Abstract
Obesity, especially central obesity, is a hereditable trait associated with a high risk for development of diabetes and metabolic disorders. Combined gene expression analysis of adipocyte- and preadipocyte-containing fractions from intraabdominal and subcutaneous adipose tissue of mice revealed coordinated depot-specific differences in expression of multiple genes involved in embryonic development and pattern specification. These differences were intrinsic and persisted during in vitro culture and differentiation. Similar depot-specific differences in expression of developmental genes were observed in human subcutaneous versus visceral adipose tissue. Furthermore, in humans, several genes exhibited changes in expression that correlated closely with body mass index and/or waist/hip ratio. Together, these data suggest that genetically programmed developmental differences in adipocytes and their precursors in different regions of the body play an important role in obesity, body fat distribution, and potential functional differences between internal and subcutaneous adipose tissue.
Keywords: adipose tissue, gene expression, subcutaneous, intraabdominal
Obesity is an epidemic health problem worldwide that impacts the risk and prognosis of many diseases, including diabetes, cardiovascular disease, hyperlipidemia, and cancer (1). However, not all obese patients have the same risk of developing these disorders. Individuals with peripheral obesity, i.e., fat distributed subcutaneously in the gluteofemoral region, are at little or no risk of the common medical complications of obesity, whereas individuals with central obesity, i.e., fat accumulated in visceral depots, are prone to these complications (2–5).
Although differentiation of adipocytes has been extensively characterized (6–8) and there have been considerable recent insights into the control of appetite and energy expenditure as contributing factors to obesity (9, 10), little is known about the genetic basis for determination of adipocyte number, differences in body fat distribution, or their association with metabolic disorders. Twin and population studies have revealed that both body mass index (BMI) and waist/hip ratio (WHR) are heritable traits, with genetics accounting for 25–70% of the observed variability (11, 12). In addition, it is known that some obese individuals, especially those with early-onset obesity, have increased adipocyte number, but how these are distributed and why this occurs is unknown (13). Anecdotally, it is also clear that individual humans observe differences in their own body fat distribution as they gain or lose weight, and this is extreme in some ethnic groups, such as Hottentot women, who have been noted for excessive accumulation of fat in the buttocks, a condition known as steatopygia (14). Striking differences in adipose tissue distribution can also be observed in individuals with partial lipodystrophy (15), in both its acquired and inherited forms. For example, familial partial lipodystrophy of the Dunnigan type due to mutations in the Lamin A/C gene is characterized by a marked loss of s.c. adipose tissue in the extremities and trunk, without loss of visceral, neck, or facial adipose tissue (16, 17). Some lipodystrophies even appear to have a segmental or dermatomal distribution (18).
In the present study, we have explored the hypothesis that patterns of fat distribution and, perhaps, to some degree, obesity itself may have a developmental genetic origin. Indeed, we find major differences in expression of multiple genes involved in embryonic development and pattern specification between adipocytes taken from intraabdominal and s.c. depots in rodents and humans. We also demonstrate similar differences in the stromovascular fraction (SVF)-containing preadipocytes and that these differences persist in culture. Most importantly, we demonstrate that some of these developmental genes exhibit changes in expression that are closely correlated with the level of obesity and the pattern of fat distribution.
Results
Genes Expression Differences Between Intraabdominal and s.c. Adipose Tissue of Mice.
Several studies have reported differences in gene expression (19–22) and proliferative capacity (23–28) between fat taken from different depots in rodents and humans, suggesting that genetic programming could affect specific adipose depot development. To address this hypothesis, we performed gene expression analysis of both adipocytes and SVF containing preadipocytes taken from s.c. (flank) fat and intraabdominal (epididymal) fat, using Affymetrix U74Av2 microarrays with 8,017 probe sets representing 6,174 different annotated genes (Fig. 1). Of these, 197 genes were found to have conjoint differential expression in both cell fractions between the two tissue beds by using stringent statistical criteria with a two-tailed t test value for both cell fractions <0.05 and a positive false discovery rate <0.05 (see Supporting Methods and Table 2, which are published as supporting information on the PNAS web site). This list was assessed against an a priori set of 198 annotated genes involved in embryonic development and pattern specification on the array (see Supporting Methods and Table 3, which are published as supporting information on the PNAS web site). Twelve of these developmental genes were found among the differentially expressed genes, representing a 1.9-fold enrichment (P = 0.006) compared with the 6,174 annotated genes on the array (Table 1).
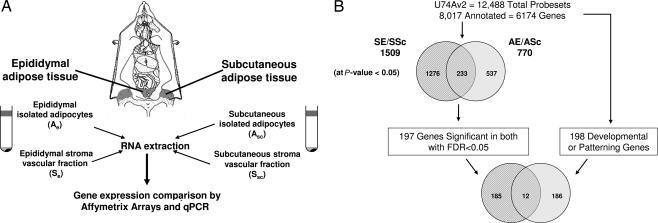
Fig. 1.
Experimental design. (A) Flank s.c. and intraabdominal (epididymal) white adipose tissue were taken from 6- to 7-week-old pooled C57BL/6 males. SVF and adipocytes were isolated after collagenase digestion of adipose tissues. Equal quantities of RNA were isolated from isolated adipocytes and SVF of each fat depot. A hybridization mixture containing 15 μg of biotinylated cRNA, adjusted for possible carryover of residual total RNA, was prepared and hybridized to mouse Affymetrix U74Av2 chips. (B) Among the 12,488 probe sets present on the U74Av2 chip, 8,017 probe sets representing 6,174 are annotated for Gene Ontology Biological Process. Significant genes with differential expression in both depots were identified by selecting genes that passed two independent filters of significance (P < 0.05, Student’s t test; positive false discovery rate < 0.05) (see Materials and Methods). The first filter (P < 0.05, Student’s t test) selected 1,276 genes differentially expressed in the SVF, 537 genes differentially expressed in isolated adipocytes, and 233 genes differentially expressed in both cell fractions. Of these 233 genes, 197 genes passed the second filter of significance (positive false discovery rate < 0.05) and were assessed against an a priori set of 198 annotated genes involved in embryonic development and pattern specification (see Materials and Methods). Twelve genes from this set were found among the differentially expressed genes.
Table 1.
Developmental and patterning genes showing differential expression in adipoctyes and SVF of adipose tissue
| Gene ID | Probe set ID | Ae | Asc | P value, Ae vs. Asc | Se | Ssc | P value, Se vs. Ssc | Gene title | Gene symbol | pFDR |
|---|---|---|---|---|---|---|---|---|---|---|
| AF041822 | 102256_at | 20.8 | 255 | 0.0012 | 23.1 | 727.1 | 0.0124 | T-box 15 | Tbx15 | 0.0026 |
| U66918 | 99042_s_at | 282.1 | 2456 | 0.0004 | 397.2 | 3,622.3 | 0.0023 | Short stature homeobox 2 | Shox2 | 0.0026 |
| L12703 | 96523_at | 53.1 | 422.9 | 0.0128 | 1,005.3 | 2,116.3 | 0.0083 | Engrailed 1 | En1 | 0.0032 |
| U88567 | 93503_at | 281.2 | 1,510.6 | 0.0405 | 621.2 | 6,317.6 | 0.0001 | Secreted frizzled-related sequence protein 2 | Sfrp2 | 0.0026 |
| X55318 | 92891_f_at | 674.4 | 1,189.3 | 0.0042 | 748.2 | 564.6 | 0.0357 | Homeobox C9 | Hoxc9 | 0.0466 |
| AW061016 | 99964_at | 27 | 53.5 | 0.0144 | 439.6 | 40.2 | 0.0278 | Vitamin D receptor | Vdr | 0.0038 |
| X83577 | 102886_at | 2,522.8 | 1,687.2 | 0.0102 | 7312 | 2151 | 0.0090 | Glypican 4 | Gpc4 | 0.0026 |
| X07439 | 93378_at | 2,484.5 | 1,597.2 | 0.0374 | 3,351.3 | 1,144.9 | 0.0003 | Homeobox C8 | Hoxc8 | 0.0028 |
| AI837110 | 96696_at | 1,628.4 | 990.7 | 0.0287 | 3,705.9 | 2,379.7 | 0.0369 | Heterogeneous nuclear ribonucleoproteins methyltransferase-like 2 | Hrmt1/2 | 0.0069 |
| Y00208 | 103086_at | 1,604.8 | 837.8 | 0.0305 | 1,657.3 | 242.6 | 0.0001 | Homeobox A5 | Hoxa5 | 0.0027 |
| X14432 | 104601_at | 2,245.4 | 861.4 | 0.0094 | 5,919.2 | 1,878.5 | 0.0005 | Thrombomodulin | Thbd | 0.0026 |
| X74134 | 102715_at | 551.1 | 101.1 | 0.0057 | 1,383.2 | 289.9 | 0.0004 | Nuclear receptor subfamily 2, group F, member 1 | Nr2f1 | 0.0026 |
Ae, epididymal isolated adipocytes; Asc, s.c. isolated adipocytes; Se, epidydimal SVF; Ssc, s.c. SVF; pFDR, positive false discovery rate. Bold represents the tissue bed with the highest level of expression.
Among these 12 genes, seven genes had higher levels of expression in intraabdominal epididymal SVF and/or adipocytes (Nr2f1, Thbd, HoxA5, HoxC8, Gpc4, Hrmt1l2, and Vdr) and five genes had higher levels of expression in s.c. SVF and/or adipocytes (Tbx15, Shox2, En1, Sfpr2, and HoxC9). Of the seven genes from intraabdominal group, we decided to focus our analysis on the five most significant genes, including two homeo box genes, HoxA5 and HoxC8; Nr2f1, nuclear receptor subfamily 2 group F member 1, also known as COUP-TFI, an orphan member of the steroid receptor superfamily thought to be involved in organogenesis (29); glypican 4 (Gpc4), a cell-surface heparan sulfate proteoglycan involved in cell division and growth regulation (30); and thrombomodulin (Thbd), a surface glycoprotein of endothelial and placental cells (31). We also studied all five genes from the s.c. group of genes including the homeobox gene HoxC9; short stature homeobox 2 (Shox2) a transcription factor with homeodomain expressed during embryonic development (32); Tbox-15 (Tbx15), a transcription factor involved in craniofacial and limb development in the mouse (33); engrailed 1 (En1), the mouse homologue of a Drosophila patterning gene (34); and secreted frizzled-related protein 2 (Sfrp2), a soluble modulator of Wnt signaling (35).
Confirmation of Interdepot Gene Expression Differences by Quantitative PCR (qPCR).
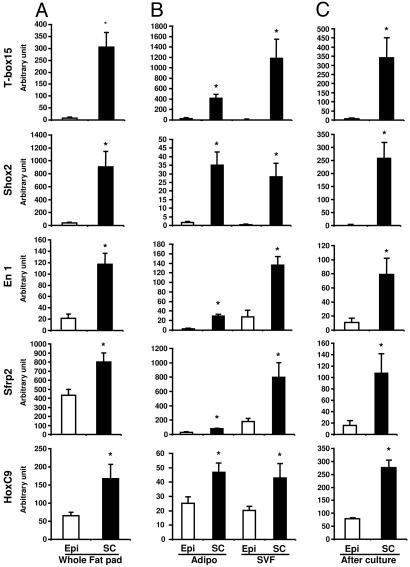
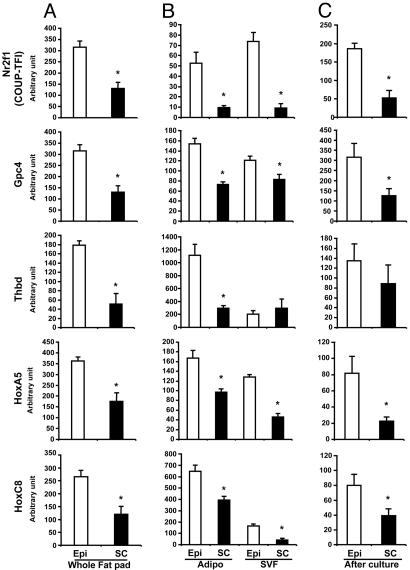
These differences of expression in genes involved in embryonic development and pattern specification were confirmed by qPCR. In whole tissue, all predominantly s.c. genes Tbx15, Shox2, En1, Sfrp2, and HoxC9 were more highly expressed in s.c. adipose tissue than in intraabdominal (epididymal) fat, with the most marked differences observed for Tbx15, Shox2, and En1 expression (39-, 23-, and 5.4-fold, respectively; P = 0.005, 0.018, and 0.008, respectively) (Fig. 2A). Conversely, all predominant intraabdominal genes, Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8, were significantly more expressed in intraabdominal adipose tissue than in s.c. adipose tissue by 2.1- to 3.5-fold (all P < 0.05) (Fig. 3A).
Fig. 2.
Differential expression of development genes Tbx15, Shox2, En1, Sfrp2, and HoxC9 in s.c. and intraabdominal adipose tissue, adipocytes, and SVF in mice. Comparison of Tbx15, Shox2, En1, Sfrp2, and HoxC9 gene expression between intraabdominal (Epi, open bars) and s.c. (SC, filled bars) adipose tissue of C57BL/6 mice was performed by using qPCR as described in Materials and Methods. These genes have a higher level of expression in s.c. in whole adipose tissue (A) (Epi versus Sc; ∗, P < 0.05), isolated adipocytes, and SVF (B) (Epi versus Sc; ∗, P < 0.05). These differences of expression are maintained when SVF taken from intraabdominal (epididymal) or s.c. adipose were placed in culture in a defined serum-free medium and subjected to in vitro differentiation (C), suggesting that these differences are independent of extrinsic factors (Epi versus Sc; ∗, P < 0.05).
Fig. 3.
Differential expression of developmental genes Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8 in s.c. and intraabdominal adipose tissue, adipocytes, and SVF. Comparison of Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8 gene expression between intraabdominal (Epi, open bars) and s.c. (SC, filled bars) adipose tissue of C57BL/6 mice was performed by using qPCR as described in Materials and Methods. These genes have a higher level of expression in intraabdominal (epidydimal) whole adipose tissue (A) (Epi versus Sc; ∗, P < 0.05), isolated adipocytes, and SVF (B) (Epi versus Sc; ∗, P < 0.05). These differences of expression are maintained when SVF taken from intraabdominal (epididymal) or s.c. adipose were placed in culture in a defined serum-free medium and subjected to in vitro differentiation (C), suggesting that these differences are independent of extrinsic factors (Epi versus Sc; ∗, P < 0.05).
Likewise, differences were confirmed in isolated adipocytes and stromovascular cells obtained from both depots by qPCR. Thus, both adipocytes and SVF cells isolated from s.c. adipose tissue expressed higher levels of all s.c. genes, Tbx15 [140- and 460-fold (P = 0.001 and 0.013)], Shox2 [20- and 205-fold (P = 0.006 and 0.012)], En1; [12.3- and 4.9-fold (P = 0.0006 and 0.0007)], Sfrp2 [2.6- and 4.5-fold (P = 0.001 and 0.04)], and HoxC9 [1.8- and 2.1-fold (P = 0.023 and 0.06)] (Fig. 2B). Conversely, adipocytes and SVF from epididymal adipose tissue expressed higher levels of intraabdominal genes Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8 [5.4- and 7.8-fold (P = 0.006 and 0.003), 2.1- and 1.5-fold (P = 0.003 and 0.05), 3.8- and 0.7-fold (P = 0.004 and 0.3), 1.6- and 2.2-fold (P = 0.04 and 0.02), and 3.8- and 1.7-fold (P = 0.009 and 0.02), respectively] (Fig. 3B).
Interdepot Differences in Gene Expression Are Independent of Extrinsic Factors.
To determine whether these differences in gene expression were cell-autonomous, preadipocytes (SVF) taken from intraabdominal (epididymal) or s.c. adipose were placed in culture in defined serum-free medium and subjected to in vitro differentiation. After 6 days, all of the predominantly s.c. genes and all of the predominantly epididymal genes maintained their interdepot differences of expression (Figs. 2C and 3C). Thus, differences of developmental gene expression between depots are independent of extrinsic factors, such as innervation, blood flow, the level of oxygenation and nutrients, or any other interstitial factors.
Interdepot Differences of Expression in Humans.
Because the striking interdepot differences for expression of these developmental genes between s.c. and intraabdominal fat in mice appeared to be intrinsic and present in both the preadipocyte and adipocyte fractions, we decided to determine whether similar differences might be present in human adipose tissue. To address this question, 53 lean subjects (22 males and 31 females with BMI <25) with normal fat distribution (WHR for males, 0.80–1.06; WHR for females, 0.62–0.87) were subjected to abdominal s.c. and visceral adipose tissue biopsies, and gene expression for the human homologues of each of these developmental genes was assessed by using qPCR.
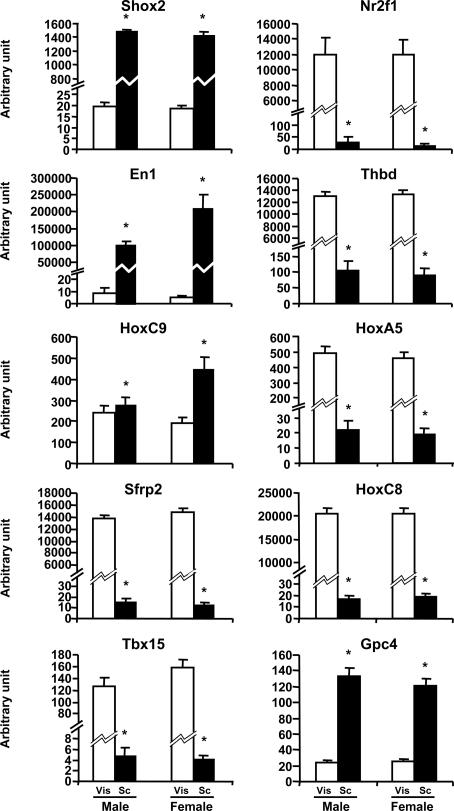
As observed in mice, Nr2f1, Thbd, HoxA5, and HoxC8, which showed higher expression in epididymal fat, showed a higher level of expression in visceral adipose tissue of humans in both males and females (Fig. 4). In addition, for these genes the magnitude of interdepot differential gene expression in humans was even greater than that in mice (Nr2f1, 461- and 894-fold; Thbd, 124- and 147-fold; HoxA5, 23- and 24-fold; HoxC8, 1,210- and 1,100-fold, for males and females, respectively). Glypican 4 (Gpc4) expression in humans also showed a strong differential expression; however, in lean humans this gene was more highly expressed in s.c. than in visceral adipose tissue, with a 5.4-fold difference in males and a 4.8-fold difference in females.
Fig. 4.
Differential expression of s.c. dominant genes and intraabdominal dominant genes in s.c. and intraabdominal adipose tissue of lean humans. Visceral (Vis, open bars) and s.c. (SC, filled bars) adipose tissue biopsies were performed on 53 lean subjects (BMI < 25; 22 males and 31 females). Tbx15, Shox2, En1, Sfrp2, HoxC9, Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8 expressions were compared in both depots by using qPCR as described in Materials and Methods (Vis versus SC; ∗, P < 0.05).
The group of s.c. genes also showed significant and differential patterns of expression between depots in humans. In this case, two of the genes, Shox2 and En1, presented a pattern of expression in humans in the same direction as in mice, and in the case of En1 the differential expression was of extreme magnitude (17,500- and 42,500-fold for males and females, respectively) (Fig. 4). As in mice, HoxC9 expression was found significantly higher in s.c. than in visceral adipose tissue (2.3-fold); however, in humans this difference was gender-specific and was not present in males. Tbx15 and Srfp2 also showed markedly different expression in humans; however, in humans these genes were more highly expressed in visceral adipose tissue than in s.c. adipose tissue in both genders (Tbx15, 27.1-fold in males and 38.7-fold in females; Sfrp2, 950-fold in males and 1,200-fold in females) (Fig. 4).
Gene Expression, BMI, and Body Fat Distribution.
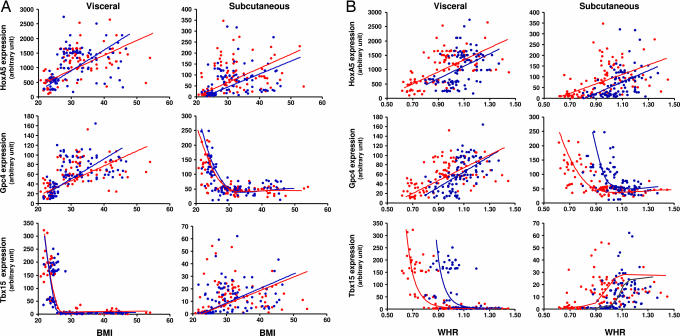
To investigate whether the genes studied were related to obesity or body fat distribution, we determined the level of gene expression in adipose tissue biopsies from this group of 53 subjects plus another group of 145 overweight or obese individuals. The final group of 198 human subjects (99 males and 99 females) ranged from lean to obese (BMI range, 21.7–46.8 for males and 20.8–54.1 for females), with variable adipose tissue distribution (WHR, 0.8–1.37 for males and 0.62–1.45 for females) (Table 4, which is published as supporting information on the PNAS web site). Three of the 10 developmental genes showed significant relationships to BMI or WHR. HoxA5 expression in both visceral and s.c. adipose tissue significantly increased with BMI in males (R = 0.448, P < 0.0001; R = 0.292, P = 0.0034, respectively) and females (R = 0.535, P < 0.0001; R = 0.361, P = 0.0002, respectively) (Fig. 5A). This correlation was more marked in visceral than in s.c. adipose tissue in both genders. In addition, there was a significant positive correlation of HoxA5 expression with WHR in visceral and s.c. adipose tissue for both males (R = 0.446, P < 0.0001; R = 0.479, P < 0.0001, respectively) and females (R = 0.580, P < 0.0001; R = 0.449, P < 0.0001, respectively) (Fig. 5B).
Fig. 5.
Expression of HoxA5, Gpc4 and Tbx15 in s.c. and visceral adipose tissue in humans are correlated with adiposity and fat distribution. One hundred ninety-eight subjects (99 males and 99 females) ranging from lean to obese with variable BMI (A) and fat distribution (WHR) (B) were subjected to visceral (Vis, open bars) and s.c. (SC, filled bars) adipose tissue biopsies. Gene expression of HoxA5, Gpc4, and Tbx15 was assessed in both fat depots by qPCR as described in Materials and Methods. Correlation significances were determined by using statview software either as linear correlations or, in the case of nonlinear correlations, by exponential or lowest curve fitting.
In human adipose, there were very strong correlations of Gpc4 expression with BMI and WHR in both males and females. In this case, the correlation in the two depots was in opposite directions, with decreasing Gpc4 expression in s.c. adipose tissue with increasing BMI (males, R = 0.74, P < 0.0001; females, R = 0.735, P < 0.0001) and WHR (males, R = 0.575, P < 0.0001; females, R = 0.730, P < 0.0001) and increasing Gpc4 expression in visceral adipose tissue with increasing BMI (males, R = 0.525, P < 0.0001; females, R = 0.507, P < 0.0001) and WHR (males, R = 0.598, P < 0.0001; females, R = 0.5, P < 0.0001) (Fig. 5). In addition, the shape of the relationship was different, being fairly linear in visceral adipose tissue, whereas in s.c. adipose tissue Gpc4 expression decreased abruptly as individuals went from normal BMI (20–25) to overweight (BMI > 25) or obese (BMI > 30) levels. Likewise, in s.c. adipose tissue Gpc4 expression displayed a curvilinear negative correlation with very low levels in males with WHR >1.1 and females with WHR >0.95.
The most profound correlations with BMI and WHR were observed for Tbx15 expression in visceral adipose tissue. As with Gpc4, there was a strong exponential negative relationship with a marked decrease in Tbx15 expression as BMI progressed from normal to overweight or obese levels. This was true in both males (R = 0.706, P < 0.0001) and females (R = 0.852, P < 0.0001) (Fig. 5A). There was also a strong exponential negative relationship between Tbox15 expression and WHR in visceral adipose tissue with marked declines above WHR of 1.05 for males (R = 0.604, P < 0.0001) and 0.95 for females (R = 0.817, P < 0.0001) (Fig. 5B). By contrast, Tbx15 expression showed a more modest positive correlation with both BMI and WHR in s.c. adipose tissue of both males (R = 0.282, P = 0.0047; R = 0.406, P < 0.0001) and females (R = 0.191, P = 0.0587; R = 0.345, P = 0.0005). However, in all cases, expression of Tbx15 in s.c. tissue was much lower than the level of expression in visceral adipose tissue of lean individuals. Thus, HoxA5, Gpc4, and Tbx15 expression in adipose tissue were strongly correlated with the level of obesity, as well as adipose tissue distribution, especially Tbx15 expression in visceral fat.
Discussion
Obesity is a multifactorial disorder influenced by a mixture of genetic and environmental factors, including control of appetite and energy expenditure, availability and nutritional content of food, and development of adipocyte cell mass. Furthermore, obesity occurs with different degrees of fat accumulation in different depots, and these are associated with different metabolic consequences with intraabdominal (visceral) accumulation of fat producing a much greater risk of diabetes, dyslipidemia, and accelerated atherosclerosis than s.c. (peripheral) fat.
Although obesity and body fat distribution are clearly hereditable traits, the role of developmental genes in obesity and fat distribution has received surprisingly little attention. Previous work has shown that SVF taken from different adipose depots (23–28) and from obese versus lean individuals show different propensities to differentiate when placed in tissue culture in vitro (36). In addition, the rate of lipolysis in adipose tissue taken from s.c. sites is lower than that of adipose tissue from visceral or omental sites (37). Furthermore, the lipolytic effect of catecholamines is weaker and the antilipolytic effect of insulin is more pronounced in s.c. than in visceral adipose tissue (38, 39).
Characterization of differences in gene expression between human s.c. and visceral adipose tissue also suggest genetic/developmental heterogeneity. Acylation-stimulating protein and angiotensinogen mRNA levels are higher in visceral adipose, whereas the levels of leptin, peroxisome proliferator-activated receptor γ, glucose transporter 4, glycogen synthase, and cholesterol ester transfer protein are higher in the s.c. depot (40, 41). In a survey of genes differentially expressed in s.c. and visceral adipose tissue in men, Vohl et al. (21) also noted differences in genes involved in lipolysis, cytokine secretion, Wnt signaling, C/EPBα, and some HOX genes. We also observed differences in large and small adipocytes taken from normal and fat insulin receptor knockout mice with regard to function and gene and protein expression (42–44). In the present study, therefore, we explored the hypothesis that developmental genes might play an important role in obesity and body fat distribution in both rodents and humans.
Using microarray and qPCR analysis, we demonstrated that 197 genes are differentially expressed in both adipocytes and SVF containing preadipocytes from s.c. and intraabdominal depots of the mouse and that at least 12 are genes known to play a role in early development and pattern specification. Of these, Tbx15, Shox2, En1, Sfrp2, and HoxC9 were more highly expressed in cells of s.c. adipose tissue, whereas Nr2f1, Gpc4, Thbd, HoxA5, and HoxC8 were more expressed in intraabdominal adipose tissue. These differences in gene expression are intrinsic and persist during in vitro culture and differentiation, indicating that they are cell-autonomous and independent of tissue microenvironment. Because the expression of these developmental genes emerges during embryogenesis, before any white adipose tissue can be detected, and is maintained during adult life, this would suggest that different adipocyte precursors are responsible for a specific adipose depot development and may participate later in the functional differences observed between internal and s.c. adipose depots.
Although all of the genes that were differential in rodents were also differential in humans, in some cases the direction of difference was different in the two species. This difference of direction may reflect the fact that fat was not taken from identical depots in the two species or may simply represent differences between development in these two species. Other differences in gene expression have also been observed between humans and rodents. Thus, leptin exhibits a higher expression in s.c. than omental adipose in humans (40, 41), whereas, in mice, leptin expression is higher in intraabdominal (epididymal) fat than s.c. fat (45). Likewise, the differential expression of α2-adrenergic receptor expression observed in humans (higher in s.c. adipose than in omental) (38) is not observed at all in mice, which do not express α2-adrenergic receptors in adipose tissue (46). Conversely, β3-adrenergic receptors are widely expressed in mouse adipose tissue, whereas little or no expression has been reported in human adipose (47). In our case, the interdepot differences of expression for developmental genes Shox2, En1, Nr2f1, HoxA5, HoxC8, and Thbd were preserved from mice to humans independent of gender, whereas interdepot differential expression of HoxC9 in humans occurred only in females, and Tbx15, Sfrp2, and Gpc4 exhibited opposite directions of differential expression in mice and humans. In both species, what is clear is that multiple developmental genes, including those involved in anteroposterior or dorsoventral patterning, exhibit dramatic differences in the level of expression in adipose and preadipose from different regions of the body.
One of the most striking features of the expression of HoxA5, Gpc4, and Tbx15 in human adipose is not only their differential expression between depots, but also their strong correlation with BMI. This correlation is particularly true for Tbx15 in visceral fat and Gpc4 in s.c. fat, such that both genes show dramatic changes in expression as BMI goes from the normal range (BMI = 20–25) to either overweight (BMI = 25–30) or obese (BMI > 30). No other parameter related to obesity or fat mass, including serum leptin, adiponectin, or insulin, shows such a distinct change at this transition point. Indeed, if the physiological separation between lean and overweight/obese had not been previously defined by epidemiological criteria, one could define the overweight population by the expression level of these genes, suggesting that expression of these genes could be related to the pathogenesis of obesity.
Distribution of adipose tissue (WHR) also has a strong heritable component (12) and has been shown to better correlate with risk of diabetes and atherosclerosis than BMI (48). Increased WHR, i.e., visceral/central or “apple-shaped” obesity, is associated with higher risks for metabolic and cardiovascular complications (2–5). We find that HoxA5, Gpc4, and Tbx15 expression also vary with fat distribution and that expression of the latter two is an excellent marker for visceral fat accumulation. Thus, high levels of Tbx15and Gpc4expression in s.c. adipose tissue and low levels of expression in visceral adipose tissue appear to be linked with high WHR and by extension should be correlated with higher risks for metabolic and cardiovascular complications.
Although the exact role of each of these genes in development and distribution of fat needs to be explored, the Hox, Shox, and Tbx genes are important in dorsal–ventral and anterior–posterior patterning in many species (49–51). Furthermore, early embryonic expression of Tbx15 in dorsal mesenchyme is complementary to Agouti expression in ventral mesenchyme and is thus thought to provide an instructional cue that might underlie region-specific differences in body morphology. Also, in humans with the Simpson–Golabi–Behmel syndrome, glypican 4 is absent because of a partial chromosomal deletion (52), and fibroblasts taken from these individuals have a higher rate of conversion to adipocytes in culture than fibroblasts from normal individuals (53).
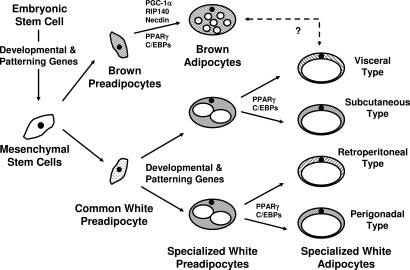
Taken together, our data suggest that genes involved in embryonic development and pattern specification in mice and humans play potentially important roles in adipocyte development and fat distribution. These results suggest that different adipocyte precursors are responsible for a specific adipose depot development in a manner similar to the scheme of differentiation defined for blood cells and other lineages (Fig. 6). These findings open an avenue of understanding fat accumulation and distribution and present therapeutic targets for this epidemic disorder.
Fig. 6.
Hypothetical scheme of adipocyte development. PPAR, peroxisome proliferator-activated receptor.
Materials and Methods
Details are presented in Supporting Methods. In brief, adipocytes and SVF were isolated from epididymal and flank s.c. adipose tissue of C57BL/6 mice. RNA from adipose tissue, isolated adipocytes, and SVF were isolated by using an RNeasy kit (Qiagen, Valencia, CA). Microarray experiments and analysis were performed as described in Fig. 1.
Paired samples of visceral and s.c. human adipose tissue were obtained from 198 Caucasian men (n = 99) and women (n = 99) who underwent open abdominal surgery with BMI ranging from 21.7 to 46.8 kg/m2 for males and 20.8 to 54.1 kg/m2 for females at the University of Leipzig. All subjects gave written informed consent before taking part in the study. Expression of murine and human genes of particular interest based on the microarray analysis was further assessed by qPCR.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant R01 DK33201, Diabetes Genome Anatomy Project Grants DK60837 (to C.R.K.) and K08DK064906 (to A.W.N.), American Diabetes Association Grant HD027748 (to C.R.K.), and Deutsche Forschungsgemeinschaft Grant BL 580/3-1 (to M.B.).
Abbreviations
- SVF
stromovascular fraction
- WHR
waist/hip ratio
- BMI
body mass index
- Thbd
thrombomodulin
- qPCR
quantitative PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lean M. E. Proc. Nutr. Soc.; 2000. pp. 331–336. [DOI] [PubMed] [Google Scholar]
- 2.Mauriege P., Despres J. P., Moorjani S., Prud’Homme D., Lamarche B., Bouchard C., Nadeau A., Tremblay A., Lupien P. J. Eur. J. Clin. Invest. 1993;23:729–740. doi: 10.1111/j.1365-2362.1993.tb01293.x. [DOI] [PubMed] [Google Scholar]
- 3.Gillum R. F. J. Chronic Dis. 1987;40:421–428. doi: 10.1016/0021-9681(87)90175-5. [DOI] [PubMed] [Google Scholar]
- 4.Kissebah A. H., Krakower G. R. Physiol. Rev. 1994;74:761–811. doi: 10.1152/physrev.1994.74.4.761. [DOI] [PubMed] [Google Scholar]
- 5.Abate N., Garg A. Prog. Lipid Res. 1995;34:53–70. doi: 10.1016/0163-7827(94)00006-8. [DOI] [PubMed] [Google Scholar]
- 6.Gregoire F. M. Exp. Biol. Med. (Maywood) 2001;226:997–1002. doi: 10.1177/153537020122601106. [DOI] [PubMed] [Google Scholar]
- 7.Koutnikova H., Auwerx J. Ann. Med. 2001;33:556–561. doi: 10.3109/07853890108995966. [DOI] [PubMed] [Google Scholar]
- 8.Tong Q., Hotamisligil G. S. Rev. Endocr. Metab. Disord. 2001;2:349–355. doi: 10.1023/a:1011863414321. [DOI] [PubMed] [Google Scholar]
- 9.Wynne K., Stanley S., McGowan B., Bloom S. J. Endocrinol. 2005;184:291–318. doi: 10.1677/joe.1.05866. [DOI] [PubMed] [Google Scholar]
- 10.Ricquier D. Proc. Nutr. Soc.; 2005. pp. 47–52. [DOI] [PubMed] [Google Scholar]
- 11.Nelson T. L., Vogler G. P., Pedersen N. L., Hong Y., Miles T. P. Twin Res. 2000;3:43–50. doi: 10.1375/136905200320565689. [DOI] [PubMed] [Google Scholar]
- 12.Baker M., Gaukrodger N., Mayosi B. M., Imrie H., Farrall M., Watkins H., Connell J. M., Avery P. J., Keavney B. Diabetes. 2005;54:2492–2496. doi: 10.2337/diabetes.54.8.2492. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch J., Batchelor B. Clin. Endocrinol. Metab. 1976;5:299–311. doi: 10.1016/s0300-595x(76)80023-0. [DOI] [PubMed] [Google Scholar]
- 14.Ersek R. A., Bell H. N., IV, Salisbury A. V. Aesthetic Plast. Surg. 1994;18:279–282. doi: 10.1007/BF00449795. [DOI] [PubMed] [Google Scholar]
- 15.Garg A., Misra A. Endocrinol. Metab. Clin. North Am. 2004;33:305–331. doi: 10.1016/j.ecl.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Garg A., Peshock R. M., Fleckenstein J. L. J. Clin. Endocrinol. Metab. 1999;84:170–174. doi: 10.1210/jcem.84.1.5383. [DOI] [PubMed] [Google Scholar]
- 17.Shackleton S., Lloyd D. J., Jackson S. N., Evans R., Niermeijer M. F., Singh B. M., Schmidt H., Brabant G., Kumar S., Durrington P. N., et al. Nat. Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 18.Shelley W. B., Izumi A. K. Arch. Dermatol. 1970;102:326–329. [PubMed] [Google Scholar]
- 19.Atzmon G., Yang X. M., Muzumdar R., Ma X. H., Gabriely I., Barzilai N. Horm. Metab. Res. 2002;34:622–628. doi: 10.1055/s-2002-38250. [DOI] [PubMed] [Google Scholar]
- 20.Linder K., Arner P., Flores-Morales A., Tollet-Egnell P., Norstedt G. J. Lipid Res. 2004;45:148–154. doi: 10.1194/jlr.M300256-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Vohl M. C., Sladek R., Robitaille J., Gurd S., Marceau P., Richard D., Hudson T. J., Tchernof A. Obes. Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- 22.von Eyben F. E., Kroustrup J. P., Larsen J. F., Celis J. Ann. N.Y. Acad. Sci. 2004;1030:508–536. doi: 10.1196/annals.1329.063. [DOI] [PubMed] [Google Scholar]
- 23.Djian P., Roncari A. K., Hollenberg C. H. J. Clin. Invest. 1983;72:1200–1208. doi: 10.1172/JCI111075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams M., Montague C. T., Prins J. B., Holder J. C., Smith S. A., Sanders L., Digby J. E., Sewter C. P., Lazar M. A., Chatterjee V. K., O’Rahilly S. J. Clin. Invest. 1997;100:3149–3153. doi: 10.1172/JCI119870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkland J. L., Hollenberg C. H., Gillon W. S. Am. J. Physiol. 1990;258:C206–C210. doi: 10.1152/ajpcell.1990.258.2.C206. [DOI] [PubMed] [Google Scholar]
- 26.Hauner H., Entenmann G. Int. J. Obes. 1991;15:121–126. [PubMed] [Google Scholar]
- 27.Tchkonia T., Giorgadze N., Pirtskhalava T., Tchoukalova Y., Karagiannides I., Forse R. A., DePonte M., Stevenson M., Guo W., Han J., et al. Am. J. Physiol. 2002;282:R1286–R1296. doi: 10.1152/ajpregu.00653.2001. [DOI] [PubMed] [Google Scholar]
- 28.Tchkonia T., Tchoukalova Y. D., Giorgadze N., Pirtskhalava T., Karagiannides I., Forse R. A., Koo A., Stevenson M., Chinnappan D., Cartwright A., et al. Am. J. Physiol. 2005;288:E267–E277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- 29.Pereira F. A., Qiu Y., Tsai M. J., Tsai S. Y. J. Steroid Biochem. Mol. Biol. 1995;53:503–508. doi: 10.1016/0960-0760(95)00097-j. [DOI] [PubMed] [Google Scholar]
- 30.De Cat B., David G. Semin. Cell Dev. Biol. 2001;12:117–125. doi: 10.1006/scdb.2000.0240. [DOI] [PubMed] [Google Scholar]
- 31.Weiler H., Isermann B. H. J. Thromb. Haemostasis. 2003;1:1515–1524. doi: 10.1046/j.1538-7836.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 32.Blaschke R. J., Monaghan A. P., Schiller S., Schechinger B., Rao E., Padilla-Nash H., Ried T., Rappold G. A. Proc. Natl. Acad. Sci. USA. 1998;95:2406–2411. doi: 10.1073/pnas.95.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh M. K., Petry M., Haenig B., Lescher B., Leitges M., Kispert A. Mech. Dev. 2005;122:131–144. doi: 10.1016/j.mod.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Joyner A. L., Martin G. R. Genes Dev. 1987;1:29–38. doi: 10.1101/gad.1.1.29. [DOI] [PubMed] [Google Scholar]
- 35.Leimeister C., Bach A., Gessler M. Mech. Dev. 1998;75:29–42. doi: 10.1016/s0925-4773(98)00072-0. [DOI] [PubMed] [Google Scholar]
- 36.van Harmelen V., Skurk T., Rohrig K., Lee Y. M., Halbleib M., Aprath-Husmann I., Hauner H. Int. J. Obes. Relat. Metab. Disord. 2003;27:889–895. doi: 10.1038/sj.ijo.0802314. [DOI] [PubMed] [Google Scholar]
- 37.Arner P. Ann. Med. 1995;27:435–438. doi: 10.3109/07853899709002451. [DOI] [PubMed] [Google Scholar]
- 38.Mauriege P., Galitzky J., Berlan M., Lafontan M. Eur J. Clin. Invest. 1987;17:156–165. doi: 10.1111/j.1365-2362.1987.tb02395.x. [DOI] [PubMed] [Google Scholar]
- 39.Bolinder J., Kager L., Ostman J., Arner P. Diabetes. 1983;32:117–123. doi: 10.2337/diab.32.2.117. [DOI] [PubMed] [Google Scholar]
- 40.Lefebvre A. M., Laville M., Vega N., Riou J. P., van Gaal L., Auwerx J., Vidal H. Diabetes. 1998;47:98–103. doi: 10.2337/diab.47.1.98. [DOI] [PubMed] [Google Scholar]
- 41.Dusserre E., Moulin P., Vidal H. Biochim. Biophys. Acta. 2000;1500:88–96. doi: 10.1016/s0925-4439(99)00091-5. [DOI] [PubMed] [Google Scholar]
- 42.Bluher M., Michael M. D., Peroni O. D., Ueki K., Carter N., Kahn B. B., Kahn C. R. Dev. Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 43.Bluher M., Patti M. E., Gesta S., Kahn B. B., Kahn C. R. J. Biol. Chem. 2004;279:31891–31901. doi: 10.1074/jbc.M404569200. [DOI] [PubMed] [Google Scholar]
- 44.Bluher M., Wilson-Fritch L., Leszyk J., Laustsen P. G., Corvera S., Kahn C. R. J. Biol. Chem. 2004;279:31902–31909. doi: 10.1074/jbc.M404570200. [DOI] [PubMed] [Google Scholar]
- 45.Trayhurn P., Thomas M. E., Duncan J. S., Rayner D. V. FEBS Lett. 1995;368:488–490. doi: 10.1016/0014-5793(95)00719-p. [DOI] [PubMed] [Google Scholar]
- 46.Castan I., Valet P., Quideau N., Voisin T., Ambid L., Laburthe M., Lafontan M., Carpene C. Am. J. Physiol. 1994;266:R1141–R1147. doi: 10.1152/ajpregu.1994.266.4.R1141. [DOI] [PubMed] [Google Scholar]
- 47.Lafontan M. Cell. Signalling. 1994;6:363–392. doi: 10.1016/0898-6568(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 48.Ohlson L. O., Larsson B., Svardsudd K., Welin L., Eriksson H., Wilhelmsen L., Bjorntorp P., Tibblin G. Diabetes. 1985;34:1055–1058. doi: 10.2337/diab.34.10.1055. [DOI] [PubMed] [Google Scholar]
- 49.Deschamps J., van Nes J. Development (Cambridge, U.K.) 2005;132:2931–2942. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 50.Yu L., Gu S., Alappat S., Song Y., Yan M., Zhang X., Zhang G., Jiang Y., Zhang Z., Zhang Y., Chen Y. Development (Cambridge, U.K.) 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- 51.Candille S. I., Van Raamsdonk C. D., Chen C., Kuijper S., Chen-Tsai Y., Russ A., Meijlink F., Barsh G. S. PLoS Biol. 2004;2:E3. doi: 10.1371/journal.pbio.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veugelers M., Vermeesch J., Watanabe K., Yamaguchi Y., Marynen P., David G. Genomics. 1998;53:1–11. doi: 10.1006/geno.1998.5465. [DOI] [PubMed] [Google Scholar]
- 53.Wabitsch M., Brenner R. E., Melzner I., Braun M., Moller P., Heinze E., Debatin K. M., Hauner H. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.