Abstract
The first step in molybdenum cofactor biosynthesis, the conversion of 5′-GTP to precursor Z, an oxygen-sensitive tetrahydropyranopterin is catalyzed by the S-adenosylmethionine (SAM)-dependent enzyme MoaA and the accessory protein MoaC. This reaction involves the radical-initiated intramolecular rearrangement of the guanine C8 atom. MoaA harbors an N-terminal [4Fe–4S] cluster, which is involved in the reductive cleavage of SAM and generates a 5′-deoxyadenosyl radical (5′-dA•), and a C-terminal [4Fe–4S] cluster presumably involved in substrate binding and/or activation. Biochemical studies identified residues involved in 5′-GTP binding and the determinants of nucleotide specificity. The crystal structure of MoaA in complex with 5′-GTP confirms the biochemical data and provides valuable insights into the subsequent radical reaction. MoaA binds 5′-GTP with high affinity and interacts through its C-terminal [4Fe–4S] cluster with the guanine N1 and N2 atoms, in a yet uncharacterized binding mode. The tightly anchored triphosphate moiety prevents the escape of radical intermediates. This structure also visualizes the l-Met and 5′-dA cleavage products of SAM. Rotation of the 5′-dA ribose and/or conformational changes of the guanosine are proposed to bring the 5′-deoxyadenosyl radical into close proximity of either the ribose C2′ and C3′ or the guanine C8 carbon atoms leading to hydrogen abstraction.
Keywords: iron–sulfur, radicals, molybdenum cofactor, GTP cyclohydrolase
FeS clusters are ubiquitous and ancient prosthetic groups that are essential for fundamental biological processes. Because of their remarkable structural plasticity and versatile chemical and electronic features, FeS clusters participate in electron transfer reactions, iron/sulfur storage, the regulation of gene expression, and enzymatic transformations (1–4). In addition, noncysteinyl ligation at a unique Fe site of a [4Fe–4S] cluster can facilitate substrate binding and activation for dehydration/hydration reactions in a wide range of hydratases/dehydratases (5), of which aconitase is the best characterized example (6).
On the basis of functional, structural, and spectroscopic studies, it has recently been shown that the molybdenum cofactor (Moco) biosynthesis protein MoaA (MOCS1A in humans) assembles two oxygen-sensitive, catalytically important [4Fe–4S]2+/1+ clusters that both feature an incomplete cysteinyl ligation (7, 8). Moco biosynthesis is an evolutionarily conserved pathway comprising several novel reactions, which include the initial formation of precursor Z, its subsequent conversion to molybdopterin, and the attachment of Mo to the dithiolene moiety of molybdopterin (9, 10). In humans, genetic deficiencies of enzymes involved in Moco biosynthesis lead to the pleiotropic loss of the molybdoenzymes sulfite oxidase, aldehyde oxidase, and xanthine dehydrogenase and trigger an autosomal recessive and usually fatal disease, which is characterized by severe neurological symptoms (11–13). Mutations affecting MOCS1A account for ≈50% of all known cases of Moco deficiency, and all genetic lesions affect conserved regions important in catalysis (8).
MoaA, together with MoaC, catalyzes the first step in Moco biosynthesis (Fig. 1A), the conversion of 5′-GTP to precursor Z (7, 8, 14), an oxygen-sensitive tetrahydropyranopterin containing a cyclic phosphate (15). This unusual reaction, in which all carbon and nitrogen atoms of 5′-GTP are used, involves a rearrangement reaction of the guanine C8 atom (16, 17), and based on MoaA’s relationship to the radical S-adenosylmethionine (SAM) superfamily, this transformation has been proposed to involve radical chemistry (8).
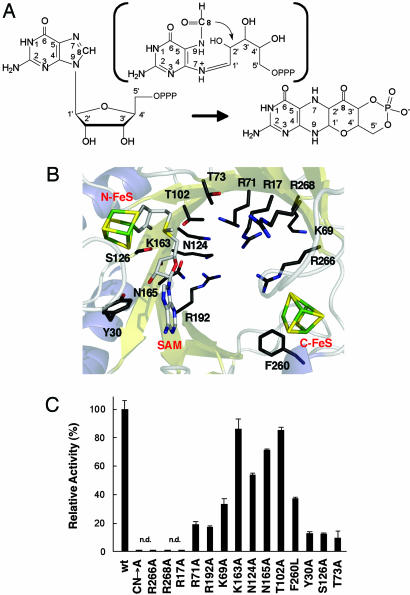
Fig. 1.
Synthesis of precursor Z and catalytic activities of MoaA active site variants. (A) Conversion of 5′-GTP to precursor Z. A proposed formamidopyrimidine-type intermediate is shown in parentheses. (B) Representation of the MoaA active site (PDB ID code 1TV8). Both [4Fe–4S] clusters, SAM, and conserved residues potentially involved in catalysis are displayed. Figs. 1B, 3, and 4 were created with pymol (www.pymol.org). (C) Synthesis of precursor Z determined by measurement of its stable oxidized derivative, compound Z, by reverse-phase HPLC. The activity of the WT protein (wt) was set to 100%. Error bars denote the SD from the mean of at least four independent experiments. n.d., not detected; CN→A, triple Cys→Ala variant of the N-terminal FeS cluster.
Radical-based enzymatic rearrangements typically require adenosylcobalamin as a coenzyme in which the reactions are initiated by homolytic cleavage of the Co–C5′ bond (18) to generate a 5′-deoxyadenosyl radical (5′-dA•). In SAM-dependent radical enzymes (19, 20), a similar approach uses SAM, which is reductively cleaved to generate a 5′-dA•. The structure of MoaA in complex with SAM (8) revealed an N-terminal [4Fe–4S] cluster that is typical of all SAM-dependent radical enzymes. This cluster anchors the cosubstrate SAM as an N/O chelate at the unique Fe site (8). A second [4Fe–4S] cluster has been identified in the C-terminal part of MoaA, and its noncysteinyl-ligated unique Fe site has been proposed to be involved in 5′-GTP binding. Both FeS clusters are on opposite sides of a hydrophilic channel that runs through the center of the incomplete TIM-barrel fold.
As in the pathways of folate, riboflavin, and biopterin synthesis (21, 22), 5′-GTP serves as the starting compound in precursor Z synthesis and an alternative GTP cyclohydrolase I (GTPCH-I)-like reaction mechanism has been proposed (16, 17). In contrast to GTPCH-I, MoaA does not release the C8 atom of 5′-GTP as formate but incorporates it in a rearrangement reaction between the C2′ and C3′ atoms of the ribose.
Here, we identify the 5′-GTP binding site by biochemical techniques and, based on a MoaA–GTP complex, describe a previously unobserved 5′-GTP-binding mode involving the unique Fe site of the C-terminal [4Fe–4S] cluster. In addition, the structural basis for the reductive cleavage of SAM and the subsequent radical chemistry catalyzed by MoaA are characterized.
Results and Discussion
Characterization of MoaA Active Site Residues.
The two FeS clusters define the general location of the MoaA active site (8). The closest distance between them is ≈17 Å, and they are on opposite sides of an active site pocket, which is interrupted by a hydrophilic channel. The active site is constructed predominantly from basic residues including five conserved Arg (R17, R71, R192, R266, and R268) and two conserved Lys residues (K69 and K163), which could compensate the negative charges of the 5′-GTP phosphate groups. To obtain further insights into MoaA function, site-directed mutagenesis of conserved active site residues (Fig. 1B) was used. Besides the triple Cys→Ala mutant of the N-terminal FeS cluster, the R17A, R266A, and R268A variants led to a complete loss of precursor Z-synthesizing activity, whereas ≈20% activity remained in the R71A and R192A variants (Fig. 1C). These results implicate all five Arg residues either in 5′-GTP binding or the catalytic mechanism.
Low activities were also observed for the Y30A and S126A variants, two residues, which hydrogen-bond to SAM in the WT protein, and the T73A variant (Fig. 1C). T73 is next to the 74GGE76 motif that coordinates the Met part of SAM. The F260A variant dramatically affected the stability of the C-terminal FeS cluster, which results in a low protein yield and altered UV-visible absorption properties, suggesting improper folding of the protein in the absence of the FeS cluster and increased susceptibility to proteolytic degradation. This observation is in agreement with a triple Cys mutant of the C-terminal FeS cluster, which also resulted in cluster destabilization (7), thus indicating, besides the proposed catalytic function, a structural role for this FeS cluster. Substitution of F260 with Leu yields a stable protein, and it therefore appears that the large hydrophobic Phe residue protects the FeS cluster.
To verify that the nondetectable levels of precursor Z synthesis of the R17A, R266A, and R268A variants were not due to misfolding, we determined the crystal structure of the R17/266/268A triple variant at a 2.25-Å resolution (see Tables 1 and 2, which are published as supporting information on the PNAS web site). The structure was refined to an Rcryst of 19.6% (Rfree = 23.7%) and revealed no major conformational changes compared to the WT in complex with SAM [Protein Data Bank (PDB) ID code 1TV8] because the Cα atoms of both models can be superimposed with an rms deviation of 0.24 Å. Surprisingly, the structure revealed that this mutant is still able to bind SAM in the same conformation as the WT, although it is no longer able to catalyze the reductive cleavage of SAM (data not shown). One possible explanation for this effect could be the reduced positive charge in the triple mutant that alters the redox potential of the N-terminal FeS cluster in such a way that it is no longer reduced by Na2S2O4 and therefore SAM is not cleaved.
5′-GTP Binding and Recognition.
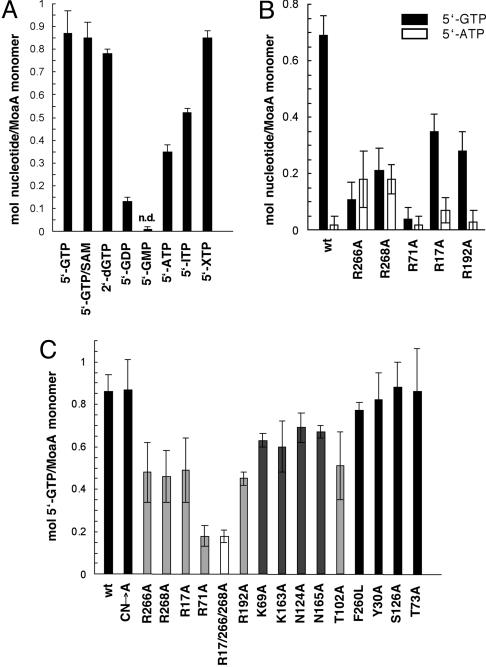
As demonstrated in ref. 8, purified MoaA and MoaC are able to catalyze the formation of precursor Z in a reaction dependent on 5′-GTP and SAM; however, neither 5′-GDP nor 5′-GMP can be used. The binding of 5′-GTP and other nucleotides to MoaA was analyzed by equilibrium dialysis, which reveals an apparent dissociation constant of 0.29 ± 0.11 μM and a binding stoichiometry of 0.71 ± 0.07 sites/monomer for 5′-GTP (see Fig. 5, which is published as supporting information on the PNAS web site). Although the reductive cleavage of SAM can be significantly increased in the presence of 5′-GTP (8), the binding of 5′-GTP is not dependent on SAM binding (Fig. 2A). Among the different purine nucleotides tested (Fig. 2A), 2′-deoxy-5′-GTP still binds with an affinity similar to 5′-GTP, indicating that the 2′-OH group is not critical for substrate recognition. The affinity of 5′-GDP, however, is already largely reduced (<15%), and no binding of 5′-GMP could be detected, indicating strong interactions between MoaA and the phosphate groups of 5′-GTP. Binding studies with three other purines revealed 40% residual binding with 5′-ATP, 60% with 5′-ITP, and 100% with 5′-XTP (Fig. 2A), demonstrating that the oxo group at position 6 (replaced by an amino group in 5′-ATP) and the amino group at position 2 (missing in 5′-ATP and 5′-ITP, but replaced by an oxo group in 5′-XTP) are important for guanine recognition. 5′-ATP is ubiquitous in the cell (intracellular concentration of ≈3 mM), whereas other nucleotides are almost an order of magnitude rarer, with the exception of 5′-GTP (≈0.9 mM) (23). Because MoaA is not able to use other purine nucleotides as alternative substrates, a mechanism must allow a molecular discrimination, especially between 5′-GTP and 5′-ATP. An in vitro competition assay with equimolar amounts of 5′-GTP and 5′-ATP revealed that MoaA selectively binds 5′-GTP (Fig. 2B). Under the same conditions, both 5′-ITP and 5′-XTP bind with ≈10-fold and ≈4-fold lower affinity than 5′-GTP (data not shown).
Fig. 2.
Determination of MoaA substrate-binding by equilibrium dialysis. (A) Nucleotide specificity. MoaA (75 μM) was equilibrated with the listed nucleotides (150 μM); n.d., not detected. (B) 5′-GTP/5′-ATP competition. WT MoaA (wt) and variants (75 μM) were dialyzed against equimolar concentrations (150 μM) of 5′-GTP (black bar) and 5′-ATP (white bar). (C) 5′-GTP binding. WT MoaA and variants (75 μM) were dialyzed against 5′-GTP (150 μM). Error bars denote the SD from the mean of at least four independent experiments. CN→A, triple Cys→Ala variant of the N-terminal FeS cluster; black bars, WT MoaA and residues not involved in 5′-GTP binding; light gray bars, residues important for 5′-GTP binding; dark gray bars, residues less important in 5′-GTP binding; white bar, R17/266/268A triple variant.
On the basis of the analysis of active site variants shown in Fig. 1B, residues involved in 5′-GTP binding have been identified (Fig. 2C). The binding site is located in the hydrophilic channel and appears to be constructed by the five Arg residues (R17, R71, R192, R266, and R268), T102, and two Lys residues (K69 and K163). The replacement of two Asn residues (N124A and N165A) also reduces 5′-GTP binding, but because these residues are not highly conserved, they are probably not essential for substrate binding. The most important residue for 5′-GTP binding is R71, which shows a similarly low affinity as the R17/266/268A triple mutant. The dissociation constants for the R71A and R17/266/268A triple mutants are ≈1,000-fold higher than WT, whereas the K69A and K163A variants are ≈100-fold higher, which corresponds to a difference in binding energy between the mutants of roughly 4 kJ/mol (1 kcal/mol).
To identify which residues confer base specificity and localize the binding site of the guanine base, all Arg variants were analyzed in a 5′-GTP/5′-ATP competition assay (Fig. 2B). The results clearly show that both R266A and R268A variants are no longer able to distinguish between 5′-GTP and 5′-ATP, thus implicating these residues in guanine binding. Because of their location (Fig. 1B), the guanine binding site appears to be in close proximity of the C-terminal FeS cluster. A frequently used mechanism for guanine binding involving Arg residues is a cation–π stacking interaction (24). One could therefore speculate that R266 and/or R268 are directly adjacent to the base with the conjugated guanidinium groups involved in π-electron stacking. Furthermore, an Arg residue can recognize a guanine through a pair of hydrogen bonds between its guanidinium group and the N7 and O6 atoms of the guanine. The remaining Arg and both Lys residues could be involved in compensating the negatively charged phosphate groups of 5′-GTP. Combining the data of the precursor Z assay (Fig. 1C) and the 5′-GTP-binding assay (Fig. 2C), it appears as if R17, R266, and R268 are crucial for the catalytic mechanism because their replacement with Ala only leads to a moderate reduction in 5′-GTP-binding, but a complete loss of precursor Z synthesis.
Structure of MoaA in Complex with 5′-GTP.
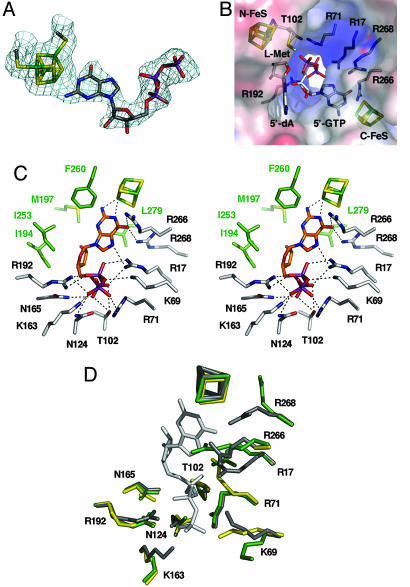
This structure was solved by isomorphous replacement at a 2.35-Å resolution (see Tables 1 and 2) and has been refined to an Rcryst of 22.0% (Rfree = 27.1%). The average B-factor is considerably higher than in the complex with SAM (8), but is in very good agreement with the Wilson plot analysis. Whereas the triphosphate moiety of 5′-GTP is well defined in the electron density maps, the ribose and base appear to be more flexible (Fig. 3A). This observation is reflected by fewer interactions of the base and ribose with the protein compared with the three phosphates (Fig. 3C). 5′-GTP binds with its purine N1 nitrogen (2.8-Å distance) and its exocyclic amino group (2.4-Å distance) in an unprecedented manner to the unique Fe site of the C-terminal [4Fe–4S] cluster (Fig. 3 A and C). In addition, R266 (Nη1–O6, 3.1 Å) and R268 (Nη2–O6, 3.3 Å) are within hydrogen-bonding distance of the exocyclic oxo group at position 6 (Fig. 3C). The observed interactions of the guanine with R266 and R268 may be important in affecting the binding of intermediates during catalysis. Interestingly, no interactions are present with the ribose indicating that high flexibility may be important for catalysis. The guanidinium group of R17 is in hydrogen-bonding distance to the N7 atom (Nη1–N7, 3.2 Å) of the imidazole ring. R17 may be involved in assisting the rearrangement reaction and/or plays a role in shielding radical intermediates from the efficient radical quencher oxygen.
Fig. 3.
Structure of MoaA in complex with 5′-GTP. (A) 2Fo–Fc map of the C-terminal [4Fe–4S] cluster with bound 5′-GTP contoured at one times the rms deviation. (B) Electrostatic potential (electropositive in blue, electronegative in red contoured at ±10 kT) surrounding the hydrophilic channel. [4Fe–4S] clusters, l-Met, 5′-dA, 5′-GTP, and residues important for 5′-GTP binding are displayed. (C) Stereoview of 5′-GTP interactions with surrounding active site residues (dashed lines). Carbon atoms of residues in hydrogen-bonding distance are in white, and carbon atoms of residues within a distance of 4 Å are in green. (D) Superposition of MoaA in complex with 5′-GTP (gray), without 5′-GTP (PDB ID code 1TV8; green) and the R17/266/268A variant (yellow).
The triphosphate part of 5′-GTP is tightly anchored within the hydrophilic channel (Fig. 3B) and is stabilized by 12 hydrogen bonds mainly contributed by R17, R71, R192, K69, and K163, which compensate the negative charges of the 5′-GTP phosphate groups (Fig. 3C and see Table 3, which is published as supporting information on the PNAS web site). Additional residues involved in binding are T102, N124, and N165, which also stabilize the guanidinium groups of R71 (Nη1–T102 Oγ1, 3.0 Å; Nη1–N124 Oδ1, 2.7 Å) and R192 (Nη1–N165 Oδ1, 3.2 Å) by hydrogen bonds. Usually Mg2+ is involved in compensating the negative charges of the phosphate groups in nucleoside triphosphates. Interestingly, biochemical assays clearly indicate that the MoaA-catalyzed reaction is not dependent on Mg2+ (8). In fact, in vitro 5′-GTP binding is reduced by ≈40% in the presence of Mg2+ (data not shown). Based on the structure there is no obvious cation-binding site and a 5′-GTP/Mg2+ complex would interfere with the positively charged residues reflecting the observed low affinity for a 5′-GTP/Mg2+ complex. The binding of the triphosphate by MoaA, involving only the assistance of positively charged residues and no additional Mg2+, is similar to the binding of 5′-GTP by GTPCH-I (25, 26). The 5′-GTP binding mode identified by structural studies nicely confirms the biochemical data (see above).
The MoaA structure in complex with 5′-GTP explains the molecular discrimination between 5′-GTP and 5′-ATP, which have distinct hydrogen bond donor and acceptor capabilities. As detailed above, the guanine exocyclic amino group at position 2 as well as the purine N1 nitrogen are in hydrogen-bonding distance to the unique Fe site of the C-terminal FeS cluster. The interaction involving the exocyclic amino group would be absent in 5′-ATP because this group is not present. Furthermore the oxo group at position 6 of 5′-GTP is in hydrogen-bonding distance to the R266 Nη1 and R268 Nη2 atoms, interactions which would not be possible with the exocyclic amino group of 5′-ATP. Nevertheless, in the WT protein 5′-ATP still binds at a level of 40%, which seems to be the result of the multiple electrostatic interactions with the triphosphate. The high-affinity interaction with 5′-XTP is also consistent with the observed interactions with 5′-GTP. The exocyclic amino group of guanine is replaced in xanthine with an oxo group that should be able to interact with the unique Fe site of the C-terminal cluster together with the purine N1 nitrogen. This interaction would in fact be analogous to the coordination of the unique Fe site of the N-terminal cluster by the Met nitrogen and oxygen atoms.
Substrate triggering is an important mechanism for radical-catalyzed reactions in which O2-sensitive catalytic radicals are formed following substrate-induced conformational changes of the enzyme (27). Indeed, the reductive cleavage of SAM is largely increased in the presence of 5′-GTP (8), but upon substrate binding, no active site closure occurs, and 5′-GTP remains solvent exposed. Nevertheless, the multiple electrostatic interactions between the positively charged Arg and Lys residues and the triphosphate presumably lock any reactive radical intermediates in the active site. Superposition of the 5′-GTP structure with the structure of apo-MoaA (PDB ID code 1TV8) and the R17/266/268A triple mutant reveal Cα rms deviations of only 0.30 Å and 0.39 Å, respectively. Nevertheless, distinct conformational changes induced by 5′-GTP binding are obvious (Fig. 3D). R17, R266, and R268, show significant differences in their side-chain conformations in the 5′-GTP complex resulting in a more open active site, which seems required for 5′-GTP binding. The side chain of R17 in the GTP-free structure would interfere with phosphate binding, and the guanidinium group of R266 would prevent binding of the guanine base. The three missing Arg residues in the R17/266/268A variant enlarge the guanine-binding site. In addition, in the absence of R17, the side chain of R71 adopts a different conformation, which would prevent phosphate binding. Although not critical for triphosphate binding, both Lys residues (K69 and K163) also show different side-chain conformations.
The Structural Basis for the Reductive Cleavage of SAM and the Subsequent Radical Reaction.
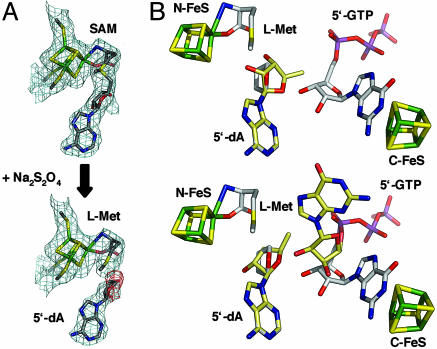
The crystal structure of MoaA in complex with 5′-GTP was obtained after reduction of the FeS clusters with Na2S2O4, which apparently resulted in the homolytic cleavage of SAM into l-Met and 5′-dA. The dithionite treatment adversely affects crystal quality and is primarily responsible for the high overall Rsym of 15.8%. To probe whether photo-induced reduction occurred during data collection, the first and second half of the data set were analyzed separately: Analysis of images 1–175 (2.25 Å; Rsym = 10.9%) and 175–350 (2.45 Å; Rsym = 18.2%) revealed in both cases good quality electron density maps (1–175: Rcryst = 21.4%, Rfree = 25.4%; 175–350: Rcryst = 21.3%, Rfree = 25.9%) including well defined electron densities for 5′-GTP and the SAM cleavage products l-Met and 5′-dA, which argues against photo-induced SAM cleavage but is consistent with the crystal decaying during data collection. The structure (Fig. 4A) showed that the Met is still bound as an N/O-chelate to the unique Fe site of the N-terminal FeS cluster and 5′-dA is bound in a similar way as in the SAM structure (PDB ID code 1TV8; ref. 8). However, the reactive C5′ group is disordered, indicating that bond cleavage increases the mobility of this group, thus allowing it to more easily engage in interactions with the radical acceptor. The 5′-dA• is a highly reactive species that immediately abstracts a hydrogen atom from the nearby substrate to form 5′-dA and a substrate radical. The presence of 5′-GTP in the crystal despite the homolytic cleavage of SAM presumably results from the high stoichiometric excess of 5′-GTP in the soaking solution, which leads to the replacement of the reaction product with fresh 5′-GTP.
Fig. 4.
Reductive cleavage of SAM and radical transfer from 5′-dA• to 5′-GTP. (A) 2Fo–Fc maps (blue; one rms deviation) of the N-terminal [4Fe–4S] cluster before (Upper) and after (Lower) reductive cleavage of SAM. Fo–Fc map (red; −2.5 rms deviations) obtained with SAM as the model. (B) Radical transfer models. Carbon atoms of 5′-dA and 5′-GTP as present in the 5′-GTP/5′-dA structure are in white, and carbon atoms of postulated models are shown as pale yellow. (Upper) 5′-dA-ribose rotation model. (Lower) 5′-dA-ribose/guanosine rotation model.
Possible sites of hydrogen abstraction in the 5′-GTP substrate of MoaA are the imidazole C8 atom and either the C2′ or C3′ atom of the ribose. In any case, both the 5′-dA• and the substrate have to be in close spatial proximity so that any radical is prevented from participating in unwanted side reactions as it moves between the radical donor and acceptor in an active site lined with polar residues. The distances between the C5′ carbon atom of SAM and the C2′, C3′, and C8 carbon atoms of 5′-GTP are 6.3, 5.3, and 8.1 Å, respectively. These distances are too far for a direct radical transfer and demand an exquisite conformational control of the 5′-dA• and/or 5′-GTP so that side reactions are averted. In contrast, in the substrate-bound structures of the radical SAM enzymes biotin synthase and lysine-2,3-aminomutase, the C5′ carbon atoms lie within 3.8–3.9 Å of the site of hydrogen abstraction in the substrate (28, 29), and this distance will decrease as the ribosyl moiety becomes rotatable around the glycosidic bond after reductive cleavage of the C5′–S bond.
The distance problem in MoaA, however, is common to all adenosylcobalamin-dependent enzymes in which the substrate-binding sites are typically 6–9 Å away from the Co–C5′ bond (18, 27). Two models of radical transfer from the coenzyme to the substrates, ribosyl rotation and ribose pseudorotation, have been proposed to explain how the 5′-dA• approaches the substrates and stereospecifically abstracts a hydrogen atom. Rotation of the MoaA ribose moiety (ribose rotation model) of the 5′-dA•, directing the C5′ atom toward the 5′-GTP guanosine part (Fig. 4B Upper), results in distances of 5.1 Å (C8) and 3.2 Å (C2′) and close distances (<2 Å) between the O3′ of 5′-GTP and the O4′ and C5′ of 5′-dA, which might be eliminated by subtle conformational changes in the 5′-dA• and the ribose. A second model in which both the 5′-dA and the guanosine change their conformation (5′-dA-ribose/guanosine rotation model) results in close distances between the C5′ of 5′-dA and C8 (3.4 Å), C2′ (2.7 Å), and C3′ (3.8 Å) of 5′-GTP (Fig. 4B Lower). Although in this model the guanosine points upwards out of the active site and is solvent-exposed, the guanidinium group of R17 would be in hydrogen-bonding distance of the ribose O4′, and the catalytically important T73 (Fig. 1 B and C) would be in close proximity of the imidazole N7 atom (<4 Å). Finally, a third model in which the 5′-dA• does not react directly with the substrate is possible. MoaAs from eubacteria and eukaryotes contain a functionally important double-Gly motif at the C terminus, and its location in the active site has been proposed in ref. 8. However, there is no interpretable electron density for the last 11 amino acids, indicating that these residues are highly mobile. Maybe, the 5′-dA• generates a glycyl radical, which in turn abstracts the substrate hydrogen, and the distance problem is solved in this way.
Mechanistic Comparison of MoaA and GTPCH-I.
On the basis of the similarity of precursor Z synthesis to other pteridine biosynthetic pathways, it has been proposed that precursor Z synthesis follows an alternative GTPCH-I-like mechanism (16, 17). In contrast to GTPCH-I, the MoaA C8 atom of 5′-GTP is not released as formate but is incorporated between the C2′ and C3′ atoms of the ribose (Fig. 1A). GTPCH-I is involved in biopterin and folate biosynthesis and catalyzes the formation of dihydroneopterin triphosphate from 5′-GTP. It initiates catalysis via a nucleophilic attack of a Zn-activated water molecule on the 5′-GTP C8 atom, which leads to hydrolytic opening of the imidazole ring resulting in a formamidopyrimidine-type intermediate (refs. 25 and 26 and references therein). The subsequent Zn-assisted release of formate generates a diaminopyrimidine-type intermediate. The formamidopyrimidine-type intermediate has been identified in the Escherichia coli GTPCH-I H179A active site mutant after acid hydrolysis via its diaminopyrimidine derivative (30). Based on the structure of MoaA, there are no conserved Cys or His residues potentially involved in Zn binding. Furthermore, an inductively coupled plasma optical emission spectroscopy analysis of MoaA for metals frequently found in proteins was negative, thus indicating that MoaA uses a different approach to this type of reaction. To get insights whether MoaA catalyzes the formation of a diaminopyrimidine-type product, MoaA was incubated with 5′-GTP and SAM, and the reaction product was converted to 6,7-dimethylpterin. Indeed, MoaA, like the E. coli GTPCH-I H179A variant, catalyzes the formation of a diaminopyrimidine, which was identified by coelution of their 6,7-dimethylpterin derivatization products (data not shown). Addition of MoaC to the reaction mixture revealed that this product is not accumulating and is immediately converted to precursor Z. The diaminopyrimidine product is not formed spontaneously; it requires SAM and the reduction of the FeS clusters with Na2S2O4. During precursor Z synthesis, pyrophosphate is released, which is not catalyzed by MoaA (data not shown), because it only occurs after addition of MoaC.
This result indicates that the reductive cleavage of SAM and subsequent formation of a 5′-dA• are essential for the formation of a diaminopyrimidine-type product, which still contains the triphosphate moiety (Fig. 1A). A radical-based approach for this type of reaction is clearly different from the GTPCH-I-catalyzed reaction with its Zn-assisted release of formate and hence indicates a novel route for pterin synthesis. The most difficult chemical step in precursor Z synthesis, which presumably requires a radical-based mechanism, is the fragmentation of the C2′–C3′ bond and the insertion of the C8 atom. In contrast, opening of the imidazole ring of the guanine could be achieved via base catalysis and a suitably oriented water molecule. The most important unresolved questions are: (i) which hydrogen atom is transferred to 5′-dA•, and (ii) is hydrogen atom abstraction required for opening of the imidazole ring? It should be pointed out that initial hydrogen abstraction from either the C2′ or C3′ atom appears feasible based on the radical transfer models discussed above. Such an attack would be similar to the reaction catalyzed by ribonucleotide reductases (ref. 31 and references therein) and could aid in the rearrangement of the C8 atom. Another scenario could be that after opening of the imidazole ring, the C8 hydrogen atom is abstracted, thus initiating the radical reaction. To resolve these questions, future studies using specifically deuterated forms of 5′-GTP will be required.
Proposed Functions of the C-Terminal FeS Cluster in Catalysis.
It is well known that noncysteinyl ligation at a unique Fe site of a [4Fe–4S] cluster can facilitate substrate binding and activation for dehydration/hydration reactions in a wide range of hydratases/dehydratases (5), including aconitase (6). Common to proteins involving FeS clusters in these reactions is that the unique Fe site acts as a Lewis acid to activate the substrate for nucleophilic attack and functions in the elimination of hydroxyl groups. There is, however, no obvious similar reaction step during precursor Z synthesis. MoaA is not able to catalyze the release of pyrophosphate, which indicates that during catalysis, 5′-GTP and its reactive radical intermediates are tightly anchored by the triphosphate moiety to prevent their escape. The six-membered ring of the guanine, which is not involved in catalysis, interacts with the unique Fe site of the C-terminal cluster. This coordination is certainly important for positioning 5′-GTP with its reactive imidazole ring in a proper conformation for catalysis. However, additional function(s) of the C-terminal cluster seem likely because a simple anchoring of the 5′-GTP could also be accomplished by amino acids. The C-terminal FeS cluster has been shown to be redox active (7). This cluster might therefore be involved in electron transfer, which is mediated by the six-membered ring and thus functions as an electron donor/acceptor system facilitating oxidation/reduction reactions during catalysis. In addition, the data presented here do not rule out that after opening of the imidazole ring and the ribose an intermediate is formed, which binds with a reactive group to the FeS cluster. A combination of the above mentioned roles is also possible because FeS clusters are known to perform different functions within the same protein (5). The structural and functional data presented here indicate that the MoaA redox active C-terminal [4Fe–4S] cluster is at least involved in substrate recognition and substrate anchoring and is essential for the structural integrity of the protein, thus underscoring the exciting functional diversity that is possible with these cofactors.
Methods
Cloning, Expression, and Purification.
See Supporting Methods, which is published as supporting information on the PNAS web site.
Crystallization, Data Collection, and Structure Determination.
Crystallization of WT MoaA under anaerobic conditions was performed as described in ref. 8. The MoaA-5′-dA/5′-GTP complex structure was obtained by soaking crystals that were cocrystallized with 10 mM 5′-GTP in a precipitant solution containing 20 mM 5′-GTP, 10 mM SAM, 1 mM Na2S2O4, and 30% (vol/vol) glycerol for only 15 min to minimize crystal damage that is caused by the addition of Na2S2O4. Crystals of the MoaA-R17/266/268A variant were grown from 300 mM Na Hepes, pH 7.5/3.15 M Na formate/3% (vol/vol) DMSO. The structure was obtained by soaking crystals cocrystallized with 10 mM SAM in precipitant solution containing 10 mM SAM and 30% (vol/vol) glycerol for 30 min. Data were collected at 100 K on an ADSC Quantum 4 detector on beamline X26C at the National Synchrotron Light Source (NSLS; Brookhaven National Laboratory, Upton, NY) at a wavelength of 1.1 Å. Three hundred and fifty images were collected in 0.5° oscillations for both data sets. All data were indexed, integrated, and scaled with the hkl software (32). The structures were solved by difference Fourier techniques by using the MoaA–SAM complex structure (PDB ID code 1TV8) and were refined with refmac5 incorporating translation, libration, and screw-rotation displacement (TLS) refinement (33, 34). The refined model for the structure in complex with 5′-dA/5′-GTP revealed electron density for l-Met, 5′-dA, and 5′-GTP in chain A and l-Met and pyrophosphate in chain B.
Enzyme Assays.
In vitro assays were performed under anaerobic conditions at room temperature for 45 min in a total volume of 180 μl of 100 mM Tris·HCl, pH 9.0/300 mM NaCl in the presence of 2 mM DTT, a 10-fold molar excess of SAM, and substrate as well as a 20-fold molar excess of Na2S2O4. Assays of precursor Z-synthesizing activity and reductive cleavage of SAM were conducted as described in ref. 8. Analysis of a MoaA ortho-diaminopyrimidine-type product is described in Supporting Methods. Pyrophosphate was quantified by using the Biomol Green phosphate detection reagent (Plymouth Meeting, PA) after addition of 2 units of inorganic pyrophosphatase (Sigma).
Equilibrium Binding of Substrates to MoaA.
Substrate binding was assessed by microdialysis (35, 36) in an anaerobic chamber. MoaA (70 μl at a monomer concentration of 75 μM) was dialyzed against 100 μl of substrate at concentrations ranging from 0.01 to 5 mM (or 150 μM in a standard assay) for 16 h at room temperature or 24 h at 4°C. A sample of the dialysis buffer was analyzed to determine the free substrate concentration, whereas the protein/substrate mixture was analyzed to determine the total substrate. The amount of bound 5′-GTP was inferred as the difference between these values. Proteins were precipitated with 40% TCA (wt/vol) or 10% perchloric acid (vol/vol) and removed by centrifugation. Nucleotides were analyzed by HPLC (260-nm detection) by using a C18 column equilibrated with 50 mM NH4H2PO4, pH 2.5/5% (vol/vol) methanol at a flow rate of 1 ml·min−1 and compared with nucleotide standards.
Supplementary Material
Acknowledgments
We thank Silke Leimkühler (Universität Potsdam, Potsdam, Germany) for help with inductively coupled plasma optical emission spectroscopy. This work was supported by National Institutes of Health (NIH) Grant DK54835 (to H.S.). The National Synchrotron Light Source is supported by the Department of Energy and NIH, and beamline X26C is supported in part by the Research Foundation of Stony Brook University.
Abbreviations
- Moco
molybdenum cofactor
- SAM
S-adenosylmethionine
- 5′-dA•
5′-deoxyadenosyl radical
- GTPCH-I
GTP cyclohydrolase I
- PDB
Protein Data Bank.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2FB2 and 2FB3).
References
- 1.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Johnson M. K. Curr. Opin. Chem. Biol. 1998;2:173–181. doi: 10.1016/s1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- 3.Beinert H., Holm R. H., Munck E. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 4.Beinert H. J. Biol. Inorg. Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 5.Flint D. H., Allen R. M. Chem. Rev. 1996;96:2315–2334. doi: 10.1021/cr950041r. [DOI] [PubMed] [Google Scholar]
- 6.Beinert H., Kennedy M. C., Stout C. D. Chem. Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 7.Hänzelmann P., Hernandez H. L., Menzel C., Garcia-Serres R., Huynh B. H., Johnson M. K., Mendel R. R., Schindelin H. J. Biol. Chem. 2004;279:34721–34732. doi: 10.1074/jbc.M313398200. [DOI] [PubMed] [Google Scholar]
- 8.Hänzelmann P., Schindelin H. Proc. Natl. Acad. Sci. USA. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzi M., Schindelin H. Curr. Opin. Struct. Biol. 2002;12:709–720. doi: 10.1016/s0959-440x(02)00385-8. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz G. Cell. Mol. Life Sci. 2005;62:2792–2810. doi: 10.1007/s00018-005-5269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiss J. Hum. Genet. 2000;106:157–163. doi: 10.1007/s004390051023. [DOI] [PubMed] [Google Scholar]
- 12.Reiss J., Johnson J. L. Hum. Mutat. 2003;21:569–576. doi: 10.1002/humu.10223. [DOI] [PubMed] [Google Scholar]
- 13.Leimkühler S., Charcosset M., Latour P., Dorche C., Kleppe S., Scaglia F., Szymczak I., Schupp P., Hahnewald R., Reiss J. Hum. Genet. 2005;117:565–570. doi: 10.1007/s00439-005-1341-9. [DOI] [PubMed] [Google Scholar]
- 14.Hänzelmann P., Schwarz G., Mendel R. R. J. Biol. Chem. 2002;277:18303–18312. doi: 10.1074/jbc.M200947200. [DOI] [PubMed] [Google Scholar]
- 15.Santamaria-Araujo J. A., Fischer B., Otte T., Nimtz M., Mendel R. R., Wray V., Schwarz G. J. Biol. Chem. 2004;279:15994–15999. doi: 10.1074/jbc.M311815200. [DOI] [PubMed] [Google Scholar]
- 16.Rieder C., Eisenreich W., O’Brien J., Richter G., Gotze E., Boyle P., Blanchard S., Bacher A., Simon H. Eur. J. Biochem. 1998;255:24–36. doi: 10.1046/j.1432-1327.1998.2550024.x. [DOI] [PubMed] [Google Scholar]
- 17.Wuebbens M. M., Rajagopalan K. V. J. Biol. Chem. 1995;270:1082–1087. doi: 10.1074/jbc.270.3.1082. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee R. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 19.Jarrett J. T. Curr. Opin. Chem. Biol. 2003;7:174–182. doi: 10.1016/s1367-5931(03)00022-x. [DOI] [PubMed] [Google Scholar]
- 20.Cheek J., Broderick J. B. J. Biol. Inorg. Chem. 2001;6:209–226. doi: 10.1007/s007750100210. [DOI] [PubMed] [Google Scholar]
- 21.Brown G. M. In: Folates and Pterins. Blakley R. L., Benkovics S. J., editors. Vol. 2. New York: John Wiley & Sons; 1985. pp. 299–419. [Google Scholar]
- 22.Bacher A. In: Chemistry and Biochemistry of Flavoenzymes. Muller F., editor. Vol. 1. Boca Raton, FL: CRC; 1990. pp. 215–259. [Google Scholar]
- 23.Bochner B. R., Ames B. N. J. Biol. Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 24.Biot C., Buisine E., Kwasigroch J. M., Wintjens R., Rooman M. J. Biol. Chem. 2002;277:40816–40822. doi: 10.1074/jbc.M205719200. [DOI] [PubMed] [Google Scholar]
- 25.Rebelo J., Auerbach G., Bader G., Bracher A., Nar H., Hosl C., Schramek N., Kaiser J., Bacher A., Huber R., Fischer M. J. Mol. Biol. 2003;326:503–516. doi: 10.1016/s0022-2836(02)01303-7. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka Y., Nakagawa N., Kuramitsu S., Yokoyama S., Masui R. J. Biochem. (Tokyo) 2005;138:263–275. doi: 10.1093/jb/mvi120. [DOI] [PubMed] [Google Scholar]
- 27.Toraya T. Chem. Rev. 2003;103:2095–3127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 28.Berkovitch F., Nicolet Y., Wan J. T., Jarrett J. T., Drennan C. L. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lepore B. W., Ruzicka F. J., Frey P. A., Ringe D. Proc. Natl. Acad. Sci. USA. 2005;102:13819–13824. doi: 10.1073/pnas.0505726102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bracher A., Fischer M., Eisenreich W., Ritz H., Schramek N., Boyle P., Gentili P., Huber R., Nar H., Auerbach G., Bacher A. J. Biol. Chem. 1999;274:16727–16735. doi: 10.1074/jbc.274.24.16727. [DOI] [PubMed] [Google Scholar]
- 31.Kolberg M., Strand K. R., Graff P., Andersson K. K. Biochim. Biophys. Acta. 2004;1699:1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Winn M. D., Isupov M. N., Murshudov G. N. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 35.Jarrett J. T., Choi C. Y., Matthews R. G. Biochemistry. 1997;36:15739–15748. doi: 10.1021/bi971987t. [DOI] [PubMed] [Google Scholar]
- 36.Ugulava N. B., Frederick K. K., Jarrett J. T. Biochemistry. 2003;42:2708–2719. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.