Abstract
The heart of the H+ conductance mechanism in the homotetrameric M2 H+ channel from influenza A is a set of four histidine side chains. Here, we show that protonation of the third of these imidazoles coincides with acid activation of this transmembrane channel and that, at physiological pH, the channel is closed by two imidazole–imidazolium dimers, each sharing a low-barrier hydrogen bond. This unique construct succeeds in distributing a pair of charges over four rings and many atoms in a low dielectric environment to minimize charge repulsion. These dimers form with identical pKas of 8.2 ± 0.2, suggesting cooperative H+ binding and clearly illustrating high H+ affinity for this channel. The protonation behavior of the histidine side chains has been characterized by using solid-state NMR spectroscopy on the M2 transmembrane domain in fully hydrated lipid bilayers where the tetrameric backbone structure is known. Furthermore, electrophysiological measurements of multichannel and single-channel experiments confirm that these protein constructs are functional.
Keywords: M2 channel, proton channel, solid-state NMR, low-barrier hydrogen bond, histidine ionization constants
A histidine tetrad in the pore of the tetrameric M2 protein has long been associated with key channel features of H+ selectivity, pH activation, gating, inhibition, and the specific conductance mechanism. M2 protein from influenza A virus conducts protons into the viral core after endocytosis, which leads to the uncoating and release of genetic material into the cytoplasm after fusion of the viral coat with the endosomal wall (1, 2). Much is known about this system from its tetrameric state (2–4), the backbone structure of the transmembrane (TM) domain (5), and numerous electrophysiological (6, 7), biophysical (8–10), and modeling (11) studies that have cast a fascinating tale for this important influenza drug target and the only proton channel of its kind to be characterized in such detail. However, the specific role of His-37 in the tetrameric protein has not been elucidated. Here, we have characterized the pKas associated with this cluster of four histidine residues in the hydrophobic interstices of the membrane. These pKa values have led us to substantial mechanistic conclusions.
There are many lines of evidence, reviewed by Kelly et al. (6), that support the conclusion that M2 is responsible for viral acidification. In vivo ion conductance recordings have shown pH sensitive conductance resulting in rapid acidification of the Xenopus oocytes (12, 13) and mammalian cells (13–15) containing M2 protein. Preparations of purified M2 protein have also been used to show proton conductance in synthetic lipid bilayers (16, 17). Single-channel conductance measurements with membranes containing M2 protein give clear evidence that it is H+ conductance, not counterion conductance, that is observed. Furthermore, the channel conductance is unchanged by addition of an excess of NaCl (18). Conductance measurements for the isolated TM domain of M2 protein have also been demonstrated previously (19) and confirmed with our preparations (see below).
The M2 channel is a 96-residue protein with a single TM helix having three hydrophilic residues, Ser-31, His-37, and Trp-41, all of which are pore-lining based on cysteine mutagenesis (20, 21) and structural characterization of the TM domain (5). In a comparison of 42 M2 protein amino acid sequences from influenza A viruses (22), His-37 and Trp-41 are absolutely conserved. Both of these residues have been shown to have important functional roles. Replacement of His-37 by a variety of other amino acids has shown that both proton selectivity and the protein’s activation by low pH are lost (23). Elegant chemical rescue experiments have shown that imidazole in the buffer rescues proton selectivity for H37G, H37S, and H37T mutants (24). Imidazole also enhances proton transport, improves pH sensitivity in these mutants, and rescues Cu(II) inhibition of conductance. The chemical rescue was very specific for imidazole, clearly demonstrating the central role for the chemical properties of the histidine tetrad.
The Trp-41 residue is also functionally important and located just one helical turn from His-37. Indeed, their close proximity has been shown by solid-state NMR spectroscopy (5) and UV resonance Raman spectroscopy (25). This latter study suggests a pH-dependent change in the interaction between His-37 and Trp-41, potentially involving a cation (His-37)–π(Trp-41) interaction. Fluorescence quenching of Trp-41 at pH values <6.0 implicated a protonated His-37 as the quencher (8). Moreover, mutations of Trp-41 to less bulky side chains permitted outward flow of H+ under favorable conditions, whereas the presence of Trp-41 prevented such conductance (26). In addition, it was shown that Cu2+ injected into oocytes inhibited conductance by the W41A mutant, but not by the wild type, further supporting the contention that the indole side chain forms a steric block or gate for H+ conductance.
The protonation state of the histidine tetrad is critical to our understanding of the role of the His-37 residues in H+ conductance and its interaction with Trp-41. Proton NMR spectroscopy of the monomeric TM peptide of M2 protein in dodecylphosphatidyl-choline micelles was analyzed to yield a pKa of 6.8 (27). Resonance Raman spectroscopy of the TM domain has been used to observe the charged imidazole state, and, based on the intensity of this band as a function of pH, a pKa of 5.7 was inferred (25). In addition, an indole band showed considerable pH dependence with the same pKa, supporting the His-37 titration results. Furthermore, the charged imidazole band showed an intensity in liposomes at pH 4.0 equivalent to that in SDS micelles at the same pH, suggesting that all four histidines have titrated with the same pKa. Detailed modeling efforts by Lear (10) suggest that, because of excessive cooperativity, the electrophysiological data do not fit when two or three pKas are identical. Many of these results do not appear to be consistent with those presented here, but, on closer inspection, much of the data can be rationalized.
Isotropic 15N chemical shift frequencies have been very effective in characterizing the protonation and charged state of histidine residues (28). Magic-angle spinning solid-state NMR spectroscopy has been even more useful in characterizing these states; dynamic averaging is reduced when isotropic tumbling of the sample is eliminated (29). The histidine side chain can adopt three protonation states for the Nδ1 and Nε2 sites, with either of the two sites protonated or with both sites protonated. Only the latter state results in a net positive charge on the side chain. For both the δ1 and ε2 15N sites, protonated resonances occur in the vicinity of 150 ppm, whereas nonprotonated resonances are near 230 ppm (29, 30). However, hydrogen bonding involving these sites can result in significant changes for the isotropic chemical shifts, reflecting the rich chemistry with which histidine residues are associated in proteins (e.g., refs. 31 and 32).
Strong, low-barrier hydrogen bonds (33, 34) have been described in which the heteroatoms sharing a hydrogen are separated by an unusually short distance and the pKas of the two heteroatoms are very similar. In other words, the heteroatom affinity for the proton is similar for each heteroatom. In addition, low-barrier hydrogen bonds are sequestered from protic solvents that could compete for the hydrogen bond (35, 36). Histidine residues have been associated with numerous low-barrier hydrogen bonds, such as in serine proteases (31, 37, 38), triosephosphate isomerase (39), and citrate synthase (35). Histidine pairs are observed in a number of proteins, such as myoglobin (40). In ribonuclease, a strong hydrogen bond between two histidine residues has been described (41). Such interactions have been studied by using imidazole and imidazolium, which form cocrystals displaying a strong hydrogen bond (42), and modeling them computationally (43). In addition, this imidazole–imidazolium sample and other complexes (44) have been extensively studied by solid-state NMR (45, 46). The presence of a low-barrier hydrogen bond is characterized by an increase in the frequency of the protonated 15N resonance, a decrease in the nonprotonated resonant frequency, and broadening of the resonances. Such characterizations are associated with the lengthening of the covalent bond, shortening of the hydrogen bond, and chemical exchange, respectively, as the hydrogen jumps across the low potential barrier.
Results
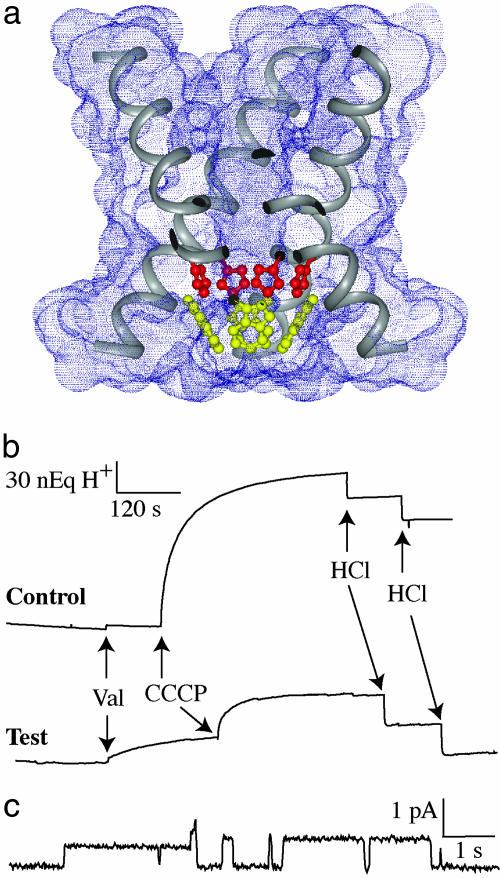
Fig. 1a displays the TM domain structure of M2 protein (Protein Data Bank ID code 1NYJ) characterized by solid-state NMR in planar lipid bilayers. Highlighted in the figure are the four histidine (red, His-37) and four tryptophan (yellow, Trp-41) residues in the pore of this H+ channel. Extravesicular pH perturbation studies (Fig. 1b) show that our preparation of the M2 TM domain also forms H+ selective channels in small unilamellar vesicles at neutral pH. Here, the TM domain channels are present at the outset but do not transport protons into the vesicles until an electrochemical driving force is created by the addition of valinomycin, specifically allowing K+ to flow out of the vesicles down its (100-fold) concentration gradient. In response to the electrochemical potential, H+ flows into the vesicles, acidifying their interior. These recordings show that a significant fraction of the peptide molecules form H+ selective channels at neutral pH. The addition of the H+ ionophore, carbonyl cyanide m-chlorophenylhydrazone, to the solution shows that not all of the vesicles had M2 TM domain channels. In addition, single-channel conductance of the M2 TM domain preparations is demonstrated in POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine)/POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) planar bilayers under acidic conditions (Fig. 1c).
Fig. 1.
Proton conduction of the transmembrane domain of M2 protein. (a) Tetrameric structure of the M2 TM domain (Protein Data Bank entry 1NYJ) corresponding to residues Ser-22–Leu-46. In this cross-sectional view, portions of the four TM helices are illustrated, and the four His-37 and Trp-41 residues are highlighted in red and yellow, respectively. (b) Multichannel electrophysiology experiments of M2 TM domain loaded vesicles. Valinomycin produces a voltage gradient that drives H+ uptake by the vesicles. Carbonyl cyanide m-chlorophenylhydrazone is added to activate those vesicles that have no M2 TM domain channel activity. (c) Single-channel currents from M2 TM domain (0.3 mg/ml) delivered in mixed-lipid liposomes (5.7 mg/ml) in phosphate buffer (pH 7) to a lipid bilayer bathed in 50 mM glycine (pH 2.3) at 100 mV and 23°C. Further details of the conductance studies are presented in the Supporting Methods.
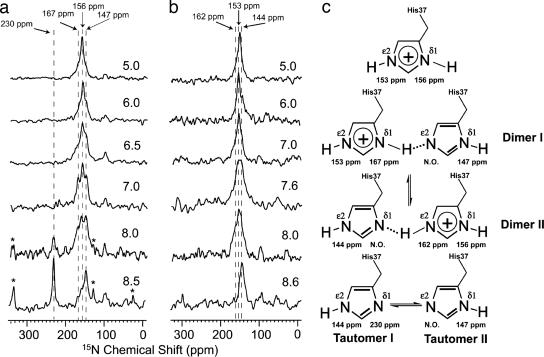
Isotropic solid-state NMR spectra of single-site 15Nδ1 and single-site 15Nε2 His-37-labeled M2 TM domain in gel state liposomes are shown in Fig. 2 as a function of pH. The 230-ppm resonance is a definitive marker for the neutral electronic states of histidine, which may be protonated at either δ1 (tautomer II, 147 ppm) or ε2 (tautomer I, 144 ppm). When the histidine is charged, both 15N sites are protonated, generating resonances at 156 ppm for δ1 and 153 ppm for ε2. In addition, two signals from a protonated state (167 ppm for δ1 and 162 ppm for ε2) are observed with linewidths nearly double those of the other protonated resonances.
Fig. 2.
Thirty-megahertz 15N cross-polarized magic-angle spinning NMR spectra of the M2 TM domain in fully hydrated 1,2-dimyristoyl-sn-3-phosphocholine/sodium 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (4:1 molar ratio) liposomes at 4°C as a function of pH. (a) 15Nδ1 His-37-labeled domain. (b) 15Nε2 His-37-labeled domain. Chemical shifts are relative to a saturated solution of 15NH4NO3 at 0 ppm. The weak signal at ≈98 ppm is due to natural abundance signals from the protein backbone. The spinning side bands are marked with an asterisk. (c) Chemical states of the histidine tetrad in the M2 TM domain and associated 15N resonance frequencies. The resonances that were not observable are labeled “N.O.”
These resonances show the evolution of histidine chemical states as the sample pH is varied. At pH 5.0, the histidines are primarily charged, and at pH 8.6, they are primarily neutral. As the spectra are viewed from high to low pH, the nonprotonated signal (230 ppm) completely disappears at a surprisingly high pH, well above the typical pKa for histidine side chains (pH 6.5). To quantitatively interpret the data, the signal intensities were corrected for the different cross-polarization kinetics of the protonated and nonprotonated signals (47) based on data obtained from the M2 TM domain as a function of spin-lock and cross-polarization time (data not shown). In addition, because the anisotropy of the nonprotonated 15N resonance is much larger than that for the protonated 15N site, the resonance intensity includes both the central (isotropic) transition at 230 ppm and the intensity of the spinning side bands marked with an asterisk in Fig. 2. The reduced intensity for the central transition due to its distribution over the spinning side bands prevents resonance observation when the fractional occupancy for the nonprotonated site is <20%. Furthermore, the spectral analysis is complicated by the broad resonance linewidths that are likely due to dynamics of the histidine side chain. Interestingly, these 15N linewidths are a factor of two narrower when the samples are observed in the presence of the antiviral drug amantadine (data not shown). Because of the broad overlapping resonances, deconvolution was used to estimate the resonance intensities in all of the data sets, resulting in a consistent set of linewidths for each resonance in the deconvolution analysis (see Supporting Methods, which is published as supporting information on the PNAS web site) as a function of pH.
The signals at 167 and 162 ppm (Fig. 3a and b) for δ1 and ε2 represent an 11- and 9-ppm shift, respectively, in the isotropic frequencies relative to the typical, charged imidazole state. Such resonance frequencies have previously been characterized as reflecting the presence of a strong hydrogen bond in imidazole–imidazolium cocrystals (46). Furthermore, the linewidths from the M2 TM domain resonances, based on the deconvolution results (48) for the entire pH titration, are nearly twice as broad as the resonances for the same sites when they are not involved in a strong hydrogen bond. Such exchange broadening clearly suggests that the proton is jumping across a low potential barrier and is therefore a low-barrier hydrogen bond. Unfortunately, the combination of relatively low fractional occupancy, a distribution of intensity in the spinning side bands, and increased linewidths has resulted in no observation of the nonprotonated site in the low-barrier hydrogen bond. Despite this, the resonances at 167 and 162 ppm dominate the middle of the titration along with the other resonances associated with the histidine dimers (Fig. 3c). As in the imidazole–imidazolium model system, it is reasonable to model these low-barrier hydrogen bonds in the M2 TM domain as being between the histidine side chains that are known to be oriented toward the pore (5, 20, 21) of this symmetric (at high pH) tetramer. Furthermore, because signals for both δ1 and ε2 sites involved in strong hydrogen bonds are observed, these bonds are formed by a bridge between δ1 and ε2 sites (Fig. 2c), as opposed to a bridge between two δ1 or two ε2 sites. In fact, the population distribution between the two Nδ1-H—Nε2 and Nδ1—H-Nε2 dimers is nearly equal, thereby optimizing charge delocalization and characterizing this potential double well as being nearly symmetric.
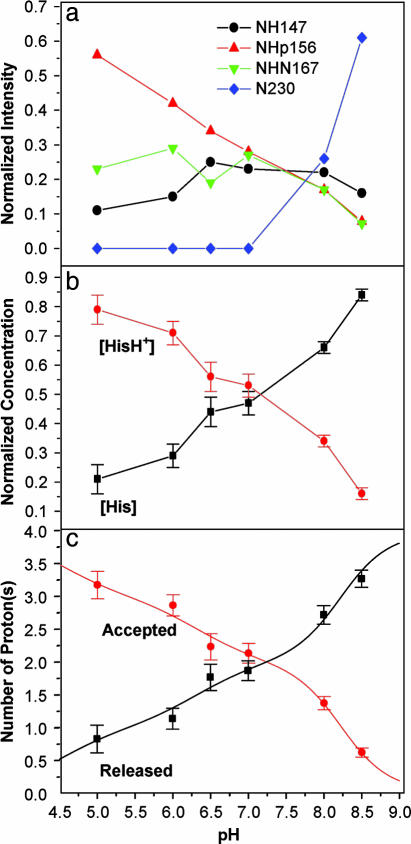
Fig. 3.
Analysis from spectral deconvolution of the pH titration data for the 15Nδ1 His-37-labeled M2 TM domain after corrections for cross-polarization kinetics. (a) Resonance intensities as a function of pH. Nonprotonated nitrogens were not observable unless their mole fraction was >20%. (b) Mole fractions were summed to present an overview of the charged [HisH+] vs. uncharged [His] imidazole rings. Error bars were calculated from the deconvolution analysis. (c) Titration curves were generated from equations presented in Supporting Methods that give protonation and deprotonation curves for the four histidines from the [His]/[HisH+] results.
The four imidazoles in the M2 channel are clustered closely together, so they cannot be considered as independently titrating groups. Four equivalent sites can accept the first H+, implying that all four histidines have the same affinity, and hence pKa values, for the first H+. The result of binding the first H+ is the formation of an imidazole–imidazolium dimer bridged by a strong hydrogen bond. In Fig. 3a, where the analysis of the δ1 site is presented, the formation of a dimer is apparent, because the 156-ppm resonance (charged dimer or monomer state) and the 167-ppm resonance (a uniquely dimer state) increase simultaneously with decreasing pH. The second H+ also forms a dimer, and at pH 7.3, >90% of the His-37 side chains are in dimeric form (Fig. 3a). At lower pH, a third H+ binds, leaving a single neutral imidazole at one of four equivalent sites. For this third H+ to be absorbed by the histidine cluster, a dimer must be disrupted, resulting in a potentially metastable state with positive charges in side-by-side residues, less charge delocalization, and broken twofold symmetry.
The intensities shown in Fig. 3a can be converted to a relative distribution of charged and uncharged histidine side chains, as shown in Fig. 3b, with error bars from the deconvolution analysis. This curve is consistent with pKas: 8.2 ± 0.2, 8.2 ± 0.2, 6.3 ± 0.3, and <5.0 (Fig. 3c). The two high pKas conclusively demonstrate that the M2 H+ channel has a very high affinity for the first two H+s. Despite the low dielectric in the TM environment, a [H+] nearly two orders of magnitude lower than that necessary to protonate histidine in bulk aqueous solution is adequate for inducing this histidine tetrad to absorb two protons.
Discussion
The first two charges added to the histidine tetrad are expected to form a repulsive interaction even though there is substantial charge delocalization, because their separation is shown to be <10 Å (5) and the effective dielectric coefficient in the histidine environs is certainly less than that in a bulk aqueous environment (49, 50). For the second H+ to bind with the same affinity as the first, the repulsive interaction must be countered with a substantially favorable interaction. The formation of a strong hydrogen-bonded imidazole dimer breaks the fourfold symmetry and may result in substantial structural distortion. Potentially, considerable stabilization energy is generated when twofold symmetry is restored through the formation of a second imidazole dimer, suggesting cooperative H+ binding. In other words, the M2 H+ channel displays not only a high affinity for H+ but also potentially cooperative binding of H+ with this high affinity.
Charge delocalization, minimizing charge repulsion, and potentially structural rearrangements of the helical bundle contribute to the energetics that are essential for achieving a construct with high affinity for H+. Two significant studies have previously characterized the His-37 pKas. For a peptide monomer in a detergent micelle (27), it is likely that the imidazole is exposed to the micellar surface or interfacial region, and, hence, a pKa characterization (6.8) typical of aqueous exposed histidine residues is not surprising. More interestingly, the UV resonance Raman study (9, 25) attempted to characterize the His-37 pKas in the M2 TM domain tetrameric state. Unfortunately, the only imidazole band characteristic of a chemical state that was observable is that of the charged state (presumably the monomeric charged state), which does not occur until below pH 7 (Fig. 3a), consistent with the UV resonance Raman imidazole data. The Raman experiments were not able to observe the neutral states and apparently not able to observe the imidazole–imidazolium dimeric states for which the energetics and vibrational frequencies have been calculated (43). In addition, both Raman (25) and fluorescence (8) studies have suggested that a pKa near 6 can be indirectly observed for Trp-41. This pKa again appears to reflect on the formation of a net positive charge on a single histidine: that is, the absorption of the third H+ by the histidine tetrad.
Although we are able to observe numerous resonances reflecting different chemical states for histidine in the histidine tetrad, we were not able to observe the nonprotonated nitrogens of the imidazole–imidazolium dimeric state. However, the missing intensity is dictated by the resonances that we were able to observe. The complete loss of the nonprotonated intensity by pH 7.0 would otherwise suggest that all four pKas are above pH 7.5, yet it is clear that the resonance intensity for the protonated and charged state continues to build below pH 6.5. This conflict is eliminated by recognizing that the broad protonated resonance (162 and 167 ppm) must be partnered with a broad nonprotonated resonance, as in model systems (46) that will still retain a large chemical shift anisotropy and poor cross-polarization kinetics, all of which work to suppress the observation of this signal. Fortunately, the observation of the resonances from both the ε2 and δ1 sites generate redundancy, so observation of the nonprotonated resonances is not essential for understanding the titration kinetics of the histidine tetrad.
Broad lines associated with a strong hydrogen bond indicate that a low-barrier hydrogen bond exists. The broadening is due to chemical exchange from hydrogen crossing the barrier at a rate that is less than the chemical shift difference between the two resonances, or ≈2 kHz. It has been argued by Cleland and Kreevoy (35) that low-barrier hydrogen bonds require, in addition to a short heteroatom distance, a nonprotic solvent environment, because protic solvents would compete for the hydrogen bond. The histidine tetrad exists in an aqueous pore and, although the polarizability of water in such a confined space will be greatly reduced (49, 50), water is still present. Pinto and coworkers (26) have shown that Trp-41 prevents access of Cu2+ to the histidines from the C-terminal side and that Trp-41 residues form a conductance gate. Potentially, these indoles, as illustrated in the solid-state NMR backbone structure of the M2 TM domain, also prevent aqueous access to the histidine tetrad from the C-terminal side.
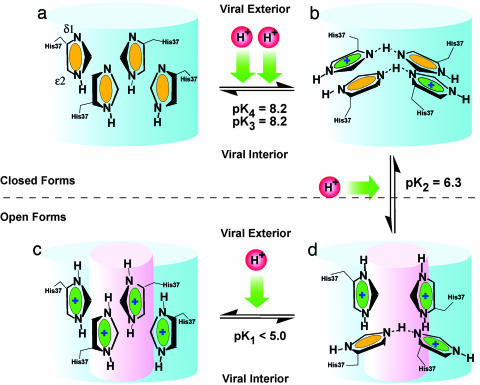
The M2 channel becomes pH-activated below pH 7 (7, 12, 13, 23, 51). The dimer-of-dimer state peaks in concentration at pH 7.3 (Fig. 3b) when [HisH+] equals [His]. We refer to this state as the “histidine-locked” state that occludes the pore. It has long been suspected that a titratable histidine is responsible for the acid gating of the channel. Now it is clear that this gating is associated with the titration of the third histidine. Disrupting one of the dimer structures appears to result in a high-energy metastable state in which the system is willing to give up a H+ and return to the paired dimer state. This hypothesis (Fig. 4) is supported by the fact that the second and third pKas are separated by nearly 2 pH units, suggesting how difficult it is to disrupt the stable histidine-locked conformation. Much is yet to be understood about this proton conductance mechanism, but the central role of the histidine tetrad is beginning to unfold, displaying unique chemistry.
Fig. 4.
A model illustrating the opening of the M2 proton channel. Only the four-histidine portion of the channel lumen is presented (as a blue cylinder). Four stages of protonation are illustrated: all four histidines are neutral (a), two charges (b), three charges (c), and fully protonated histidine tetrad (d). Neutral and positively charged histidine residues are differentiated in green and yellow, respectively. The open state is illustrated by a small pink cylinder in the middle of the blue cylinder. Dashed lines in histidine dimers represent strong hydrogen bonds, but no effort has been made here to display the actual geometry of the histidines in the histidine tetrad as a function of pH.
Materials and Methods
15Nδ1 histidine and 15Nε2 histidine were purchased from Cambridge Isotope Laboratories (Cambridge, MA) and chemically protected by a trityl (trt) and 9-fluorenylmethoxycarbonyl (Fmoc) groups following procedures described in refs. 52 and 53. The total yield of Fmoc-His-(trt)-OH was 73–81%. M2 TM domain (NH2-Ser-22-Ser-Asp-Pro-Leu-Val-Val-Ala-Ala-30-Ser-Ile-Ile-Gly-Ile-Leu-(15Nδ1 or 15Nε2)His-37-Leu-Ile-Leu-40-Trp-Ile-Leu-Asp-Arg-Leu-46-COOH) was chemically synthesized on an Applied Biosystems 430A peptide synthesizer. The peptides were purified and characterized as described in ref. 54.
15N His-37-labeled M2 TM domain was incorporated into liposomes through a detergent removal technique (55, 56). First, 10 mg of M2 TM domain, 40 mg of 1,2-dimyristoyl-sn-3-phosphocholine, 10 mg of sodium 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol, and 320 mg of 1-O-octyl-β-d-glucopyranoside were codissolved in 20 ml of 2,2,2-trifluoroethanol and chloroform (vol/vol: 4/1). The organic solvent was evaporated by using a rotary flask; then the mixture was placed under high vacuum for at least 6 h for further removal of organic solvent. Fifteen milliliters of 10 mM citrate–borate–phosphate buffer of a specific pH with 1 mM EDTA was added to the dried mixture. A clear solution was prepared after the flask was vortexed in a shaker for 20 min. This solution was dialyzed by using 3-kDa molecular-mass cutoff dialysis tubing at 4°C against the buffer. The buffer was changed at least five times in 5 days to ensure virtually complete removal of the detergent. The vesicle suspension loaded with M2 TM domain was pelleted in 2.5 h by ultracentrifugation at 196,000 × g. The pellet was packed into a 7-mm zirconia spinner (Bruker, Billerica, MA) with a sealing cap. The pH was measured from the supernatant.
Details of the electrophysiology experiments and the data analysis, including deconvolution and the equations that lead to the titration curves, are presented in the Supporting Methods.
All cross-polarized magic-angle spinning (CPMAS) NMR experiments were conducted on a Bruker DMX-300 NMR spectrometer by using a 7-mm rotor (3-kHz spinning rate) triple resonance probe. The 15N CPMAS spectra were recorded by using a 7-μs, 90° pulse width, a 2-ms mixing time, and a 5-s recycle delay throughout the series of experiments observed at 277 K and different pH values. Approximately 10,000 transients were acquired, and 100 Hz of exponential line broadening was applied in the data processing.
Supplementary Material
Acknowledgments
We thank the staff of the National High Magnetic Field Laboratory (NHMFL) (especially A. Blue), the staff of the Bioanalytical Synthesis and Services Laboratory (Department of Chemistry and Biochemistry, Florida State University) (H. Hendricks and U. Goli), and Rochelle Davison and Brad Rodgers for technical assistance. This work was supported by National Institute of Arthritis and Infectious Disease Grant AI 23007. NMR experiments were conducted at the NHMFL, which is supported by Cooperative Agreement DMR-0084173 and the State of Florida.
Abbreviation
- TM
transmembrane.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Sugrue R. J., Bahadur G., Zaambon M. C., Hall S. M., Douglas A. R., Hay A. J. EMBO J. 1990;9:3469–3476. doi: 10.1002/j.1460-2075.1990.tb07555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugrue R. J., Hay A. J. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakaguchi T., Tu Q., Pinto L. H., Lamb R. A. Proc. Natl. Acad. Sci. USA. 1997;94:5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochendoerfer G. G., Salom D., Lear J. D., Wilk-Orescan R., Kent S. B., DeGrado W. F. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura K., Kim S., Zhang L., Cross T. A. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- 6.Kelly M. L., James J. A., Brown-Augsburger P., Heinz B. A., Smith M. C., Pinto L. H. FEBS Lett. 2003;552:61–67. doi: 10.1016/s0014-5793(03)00851-2. [DOI] [PubMed] [Google Scholar]
- 7.Chizhmakov I. V., Ogden D. C., Geraghty F. M., Hayhurst A., Skinner A., Betakova T., Hay A. J. J. Physiol. (London) 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czabotar P. E., Martin S. R., Hay A. J. Virus Res. 2004;99:57–61. doi: 10.1016/j.virusres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi K., Lamb R. A. J. Virol. 1994;68:911–919. doi: 10.1128/jvi.68.2.911-919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lear J. D. FEBS Lett. 2003;552:17–22. doi: 10.1016/s0014-5793(03)00778-6. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y., Voth G. A. FEBS Lett. 2003;552:23–27. doi: 10.1016/s0014-5793(03)00779-8. [DOI] [PubMed] [Google Scholar]
- 12.Pinto L. H., Holsinger L. J., Lamb R. A. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 13.Chizhmakov I. V., Geraghty F. M., Ogden D. C., Hayhurst A., Antoniou M., Hay A. J. J. Physiol. (London) 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panayotov P. P., Schlesinger R. W. Virology. 1992;186:352–355. doi: 10.1016/0042-6822(92)90096-8. [DOI] [PubMed] [Google Scholar]
- 15.Wang C., Lamb R. A., Pinto L. H. Virology. 1994;205:133–140. doi: 10.1006/viro.1994.1628. [DOI] [PubMed] [Google Scholar]
- 16.Tosteson M. T., Pinto L. H., Holsinger L. J., Lamb R. A. J. Membr. Biol. 1994;142:117–126. doi: 10.1007/BF00233389. [DOI] [PubMed] [Google Scholar]
- 17.Lin T., Schroeder C. J. Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vijayvergiya V., Wilson R., Chorak A., Gao P. F., Cross T. A., Busath D. D. Biophys. J. 2004;87:1697–1704. doi: 10.1529/biophysj.104.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duff K. C., Ashley R. H. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 20.Pinto L. H., Dieckmann G. R., Gandhi C. S., Papworth C. G., Braman J., Shaughnessy M. A., Lear J. D., Lamb R. A., DeGrado W. F. Proc. Natl. Acad. Sci. USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer C. M., Pinto L. H., Cross T. A., Lamb R. A. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- 22.Ito T., Gorman O. T., Kawaoka K., Bean W. J., Webster R. G. J. Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C., Lamb R. A., Pinto L. H. Biophys. J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatamaran P., Lamb R. A., Pinto L. H. J. Biol. Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 25.Okada A., Miura T., Takeuchi H. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 26.Tang Y., Zaitseva F., Lamb R. A., Pinto L. H. J. Biol. Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 27.Salom D., Hill B. R., Lear J. D., DeGrado W. F. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blomberg F., Maurer W., Rüterjans H. J. Am. Chem. Soc. 1977;99:8149–8159. doi: 10.1021/ja00467a005. [DOI] [PubMed] [Google Scholar]
- 29.Munowitz M., Bachovchin W. W., Herzfeld J., Dobson C. M., Griffin R. G. J. Am. Chem. Soc. 1982;104:1192–1196. [Google Scholar]
- 30.Huang T.-H., Bachovchin W. W., Griffin R. G., Dobson C. M. Biochemistry. 1984;23:5933–5937. doi: 10.1021/bi00320a007. [DOI] [PubMed] [Google Scholar]
- 31.Smith S. O., Farr-Jones S., Griffin R. G., Bachovchin W. W. Science. 1989;244:961–964. doi: 10.1126/science.2499045. [DOI] [PubMed] [Google Scholar]
- 32.Ash E. L., Sudmeier J. L., DeFabo E. C., Bachovchin W. W. Science. 1997;278:1128–1132. doi: 10.1126/science.278.5340.1128. [DOI] [PubMed] [Google Scholar]
- 33.Cleland W. W. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]
- 34.Kreevoy M. M., Liang T.-M., Chang K.-C. J. Am. Chem. Soc. 1977;99:5207–5209. [Google Scholar]
- 35.Cleland W. W., Kreevoy M. M. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 36.Albrecht G., Zundel G. Z. Naturforsch. 1984;39a:986–992. [Google Scholar]
- 37.Frey P. A., Whitt S. A., Tobin J. B. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 38.Bachovchin W. W. Biochemistry. 1986;25:7751–7759. doi: 10.1021/bi00371a070. [DOI] [PubMed] [Google Scholar]
- 39.Lodi P. J., Knowles J. R. Biochemistry. 1991;30:6948–6956. doi: 10.1021/bi00242a020. [DOI] [PubMed] [Google Scholar]
- 40.Hennig M., Geierstanger B. H. J. Am. Chem. Soc. 1999;121:5123–5126. [Google Scholar]
- 41.Golubev N. S., Denisov G. S., Gindin V. A., Ligay S. S., Limbach H.-H., Smirnov S. N. J. Mol. Struct. 1994;322:83–91. [Google Scholar]
- 42.Quick A., Williams D. J. Can. J. Chem. 1976;54:2465–2469. [Google Scholar]
- 43.Tatara W., Wojcik M. J., Lindgren J., Probst M. J. Phys. Chem. A. 2003;107:7827–7831. [Google Scholar]
- 44.Krause J. A., Baures P. W., Eggleston D. S. Acta Crystallogr. B. 1991;47:506–511. doi: 10.1107/s0108768191000915. [DOI] [PubMed] [Google Scholar]
- 45.Song X.-J., Rienstra C. M., McDermott A. E. Magn. Reson. Chem. 2001;39:S30–S36. [Google Scholar]
- 46.Song X. J., McDermott A. E. Magn. Reson. Chem. 2001;39:S37–S43. [Google Scholar]
- 47.Fu R., Hu J., Cross T. A. J. Magn. Reson. 2004;168:8–17. doi: 10.1016/j.jmr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Hu J. Ph.D. dissertation. Tallahassee: Florida State Univ.; 2005. [Google Scholar]
- 49.Partenskii M. B., Jordan P. C. J. Phys. Chem. 1992;96:3906–3910. [Google Scholar]
- 50.Partenskii M. B., Jordan P. C. Q. Rev. Biophys. 1992;25:477–510. doi: 10.1017/s0033583500004388. [DOI] [PubMed] [Google Scholar]
- 51.Mould J. A., Drury J. E., Frings S. M., Kaupp U. B., Pekosz A., Lamb R. A., Pinto L. H. J. Biol. Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- 52.Barlos K., Papaioannou D., Theodoropoulos D. J. Org. Chem. 1982;47:1324–1326. [Google Scholar]
- 53.Chang C. D., Waki M., Ahmad M., Meiennhofer J., Lundell E. O., Haug J. D. Int. J. Pept. Protein Res. 1980;15:59–66. doi: 10.1111/j.1399-3011.1980.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 54.Kovacs F., Cross T. A. Biophys. J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mimms L. T., Zampighi G., Nozaki Y., Tanford C., Reynolds J. A. Biochemistry. 1981;20:833–840. doi: 10.1021/bi00507a028. [DOI] [PubMed] [Google Scholar]
- 56.Sanders C. R., Oxenoid K. Biochim. Biophys. Acta. 2000;1508:129–145. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.