Abstract
The mitochondrion of Trypanosoma brucei does not encode any tRNAs. This deficiency is compensated for by the import of a small fraction of nearly all of its cytosolic tRNAs. Most trypanosomal aminoacyl-tRNA synthetases are encoded by single-copy genes, suggesting the use of the same enzyme in the cytosol and mitochondrion. However, the T. brucei genome contains two distinct genes for eukaryotic tryptophanyl-tRNA synthetase (TrpRS). RNA interference analysis established that both TrpRS1 and TrpRS2 are essential for growth and required for cytosolic and mitochondrial tryptophanyl-tRNA formation, respectively. Decoding the mitochondrial tryptophan codon UGA requires mitochondria-specific C→U RNA editing in the anticodon of the imported tRNATrp. In vitro charging assays with recombinant TrpRS enzymes demonstrated that the edited anticodon and the mitochondria-specific thiolation of U33 in the imported tRNATrp act as antideterminants for the cytosolic TrpRS1. The existence of two TrpRS enzymes, therefore, can be explained by the need for a mitochondrial synthetase with extended substrate specificity to achieve aminoacylation of the imported thiolated and edited tRNATrp. Thus, the notion that, in an organism, all nuclear-encoded tRNAs assigned to a given amino acid are charged by a single aminoacyl-tRNA synthetase, is not universally valid.
Keywords: genetic code, mitochondria, RNA editing, aminoacyl-tRNA synthetase
In animal and most fungal mitochondria, the total set of tRNAs required for translation is encoded in the mitochondrial genome and, therefore, of bacterial evolutionary origin. The aminoacyl-tRNA synthetases (aaRSs) responsible for charging of mitochondrial tRNAs always are nuclear-encoded and, therefore, need to be imported into mitochondria (1). However, their evolutionary origin, just as the one of their substrate tRNAs, is in most cases bacterial. Thus eukaryotes, if we exclude all plastid-containing organisms, generally have two sets of aaRSs, one for cytosolic and a second one for mitochondrial aminoacyl-tRNA synthesis. In most eukaryotes, however, there are a few aaRSs that are targeted to both the cytosol and the mitochondria, indicating that the two sets of enzymes overlap to a limited extent (2).
Most mitochondrial genetic systems show deviations from the universal genetic code. The most common one is a reassignment of the termination codon UGA to tryptophan (3). Thus, to decode the UGA tryptophan codon, mitochondria require a nonstandard tRNATrp carrying a UCA instead of CCA anticodon. Cytosolic tRNATrpCCA is aminoacylated by a eukaryotic tryptophanyl-tRNA synthetase (TrpRS) (4), whereas the mitochondrial-encoded tRNATrpUCA is charged by a bacterial-type TrpRS (5). The CCA anticodon is a known identity element for both enzymes (6–8). It is clear, however, that the bacterial-type enzyme in mitochondria must be able to tolerate an UCA anticodon (5).
In contrast to animals and most fungi, mitochondria from protozoa and plants generally lack a variable number of mitochondrial tRNA genes. In these cases, the missing tRNAs are replaced by import of a small fraction of the corresponding nuclear-encoded cytosolic tRNAs (9). As a consequence, the imported tRNAs are always of a eukaryotic evolutionary origin. An intriguing situation is found in trypanosomatids, e.g., Trypanosoma brucei and Leishmania spp., which have lost all mitochondrial tRNA genes (10–12). Their mitochondrial translation system therefore must function exclusively with imported eukaryotic-type tRNAs.
Trypanosomatid mitochondria use UGA as tryptophan codon. However, the only tRNATrp gene in T. brucei is nuclear and carries the standard tryptophan anticodon CCA. Recent work in Leishmania revealed that trypanosomatids use RNA editing to convert the CCA anticodon of ≈40% of the imported tRNATrp to UCA; the resulting tRNA now is able to recognize UGA codons (13). Besides RNA editing, the imported tRNATrp is subjected to additional mitochondria-specific modifications; most importantly, the thiolation of U33, the “universally unmodified” uridine in all known tRNAs (14). tRNA editing is not required for thiolation of U33 but it is possible that the modification is needed for editing. In any case, the close proximity of the thiolated U33 to the anticodon suggests that it influences decoding.
Cytosolic and mitochondrial tRNAs of trypanosomatids originate from the same set of nuclear genes. Therefore, it is reasonable to assume that the same aaRSs are used in the cytosol and in mitochondria. This assumption is supported by the fact that in T. brucei, most aaRSs are represented by single genes only (15). Furthermore a dual localization in the cytosol and in mitochondria has been shown for T. brucei glutaminyl-tRNA synthetase and the glutamyl-tRNA synthetase (16). The imported trypanosomal tRNATrp, however, represents a special case, because its anticodon loop, due to the editing event and the thiolation of U33, differs from its cytosolic counterpart (14). This situation raises the question of how cytosolic and mitochondrial tryptophanyl-tRNA species are formed in T. brucei? Here we show that unlike most other trypanosomal tRNAs, because of the mitochondrial use of UGA as tryptophan codon, cytosolic and mitochondrial aminoacylation of tRNATrp requires two distinct eukaryotic-type TrpRSs.
Results
The T. brucei Genome Encodes Two Eukaryotic TrpRSs.
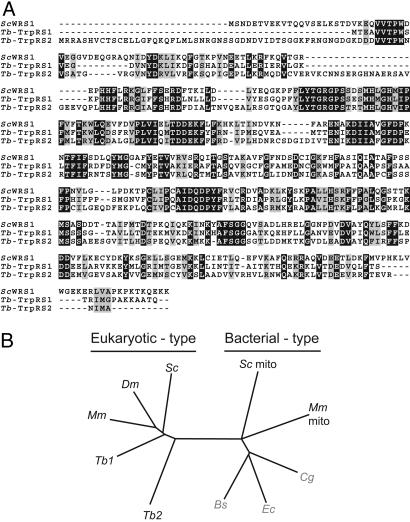
In the genome of Saccharomyces cerevisiae, probably the best characterized eukaryote, we find annotated genes for 36 different aaRSs (www.yeastgenome.org). These enzymes can be divided into 16 cytosol-specific ones, 14 of which are specific for mitochondria, and four are known to be doubly targeted to the cytosol and the mitochondria (these numbers are still, in part, based on predictions and, therefore, represent approximations). It is striking to compare yeast with T. brucei, whose genome encodes only 23 annotated aaRSs (15). This low number makes sense because the tRNAs in the cytosol and mitochondria derive from the same nuclear genes (10); it therefore can be expected that the same aaRSs are used in both compartments. Nevertheless, two distinct genes are found for aspartyl-tRNA synthetase, lysyl-tRNA synthetase, and TrpRS. Furthermore, in each case, one of the two proteins is predicted to have a mitochondrial targeting sequence. It is not obvious why T. brucei should have two aspartyl- or lysyl-tRNA synthetases, because the corresponding cytosolic and mitochondrial substrate tRNAs derived from the same nuclear genes. However, the need for two distinct TrpRSs might be explained by the fact that because of editing in the mitochondrion, cytosolic and mitochondrial tRNATrp differ in an anticodon nucleotide (13). In this study, we focus on the functional analysis of Tb-TrpRS1 and Tb-TrpRS2, the two trypanosomal TrpRS homologues. The two proteins are 41% identical, phylogenetic analysis shows that both are of the eukaryotic type, and Tb-TrpRS2 contains a predicted 50-aa mitochondrial presequence (Fig. 1). Tb-TrpRS1 shares 48–53% sequence identity to eukaryotic TrpRSs, whereas Tb-TrpRS2 is more diverged, showing an identity to other eukaryotic TrpRSs of only 39–41%. Interestingly, essentially the same situation is found in Leishmania (17). The two leishmanial proteins Lm-TrpRS1 and Lm-TrpRS2 are 76% and 59% identical to their trypanosomal counterparts.
Fig. 1.
T. brucei contains two eukaryotic TrpRSs. (A) Multiple sequence alignment of the cytosolic TrpRS from S. cerevisiae (ScWRS1) and the two T. brucei orthologues (Tb-TrpRS1 and Tb-TrpRS2). The sequences were aligned by using the clustal w program with default parameters. Strictly conserved residues and conservative replacements are shown in black and gray boxes, respectively. (B) Position of the two trypanosomal enzymes Tb-TrpRS1 (Tb1) and Tb-TrpRS2 (Tb2) in a phylogenetic tree based on a multiple sequence alignment of the cytosolic TrpRS from mouse [Mus musculus (Mm)], Drosophila melanogaster (Dm), and yeast [S. cerevisiae (Sc)]; the mitochondrial enzyme of yeast (Sc mito) and mouse (Mm mito); and, indicated by gray letters, the TrpRS from the bacteria Corynebacterium glutamicum (Cg), Bacillus subtilis (Bs), and E. coli (Ec). The tree was constructed by using treeview, which is available on http://taxonomy.zoology.gla.ac.uk/rod/treeview.html.
Intracellular Localization of Tb-TrpRS1 and Tb-TrpRS2.
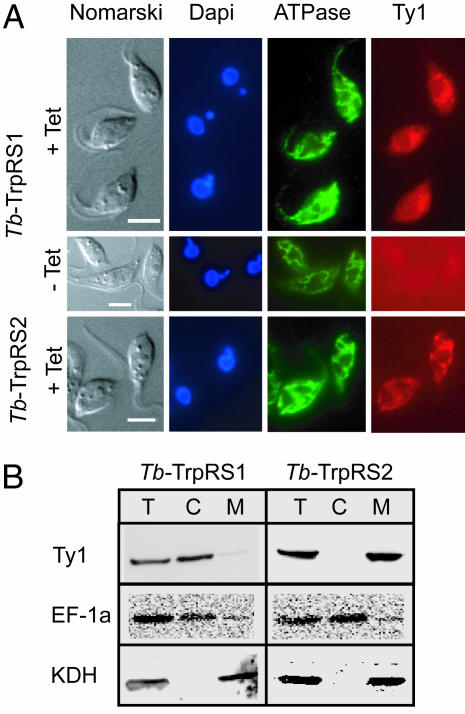
Tb-TrpRS2 is predicted to have a mitochondrial targeting signal. However, in T. brucei, such predictions are difficult because mitochondrial presequences can be very short (18). Thus, to determine the localization of the two enzymes, we prepared transgenic cell lines, allowing inducible expression of Tb-TrpRS1 and Tb-TrpRS2 versions carrying the 10-aa-long Ty1-peptide as epitope tags at their carboxyl termini (19). Immunofluorescence analysis by using an anti Ty1-antibody showed a tetracycline-inducible diffuse staining of the tagged Tb-TrpRS1, consistent with a cytosolic localization (Fig. 2A Top and Center). For tagged Tb-TrpRS2, on the other hand, a staining identical to the one seen with the mitochondrial marker was obtained (Fig. 2A Bottom). Furthermore, the two transgenic cell lines were subjected to a biochemical analysis that showed that the tagged Tb-TrpRS1 copurifies with the cytosolic marker, whereas the tagged Tb-TrpRS2 together with the mitochondrial marker is recovered in the pellet (Fig. 2B). These results are consistent with the immunofluorescence analysis and show that the two TrpRSs have a nonoverlapping intracellular distribution: Tb-TrpRS1 is exclusively cytosolic and Tb-TrpRS2 is exclusively mitochondrially localized.
Fig. 2.
Localization of trypanosomal TrpRSs. (A Top and Center) Double immunofluorescence analysis of a T. brucei cell line expressing Tb-TrpRS1 carrying the Ty1 tag at its carboxyl terminus under the control of the tetracycline-inducible (+Tet and −Tet) procyclin promoter. The cells were stained for DNA by using DAPI, for a subunit of the ATPase, serving as a mitochondrial marker and with a monoclonal antibody recognizing the Ty1-Tag. (A Bottom) Same as Top and Center, but a cell line expressing carboxyl-terminally Ty1-tagged Tb-TrpRS2 was analyzed. (Scale bars: 10 μm.) (B) Immunoblot analysis of total cellular (T), crude cytosolic (C), and crude mitochondrial extracts (M) for the presence of the Ty1-tagged Tb-TrpRS1 and Tb-TrpRS2, respectively. Elongation factor 1a (EF-1a) served as a cytosolic and α-ketoglutarate dehydrogenase (KDH) as a mitochondrial marker.
RNA Interference (RNAi)-Mediated Ablation of Tb-TrpRS1 and Tb-TrpRS2.
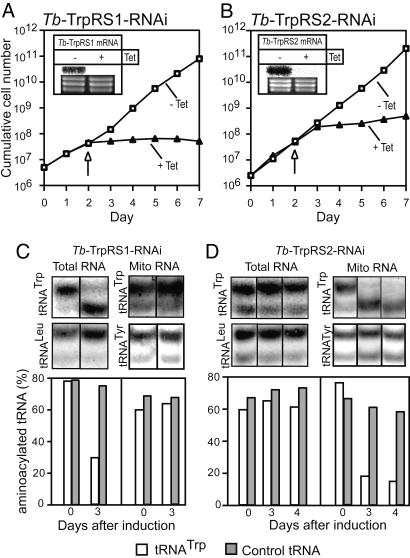
To determine the function of Tb-TrpRS1 and Tb-TrpRS2, we established two stable transgenic cell lines, which allow tetracycline-inducible ablation of each of the two enzymes. The Northern blot insets in Fig. 3A and B show that induction of RNAi in these two cell lines leads to specific degradation of the corresponding Tb-TrpRS mRNAs. Most importantly, concomitant with the depletion of the mRNAs, a growth arrest is observed 2 (for Tb-TrpRS1) and 3 days (for Tb-TrpRS2) after the addition of tetracycline (Fig. 3 A and B). Thus, Tb-TrpRS1 and Tb-TrpRS2 are both essential for growth of insect stage T. brucei.
Fig. 3.
Tb-TrpRS1 and Tb-TrpRS2 are essential for the growth of procyclic T. brucei and are responsible for formation of tryptophanyl-tRNA in the cytosol and the mitochondria, respectively (A) Growth curve in the presence and absence of tetracycline (+Tet and −Tet) of a representative clonal T. brucei RNAi cell line ablated for Tb-TrpRS1. (A Inset) A Northern blot for Tb-TrpRS1 mRNA. The time of sampling is indicated by the arrow. The rRNAs in the lower panel serve as loading controls. (B) Same as A for an RNAi cell line ablated for Tb-TrpRS2. (C) Northern blot analysis of total and mitochondrial RNA isolated under acidic conditions from the Tb-TrpRS1 RNAi cell line. The total RNA fraction only contains ≈5% of mitochondrial RNA and, thus, essentially represents cytosolic RNA. Days of induction (0 and 3) by tetracycline are indicated at the bottom. The blots were probed for the T. brucei tRNATrp as well as tRNALeu and tRNATyr, which serve as controls not affected by the RNAi. The RNA fractions were resolved on long acid urea gels, which allow separation of aminoacylated from deacylated tRNAs. The bar graph shows the quantification of the results. Relative amounts of aminoacylated tRNAs are indicated for the tRNATrp and the controls (tRNALeu and tRNATyr), respectively. For each lane, the sum of aminoacylated and deacylated tRNA was set to 100%. (D) Same as C, but analysis was done for the Tb-TrpRS2 RNAi cell line.
To determine the biochemical phenotype of the two RNAi cell lines, we isolated total and mitochondrial RNA from untreated cells and from cells grown in the presence of tetracycline. Subsequently, the RNAs were resolved on long acid urea polyacrylamide gels (20), followed by Northern blot analysis, to determine the ratio of uncharged tRNATrp to tryptophanyl-tRNAsTrp (Fig. 3 C and D). The results in Fig. 3C Left show that ablation of Tb-TrpRS1 results in the accumulation of uncharged cytosolic tRNATrp. Interestingly, in measurements of the levels of tryptophanyl-tRNATrp in the induced Tb-TrpRS2 cell line, the converse result was obtained, and a selective accumulation of deacylated mitochondrial tRNATrp was observed (Fig. 3D Right). As expected, ablation of either TrpRS had no influence on the aminoacylation levels of cytosolic tRNALeu or mitochondrial tRNATyr. These results show that, in agreement with its exclusive cytosolic localization, Tb-TrpRS1 is responsible for aminoacylation of the cytosolic tRNATrp. On the other hand Tb-TrpRS2, in line with its mitochondrial localization, is required for charging of imported mitochondrial tRNATrp.
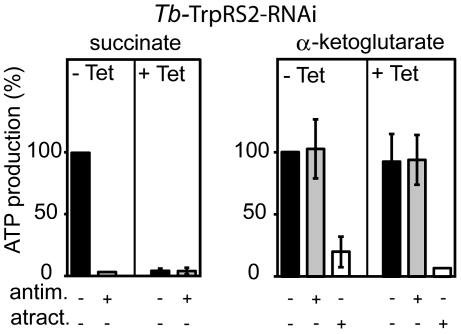
Ablation of Tb-TrpRS2 abolishes mitochondrial protein synthesis and, consequently, is expected to interfere with oxidative phosphorylation (OXPHOS). Mitochondria produce ATP, by OXPHOS and by substrate level phosphorylation linked to the citric acid cycle. We recently established an assay that allows quantitation of both modes of ATP production in isolated T. brucei mitochondria (21, 22). To measure antimycin-sensitive OXPHOS, mitochondria are incubated with ADP and succinate. α-ketoglutarate is used in the determination of the antimycin-resistant substrate level phosphorylation, whereas atractyloside treatment that prevents mitochondrial import of the added ADP is the control. The results in Fig. 4 show that ablation of Tb-TrpRS2 selectively knocks down OXPHOS that, in part, depends on mitochondrial-encoded proteins but does not interfere with substrate level phosphorylation, which depends solely on nuclear encoded proteins.
Fig. 4.
Ablation of Tb-TrpRS2 selectively abolishes OXPHOS. Succinate and α-ketoglutarate mitochondrial ATP production in crude mitochondrial fractions of the Tb-TrpRS2 RNAi cell line. Uninduced cells (−Tet) are shown on the left, and induced cells (+Tet) are shown on the right of the graphs. The substrate tested is indicated at the top, and additions of antimycin (antim.) and atractyloside (atract.) are indicated at the bottom. ATP productions in mitochondria isolated from uninduced cells tested without antimycin or atractyloside are set to 100%. The bars represent means expressed as percentages from three or more independent inductions. SEs are indicated.
Substrate Specificities of Tb-TrpRS1 and Tb-TrpRS2.
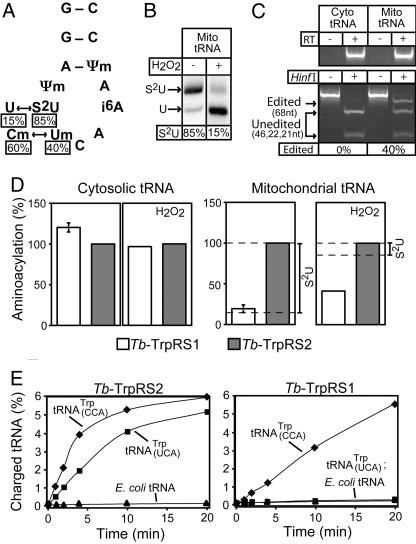
Recent work in Leishmania (14) showed that the imported tRNATrp is present in two main forms: (i) the tRNATrpCCA carrying a mitochondria-specific methylation on the C34 and (ii) the tRNATrpUCA in which the methylated C34 has been edited to a methylated U and which also contains a mitochondria-specific thiolated U33. Furthermore, it was shown that both tRNATrp forms contain a mitochondria-specific methylation on the Ψ32 (Fig. 5A). Fig. 5B shows that in T. brucei ≈85% of the imported tRNATrp is thiolated. Furthermore, RT-PCR analysis by using cytosolic and mitochondrial RNA as substrates demonstrates that ≈40% of mitochondrial tRNATrp is edited (Fig. 5C). Thus, in contrast to Leishmania, where all thiolated tRNATrp is edited (14), there is a population of T. brucei tRNATrp, which is thiolated but not edited.
Fig. 5.
Tb-TrpRS1 and Tb-TrpRS2 have distinct substrate specificities. (A) Mitochondrial editing and modification events of the anticodon loop of the tRNATrp as described for Leishmania (14). The percentages of the thiolated U (s2U) and the C→U editing as determined for T. brucei tRNATrp are indicated. (B Left) The percentage of thiomodified mitochondrial tRNATrp of T. brucei was measured by N-acryloylaminophenylmercuric chloride gel electrophoresis (33) and Northern blot hybridization. The shifted band represents thiolated tRNATrp(S2U). (B Right) The fraction of thiolated mitochondrial tRNATrp that remains thiolated after H2O2 treatment was determined as in the left lane. (C) Quantitative RT-PCR assay for edited tRNATrp. (C Upper) The cytosolic and mitochondrial tRNA fractions that were used as templates are free of DNA. (C Lower) The blot shows that RNA editing can be analyzed by a restriction digest because it destroys a HinfI site that is present in the cDNA derived from the unedited tRNATrp (14). Introduction of a synthetic HinfI plus 20 flanking nucleotides at the 5′ end of the 5′ RT-PCR primer provides an internal control for the HinfI digestion, allowing the quantitative determination of RNA editing. cDNA amplified from unedited tRNATrp contains two HinfI sites and, thus, will be digested into three fragments (46, 22, and 21 nt; unedited). The cDNA derived from edited tRNATrp contains the synthetic HinfI site only and will be digested into two fragments (68 and 21 nt; edited). Measuring the intensities of the diagnostic bands for nonedited (46 nt) and edited (68 nt) tRNATrp allows, after correcting for their different molecular mass, determination of the fraction of edited tRNATrp in T. brucei mitochondria. (D) In vitro aminoacylation assays by using [3H]tryptophan and recombinant Tb-TrpRS1 as well as Tb-TrpRS2 as enzymes. (Left and Center Left) Untreated and H2O2-treated cytosolic tRNA were charged. (Right and Center Right) Same as Left and Center Left but with untreated and H2O2-treated mitochondrial tRNAs. For each graph, the tRNA charged by the Tb-TrpRS2 was set to 100%. The percentage of mitochondrial tRNATrp that is thiolated in the untreated and treated fractions is indicated on the right. The means and SE for three independent experiments are shown for aminoacylation of untreated tRNAs. For experiments using H2O2-treated tRNAs, the mean of two experiments is shown. The two values each for cytosolic and mitochondrial tRNAs varied by 20% and 10%, respectively. (E) Aminoacylation of T. brucei tRNATrp overexpressed in E. coli by using 200 nM enzyme and 0.25 μg/μl tRNA. (Right) Tb-TrpRS1 charging of total E. coli tRNA with T. brucei tRNATrpUCA overexpressed (■), total E. coli tRNA with T. brucei tRNATrpCCA overexpressed (♦), and total E. coli tRNA (▴). (Left) Tb-TrpRS2 charging of total E. coli tRNA with T. brucei tRNATrpUCA overexpressed (■), total E. coli tRNA with T. brucei tRNATrpCCA overexpressed (♦), and total E. coli tRNA (▴).
To test their substrate specificities, recombinant proteins of Tb-TrpRS1 and Tb-TrpRS2 were overexpressed in Escherichia coli and purified to >95% homogeneity. In vitro charging assays showed that although neither enzyme was able to recognize in vitro transcripts corresponding to unedited or edited tRNATrp (data not shown), both efficiently aminoacylated isolated T. brucei cytosolic tRNA (Fig. 5D Left). Thus, the cytosolic unedited tRNATrpCCA can be charged with very similar efficiencies by both the cytosolic and the mitochondrial enzyme. Interestingly, however, when using isolated mitochondrial tRNAs as a substrate, the level of aminoacylation achieved by the cytosolic Tb-TrpRS1 dropped to 19% of that obtained with the mitochondrial enzyme (Fig. 5D Center Right). This fraction correlates with the level of nonthiolated mitochondrial tRNATrp (Fig. 5B), suggesting that, in contrast to mitochondrial Tb-TrpRS2, Tb-TrpRS1 is not able to charge thiolated tRNATrp. Fig. 5B shows that hydrogen peroxide treatment produces a mitochondrial tRNATrp population in which only 15% (instead of 85%) of the molecules contain thiolated U33. Interestingly, the H2O2-treated mitochondrial tRNA fraction can be charged by cytosolic Tb-TrpRS1 to a level that corresponds to ≈40% of the one observed with the mitochondrial enzyme (Fig. 5D Right). However, the reactivity of either enzyme did not change when tested with H2O2-treated cytosolic tRNAs, which do not contain thiolated tRNATrp (Fig. 5D Center Left). Thus, removing the thio group converts a population of mitochondrial tRNATrp into a substrate for the cytosolic Tb-TrpRS1. Interestingly, even in the absence of thiolated U33, ≈60% of the mitochondrial tRNATrp remained refractory to aminoacylation by the cytosolic enzyme. The uncharged fraction represents ≈50% of the thiolated U33 lacking tRNATrp population and, thus, is similar to the ≈40% observed for the edited population. Therefore, the most parsimonious explanation of these results is that the edited U34, just as the thiolated U33, both act as independent antideterminants for cytosolic Tb-TrpRS1. To confirm the charging results presented above, we expressed the T. brucei tRNATrpUCA and tRNATrpCCA genes in E. coli and isolated total tRNA. Aminoacylation of these tRNA samples with the two TrpRSs demonstrated (Fig. 5E) that the mitochondrial Tb-TrpRS2 enzyme charged both tRNATrp isoacceptors, whereas the cytosolic Tb-TrpRS1 acylated only tRNATrpCCA. Neither enzyme charged E. coli tRNA. Thus, Tb-TrpRS2 efficiently aminoacylates the unedited and edited fraction of mitochondrial tRNATrp. Furthermore, the in vivo aminoacylation level of total mitochondrial tRNATrp was shown to be close to 80% (Fig. 3D). These results suggest that both isoacceptors are used in mitochondrial protein synthesis, the unedited tRNATrp being restricted to decode the standard UGG tryptophan codons.
Our current understanding of the structure of eukaryotic TrpRS does not allow a prediction of the anticodon binding sites that might explain the different tRNA recognition properties. Based on structural modeling of human TrpRS (D. Kennedy and W. Yin, unpublished data), the putative anticodon-binding domain of Tb-TrpRS1 extends from G270-F373. The sequences of the two Tb-TrpRSs are quite different in this region; they share only 30% amino acid identity.
Discussion
One of the few generalizations about mitochondrial tRNA import states that only a small fraction of a given nuclear encoded tRNA is imported and that the remainder functions in cytosolic translation (9). Thus, in all organisms, imported tRNAs are always of the eukaryotic type. Mitochondrial translation, however, is of a bacterial evolutionary origin. tRNA import, therefore, can be considered as a horizontal gene product transfer between the eukaryotic and bacterial domains. There are some fundamental differences between eukaryotic and bacterial-type translation systems that, for some tRNAs, are expected to prevent the dual use in the cytosol and in mitochondria. One such difference is necessitated by mitochondrial variants of the genetic code. The most frequent code deviation is the reassignment of the stop codon UGA to tryptophan (3). Reading this reassigned codon requires a tRNATrp with a UCA anticodon. Simultaneous use of this tRNA in the cytosol and in mitochondria, however, is not possible because it would act as a nonsense suppressor in the cytosol. In line with this constraint, it was recently shown that in most organisms that import tRNAs the mitochondrial tRNATrp gene has been retained (23). Thus, it is unexpected that trypanosomatids import the tRNATrp. The way Leishmania mitochondria accommodate the change in the genetic code is by mitochondria-specific C→U editing of the first anticodon position of the imported tRNATrp (13). In the present work, we have analyzed the situation in the closely related T. brucei and shown that although editing of the imported tRNATrp is necessary, it is not sufficient to allow decoding of the mitochondrial UGA codons. The problem that arises is that the mitochondrially localized tRNATrp cannot be aminoacylated by the standard eukaryotic-type TrpRS (Tb-TrpRS1). In contrast to most other trypanosomal aaRSs, which have dual location and are used to charge cytosolic and imported tRNAs (16), Tb-TrpRS1 is exclusively found in the cytosol. A survey of the genome showed that T. brucei has a second TrpRS (Tb-TrpRS2) that is related to the cytosolic Tb-TrpRS1 but is localized exclusively in mitochondria. This enzyme has an extended substrate specificity and aminoacylates in vitro both cytosolic and mitochondrial tRNATrp. Further studies showed that both the thio-modified U33 and the C→U editing of the first nucleotide in the anticodon prevent mitochondrial tRNATrp to be recognized by the cytosolic Tb-TrpRS1. The function of the thiolated U33 in mitochondrial tRNATrp of trypanosomatids is unknown (14). However, the fact that it is localized immediately 5′ of the first position of the anticodon suggests that it influences the decoding properties of the tRNATrp or that it even may be required for tRNA editing. In most systems, the reassignment of the UGA codon is accommodated by a single mutation in the mitochondrial tRNATrp gene, which probably requires an adaptation of the mitochondrial TrpRS (3). The situation is different for mitochondria of trypanosomatids because they need (i) a mitochondria-specific tRNA editing enzyme that produces the tRNATrpUCA (13) and (ii) a separate TrpRS (Tb-TrpRS2) with an extended substrate specificity. The need for a record TrpRS is due to the fact that the C→U editing not only changes the decoding properties of the tRNATrp but also its identity toward the classic TrpRS (Tb-TrpRS1). Thus, recoding of UGA to tryptophan appears to be more costly for trypanosomatids than for other organisms.
Doubly targeted aaRSs, which are able to aminoacylate both nuclear-encoded tRNAs in the cytosol and the corresponding mitochondrial encoded tRNAs, are found in most, if not all, eukaryotes (2). In this study, we describe the converse situation in that tRNAs derived from the same nuclear gene require two distinct aaRSs for proper function. Thus, the presence of a single modification, be it the thio group on the U33 or the C→U editing of the anticodon, is sufficient to prevent aminoacylation by Tb-TrpRS1. Our results show that the notion that in an organism all nuclear-encoded tRNA isoacceptors for a given amino acid are charged by a single aaRS (4) is not universally valid.
The anticodon and the discriminator nucleotide are the major identity elements for TrpRS (6, 7). Whereas the anticodon is a phylogenetically shared identity element, the discriminator nucleotide differs between bacteria and eukaryotes (6), which explains why the two TrpRSs cannot efficiently cross-aminoacylate the corresponding tRNATrp species (24, 25). Most mitochondria decode UGA as tryptophan, thus it is clear that the bacterial-type mitochondrial TrpRS has evolved to tolerate an UCA anticodon (3, 5). Interestingly, the same applies for the eukaryotic mitochondria-localized Tb-TrpRS2 in T. brucei because, unlike the cytosolic Tb-TrpRS1, it is able to charge eukaryotic tRNATrp carrying an UCA anticodon. Thus, the existence of both bacterial and eukaryotic TrpRSs that are able to charge mitochondrial tRNATrp carrying the nonstandard UCA anticodon is a result of convergent evolution. Whether bacterial or eukaryotic TrpRS is used inside mitochondria might be determined to a great extent by the discriminator nucleotide on the corresponding tRNATrp and, therefore, depends on whether the tRNATrp is encoded in the mitochondrial DNA or imported from the cytosol.
Thus Tb-TrpRS2 represents an adaptation to the fact that the trypanosomal mitochondrial translation system has to function with eukaryotic-type tRNAs. Another adaptation of the same kind is the unusual T. brucei methionyl-tRNAMet formyltransferase (MTF), which because of the absence of a bacterial-type initiator tRNAMet, has to formylate the only tRNAMet present in mitochondria, which is the imported elongator tRNAMet (26, 27). To perform their function, the trypanosomal MTF and the Tb-TrpRS2 had to develop a substrate specificity that is distinct from their orthologues. Trypanosomal MTF completely switched its specificity and exclusively formylates elongator tRNAMet, unlike all other MTFs, which only recognize bacterial-type initiator tRNAsMet. In the case of Tb-TrpRS2, the enzyme still recognizes the standard substrate for eukaryotic TrpRS, the tRNATrpCCA, but has extended its substrate specificity to tRNAsTrp carrying a thiolated U33 and an UCA anticodon.
Studies of the adaptations of mitochondrial translation factors that interact directly with tRNAs (e.g., aaRSs, initiation factor 2 (28), or elongation factor Tu) to imported eukaryotic tRNAs should yield additional surprises and reveal more fundamental requirements of translation.
Materials and Methods
Cells.
Procyclic T. brucei, strain 29-13 (29) was grown in SDM-79, supplemented with 15% FCS/25 μg/ml hygromycin/15 μg/ml G-418 at 27°C, and harvested at 1.5–3.5 × 107 cells per ml.
Production of Transgenic Cell Lines.
As a tag to localize Tb-TrpRS1 (accession no. Tb927.3.5580) and Tb-TrpRS2 (accession no. Tb927.8.2240), we used a 10-aa epitope of the major structural protein of yeast Ty1, which is recognized by the monoclonal antibody BB2 (19). The sequences corresponding to the carboxyl-terminal Ty1-tagged Tb-TrpRS1 and Tb-TrpRS2 were cloned into a derivative of pLew-100 to allow tetracycline-inducible expression of the tagged proteins (29).
RNAi of Tb-TrpRS1 and Tb-TrpRS2 was performed by using stem loop constructs containing the puromycin resistance gene (22). As inserts, we used a 537-bp fragment (nucleotides 4–540) of the Tb-TrpRS1 gene and a 537-bp fragment (nucleotides 4–540) of the Tb-TrpRS2 gene.
Transfection of T. brucei and selection with antibiotics, cloning, and induction with tetracycline were done as described in ref. 30.
Cell Fractionation by Digitonin.
Fractionation of Ty1 epitope-tagged Tb-TrpRS1- and Tb-TrpRS2-expressing cells was done by digitonin extraction. Washed T. brucei cells (108 cells) were resuspended in 0.5 ml of SoTE (0.6 M sorbitol/20 mM Tris·HCl, pH 8/2 mM EDTA). After the addition of 0.5 ml of SoTE containing 0.03% (wt/vol) of digitonin, the samples were mixed by pipetting and incubated on ice for 5 min. The suspensions were centrifuged at 6,800 × g for 5 min at 4°C, resulting in a pellet that corresponds to a crude mitochondrial fraction and a supernatant faction. The latter was cleared by centrifugation (10 min at 21,000 × g at 4°C), yielding a crude cytosolic fraction. Finally, 0.3 × 107 cell equivalents each of total protein extract, crude cytosolic, and crude mitochondrial fractions were separated by SDS-gel electrophoresis and analyzed by immunoblotting.
Analysis of in Vivo Aminoacylation.
Mitochondria from uninduced and induced Tb-TrpRS1 and Tb-TrpRS2 RNAi cell lines were isolated by digitonin extraction and subsequent RNase treatment as described to yield a crude mitochondrial fraction that is essentially free of cytosolic RNAs (10). RNA was isolated from the crude mitochondrial fractions as well as uninduced and induced total cells by using the acid guanidinium isothiocyanate procedure (31), which allows isolation of charged tRNA. Then charged tRNA was separated from free tRNA on acid urea polyacrylamide gels as described in ref. 20 and visualized by Northern blot hybridization with labeled oligonucleotides (tRNATrp, TGAGGACTGCAGGGATTG; tRNALeuCAG, CCTCCGGAGAGATGACGA; tRNATyrGUA, TGGTCCTTCCGGCCGGAATCGAA).
Cloning, Overexpression, and Purification of Tb-TrpRS1 and Tb-TrpRS2.
The gene sequences were PCR-amplified by using the Expand High Fidelity PCR System (Roche Applied Science). Tb-TrpRS1 was cloned into NcoI/XhoI in pET20b (Novagen) with a C-terminal His tag. For Tb-TrpRS2, the N-terminal 21 amino acids (mitochondrial leader sequence) were omitted, and the sequence was cloned into the NdeI/XhoI in pET15b (Novagen). After sequence verification, the resulting plasmids were transformed into E. coli Bl-21-CodonPlus(DE3)-RIL cells (Stratagene).
Cells were grown at 37°C in LB medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml), and protein expression was autoinduced by using the Overnight Express Autoinduction System (Novagen) according to the manufacturer’s instructions. After harvest, the cells were sonicated, and the proteins were purified by Ni-NTA chromatography (Qiagen, Valencia, CA). The desired fractions were pooled, dialyzed against 50 mM Na2HPO4, pH 8.0/5 mM 2-mercaptoethanol/50% glycerol, and stored at −20°.
In Vitro Aminoacylation Assays.
(i) Using native tRNA.
Mitochondria were prepared by hypotonic lysis and Percoll gradient centrifugation as described in ref. 32. The supernatant obtained after the initial lysis was used to isolate cytosolic RNAs by repeated phenol extractions and ethanol precipitations. Mitochondrial RNA was isolated from gradient purified mitoplasts by the acid guanidinium isothiocyanate procedure (31). A cumulative 25-liter T. brucei culture yielded 6 mg of mitochondrial RNA. Total and mitochondrial RNA were deacylated in 0.3 M Tris·HCl (pH 9.0) at 30°C for 1 h. Finally, tRNAs were isolated from both fractions by using Qiagen-Tip columns as described in ref. 16. The percentage yield of tRNAs from deacylated total and mitochondrial RNAs was ≈5%. H2O2 treatment of mitochondrial and total tRNA was done at 0.2 mg/ml by using 0.21% (wt/vol) of H2O2 in 10 mM Tris·HCl (pH 7.5) for 20 h at 20°C. The reaction was stopped by adding 2-mercaptoethanol to 50 mM, and tRNAs were purified by ethanol precipitation. The presence and absence of thiolated U33 was monitored by using 8 M urea/10% polyacrylamide gels containing a 25 mM concentration of N-acryloylaminophenylmercuric chloride (33).
In vitro aminoacylation assay were performed in 50 mM Hepes, pH 7.0/10 mM Mg-acetate/2 mM ATP/4 mM DTT/0.05% (wt/vol) BSA, and a mixture of 38 μM cold and 2 μM [3H]tryptophan (32 Ci/mmol; 1 Ci = 37 GBq). The enzyme and tRNA concentrations used were as follows: 400 nM recombinant Tb-TrpRS1 or Tb-TrpRS2 and 1 mg/ml isolated total or mitochondrial tRNA as substrates. Incubation was for 10 min at 37°C, and Trp-tRNA was determined as described in ref. 34.
(ii) Using E. coli tRNA in which T. brucei tRNATrpUCA and tRNATrpCCA were overexpressed.
Reactions were carried out as described above with the following modifications: 75.5 μM cold and 4.5 μM [3H]tryptophan (32 Ci/mmol)/200 nM TrpRS/0.25 mg/ml tRNA.
Miscellaneous.
Northern blots of tRNAs and ATP production assays were done as described in refs. 10 and 22. For the RT-PCR to amplify cDNA of cytosolic and mitochondrial tRNATrp, the oligonucleotides used were as follows: forward primer (AGAGAGAGCGAGGAAGGCGAGATTCTCAGTGGTAGAGCATTGG, containing a synthetic HinfI site) and reverse primer (TGGTGAGGACTGCAGGGATTG).
Acknowledgments
We thank G. Cross (The Rockefeller University, New York) and D. Speijer (University of Amsterdam, Amsterdam) for cell lines, plasmids, and antisera and J. Rinehart for encouragement. This work was supported by Swiss National Foundation Grant 3100-067906 (to A.S.), a Fellowship of the Roche Research Foundation (to F.C.), and grants from the National Institute of General Medical Sciences (to D.S.).
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- OXPHOS
oxidative phosphorylation
- RNAi
RNA interference
- TrpRS
tryptophanyl-tRNA synthetase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bullerwell C. E., Gray M. W. Curr. Opin. Microbiol. 2004;7:528–534. doi: 10.1016/j.mib.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Small I., Wintz H., Akashi K., Mireau H. Plant Mol. Biol. 1998;38:265–277. [PubMed] [Google Scholar]
- 3.Knight R. D., Freeland S. J., Landweber L. F. Nat. Rev. Genet. 2001;2:49–58. doi: 10.1038/35047500. [DOI] [PubMed] [Google Scholar]
- 4.Woese C. R., Olsen G. J., Ibba M., Söll D. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorgensen R., Sogaard T. M. M., Rossing A. B., Martensen P. M., Justesen J. J. Biol. Chem. 2000;275:16820–16826. doi: 10.1074/jbc.275.22.16820. [DOI] [PubMed] [Google Scholar]
- 6.Xue H., Shen W., Giege R., Wong J. T.-F. J. Biol. Chem. 1993;268:9316–9322. [PubMed] [Google Scholar]
- 7.Ulmasov B., Topin A., Chen Z., He S. H., Folk W. R. Nucleic Acids Res. 1998;26:5139–5141. doi: 10.1093/nar/26.22.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yesland K. D., Nelson A. W., Feathers D. M. S., Johnson J. D. J. Biol. Chem. 1993;268:217–220. [PubMed] [Google Scholar]
- 9.Schneider A., Marechal-Drouard L. Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 10.Tan T. H. P., Pach R., Crausaz A., Ivens A., Schneider A. Mol. Cell. Biol. 2002;22:3707–3717. doi: 10.1128/MCB.22.11.3707-3716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock K., Hajduk S. L. J. Biol. Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- 12.Simpson A. M., Suyama Y., Dewes H., Campbell D. A., Simpson L. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfonzo J. D., Blanc V., Estevez A. M., Rubio M. A. T., Simpson L. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crain P. F., Alfonzo J. D., Rozenski J., Kapushoc S. T., McCloskey J. A., Simpson L. RNA. 2002;8:752–761. doi: 10.1017/s1355838202022045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., et al. Science. 2005;309:416–422. [Google Scholar]
- 16.Rinehart J., Horn E. K., Wei D., Söll D., Schneider A. J. Biol. Chem. 2004;279:1161–1166. doi: 10.1074/jbc.M310100200. [DOI] [PubMed] [Google Scholar]
- 17.Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., et al. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häusler T., Stierhof Y.-D., Blattner J., Clayton C. Eur. J. Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- 19.Bastin P., Bagherzadeh A., Matthews K. R., Gull K. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 20.Varshney U., Lee C.-P., RajBhandary U. L. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 21.Schneider A., Bouzaidi-Tiali N., Chanez A.-L., Bulliard L. Methods Mol. Biol. 2006 doi: 10.1007/978-1-59745-365-3_27. in press. [DOI] [PubMed] [Google Scholar]
- 22.Bochud-Allemann N., Schneider A. J. Biol. Chem. 2002;277:32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 23.Schneider A. Trends Genet. 2001;17:557–558. doi: 10.1016/s0168-9525(01)02439-8. [DOI] [PubMed] [Google Scholar]
- 24.Xu F., Chen X., Xin L., Chen L., Jin Y., Wang D. Nucleic Acids Res. 2001;29:4125–4133. doi: 10.1093/nar/29.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Q., Gong Q., Tong K. L., Vestergaard B., Costa A., Desgres J., Wong M., Grosjean H., Zhu G., Wong J. T., et al. J. Biol. Chem. 2002;277:14343–14349. doi: 10.1074/jbc.M111745200. [DOI] [PubMed] [Google Scholar]
- 26.Tan T. H. P., Bochud-Allemannn N., Horn E. K., Schneider A. Proc. Natl. Acad. Sci. USA. 2002;99:1152–1157. doi: 10.1073/pnas.022522299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin N. C. Proc. Natl. Acad. Sci. USA. 2002;99:1110–1112. doi: 10.1073/pnas.042011199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charrière F., Tan T. H. P., Schneider A. J. Biol. Chem. 2005;280:15659–15665. doi: 10.1074/jbc.M411581200. [DOI] [PubMed] [Google Scholar]
- 29.Wirtz E., Leal S., Ochatt C., Cross G. A. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 30.McCulloch R., Vassella E., Burton P., Boshart M., Barry J. D. Methods Mol. Biol. 2004;262:53–86. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 31.Chomczyinski P., Sacchi N. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 32.Schneider A., Charrière F., Pusnik M., Horn E. K. Methods Mol. Biol. 2006 doi: 10.1007/978-1-59745-365-3_5. in press. [DOI] [PubMed] [Google Scholar]
- 33.Igloi G. L. Anal. Biochem. 1992;206:363–368. doi: 10.1016/0003-2697(92)90379-l. [DOI] [PubMed] [Google Scholar]
- 34.Nabholz C. E., Hauser R., Schneider A. Proc. Natl. Acad. Sci. USA. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]