br. J. Cancer. 2004;90:455–462. doi: 10.1038/sj.bjc.6601535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazumdar A., Wang R. A., Mishra S. K., Adam L., Bagheri-Yarmand R., Mandal M., Vadlamudi R. K., Kumar R. Nat. Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 13.Mishra S. K., Mazumdar A., Vadlamudi R. K., Li F., Wang R. A., Yu W., Jordan V. C., Santen R. J., Kumar R. J. Biol. Chem. 2003;278:19209–19219. doi: 10.1074/jbc.M301968200. [DOI] [PubMed] [Google Scholar]
- 14.Talukder A. H., Gururaj A., Mishra S. K., Vadlamudi R. K., Kumar R. Mol. Cell. Biol. 2004;24:6581–6591. doi: 10.1128/MCB.24.15.6581-6591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagheri-Yarmand R., Talukder A. H., Wang R. A., Vadlamudi R. K., Kumar R. Development (Cambridge, U.K.) 2004;131:3469–3479. doi: 10.1242/dev.01213. [DOI] [PubMed] [Google Scholar]
- 16.Mishra S. K., Talukder A. H., Gururaj A. E., Yang Z., Singh R. R., Mahoney M. G., Franci C., Vadlamudi R. K., Kumar R. J. Biol. Chem. 2004;279:32709–32715. doi: 10.1074/jbc.M402942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinclair C. S., Rowley M., Naderi A., Couch F. J. Breast Cancer Res. Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- 18.Ryden L., Jirstrom K., Bendahl P. O., Ferno M., Nordenskjold B., Stal O., Thorstenson S., Jonsson P. E., Landberg G. J. Clin. Oncol. 2005;23:4695–4704. doi: 10.1200/JCO.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 19.Klinge C. M. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laganiere J., Deblois G., Giguere V. Mol. Endocrinol. 2005;19:1584–1592. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X. Y., DeSalle L. M., Patel J. H., Capobianco A. J., Yu D., Thomas-Tikhonenko A., McMahon S. B. Proc. Natl. Acad. Sci. USA. 2005;102:13968–13973. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vadlamudi R. K., Adam L., Wang R. A., Mandal M., Nguyen D., Sahin A., Chernoff J., Hung M. C., Kumar R. J. Biol. Chem. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 23.Vadlamudi R. K., Balasenthil S., Broaddus R. R., Gustafsson J. A., Kumar R. J. Clin. Endocrinol. Metab. 2004;89:6130–6138. doi: 10.1210/jc.2004-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.