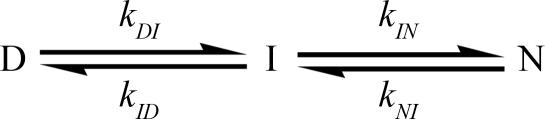Abstract
The folding kinetics of bovine acyl-CoA binding protein was studied by 15N relaxation dispersion measurements under equilibrium conditions. Relaxation dispersion profiles were measured at several concentrations of guanidine hydrochloride (GuHCl). The unfolding rate constant (ku) was determined under conditions favoring folding, for which the folding rate constant (kf) dominates the relaxation in stopped-flow kinetic measurements. Conversely, kf was determined under conditions favoring unfolding, for which ku dominates stopped-flow data. The rates determined by NMR therefore complement those from stopped-flow kinetics and define an “inverted chevron” plot. The combination of NMR relaxation and stopped-flow kinetic measurements allowed determination of kf and ku in the range from 0.48 M GuHCl to 1.28 M GuHCl. Individually, the stopped-flow and NMR data fit two-state models for folding. However, although the values of kf determined by the two methods agree, the values of ku do not. As a result, a combined analysis of all data does not comply with a two-state model but indicates that an unfolding intermediate exists on the native side of the dominant energy barrier. The denaturant and temperature dependencies of the chemical shifts and ku indicate that the intermediate state is structurally similar to the native state. Equilibrium unfolding monitored by optical spectroscopy corroborate these conclusions. The temperature dependence of the chemical shifts identifies regions of the protein that are selectively destabilized in the intermediate. These results illustrate the power of combining stopped-flow kinetics and NMR spectroscopy to analyze protein folding.
Keywords: kinetic analysis, NMR relaxation dispersion, protein folding, stopped flow
A large number of cellular processes involve complete or partial unfolding of proteins from their compact native states. In normal cells, protein unfolding occurs in gene regulation, trafficking, protein degradation, and quality control (1, 2). Disease-related aggregation of proteins also involves partial unfolding and subsequent oligomerization (3, 4). In many of the latter cases, it is believed that the cytotoxic species is an intermediate state that exists before aggregation or fibril formation (5, 6). A full understanding of these processes requires detailed knowledge of the protein folding mechanisms. Critical insights have been obtained from the study of small proteins, which often display apparent two-state kinetics. However, detailed analyses have shown that folding intermediates also exist for proteins following apparent two-state kinetics (7). The balance between two- and three-state folding is delicate and may be shifted by changes in the folding conditions or by mutations (8–10).
To study the folding process, it is necessary to populate the unfolded state, commonly achieved by the addition of denaturants, such as guanidine hydrochloride (GuHCl) or urea, which results in preferential stabilization of expanded conformations that expose hydrophobic surfaces (11). Traditionally, folding kinetics has been studied by monitoring the relaxation toward equilibrium after rapid perturbation of the system. For example, the response to a rapid change in denaturant concentration can be monitored by using stopped-flow methods. In this fashion, folding (unfolding) rates can be measured sensitively only at denaturant concentrations below (above) the midpoint of the folding transition, where relaxation to the new equilibrium is dominated by the folding (unfolding) rate constant. Hence, long extrapolations are usually necessary to extract the unfolding rates under physiological conditions at zero denaturant concentration. Protein unfolding under conditions favoring the native folded state has been studied previously by indirect techniques, such as proteolytic degradation (12), hydrogen exchange (13, 14), and site-directed chemical labeling (15). However, it may not always be straightforward to implement these techniques or to analyze the results.
Recent advances in spin relaxation dispersion methods have set the scene for detailed studies of conformational exchange processes on the microsecond to millisecond time-scales (16–19). One of the great advantages of these techniques is the ability to measure conformational exchange between states with heavily skewed populations. Indeed, minor species with a population of only 1% can be detected, which makes spin relaxation dispersion ideally suited for the study of transient intermediates in protein folding and for studying protein unfolding under native-like conditions (16, 19).
The folding process of the 86-residue acyl-CoA binding protein (ACBP) has been investigated extensively by a range of biophysical techniques. The unfolded state has been studied thoroughly by various NMR experiments, demonstrating that it is far from random-coil like and that it becomes increasingly native-like under less denaturing conditions (20–22). Although the folding process of ACBP appears to follow two-state behavior (23), an early folding intermediate with native-like distances between the C terminus and helix 3 but with few native contacts has been observed by fluorescence resonance energy transfer in combination with rapid mixing (7). Hydrogen exchange pulse labeling has shown that helical structure forms early in the folding process, especially in the C-terminal helix 4 (24). The major folding transition state has been characterized in detail by φ value analyses of a large number of mutants, revealing that interactions between helices 1, 2, and 4 exist in this state (23, 25).
Here, we have applied 15N Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiments to measure unfolding under conditions favoring folding and vice versa. By comparing the rates measured by spin relaxation dispersion with those from stopped-flow experiments, we detected a native-like unfolding intermediate under near-native conditions.
Results
Spin Relaxation Dispersions.
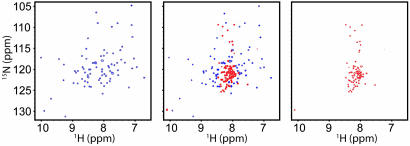
The protein folding process of ACBP was characterized by 15N relaxation dispersion experiments at eight different GuHCl concentrations from 0.48 to 1.28 M at 40°C and at three different temperatures (35°C, 40°C, and 43°C) at 0.55 M GuHCl. Under all conditions, the folding equilibrium falls in the slow exchange regime, as verified below. Near the transition midpoint, two distinct sets of peaks from the folded and unfolded states are observed that move only slightly relative to one another as the denaturant concentration is varied (Fig. 1). For each cross peak, R2eff was determined as a function of νCPMG, as exemplified in Fig. 2. At concentrations of GuHCl below 0.9 M, cross peaks are observed only from folded ACBP, allowing determination of only the rate constant for unfolding, ku. Conversely, at 1.28 M GuHCl, only the unfolded state is observed, and only the rate constant for folding, kf, can be determined. At 1 M GuHCl, cross peaks are observed from the folded and denatured states, enabling simultaneous measurement of ku and kf.
Fig. 1.
1H–15N correlated spectra from the 15N CPMG relaxation dispersion measurements of ACBP at pH 5.3 and 40°C in 0.55 M (Left), 1 M (Center), and 1.28 M (Right) GuHCl. Under these conditions, the population of unfolded protein is ≈5%, 50%, and 95%. The contour levels of the spectra are adjusted to show only the major components (i.e., the native state at 0.55 M and the unfolded state at 1.28 M).
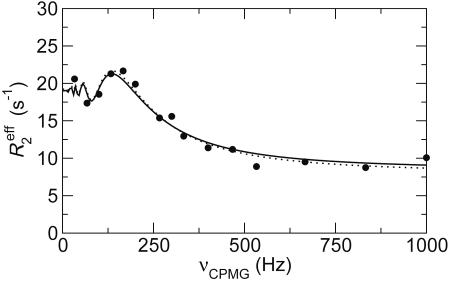
Fig. 2.
Representative 15N CPMG relaxation dispersion curve measured on the cross peaks from residue L61 in folded ACBP at pH 5.3, 1 M GuHCl, and 40°C. The dotted line represents a residue-specific fit of all parameters in Eq. 1: ka = 11.3 ± 0.7 s−1, Δω = (2.45 ± 0.09) × 103 s−1, R2 = 8.0 ± 0.5 s−1. The solid line represents a global fit of ka to all protein residues and a residue-specific fit of Δω and R2: ka = 10.55 ± 0.08 s−1, Δω = (2.44 ± 0.08) × 103 s−1, R2 = 8.4 ± 0.3 s−1.
At each denaturant concentration, the parameters of Eq. 1 (namely, ka, Δω, and R2) were fitted to the CPMG relaxation dispersion curve, employing both residue-specific and global optimizations of ka (in both cases Δω and R2 were fitted individually to each residue). The quality of the fits and the values of Δω and R2 were not significantly different between the two fitting procedures (Fig. 2); the mean differences in these parameters at 1 M GuHCl were 〈δΔω〉 = 0.0 ± 0.2 ppm and 〈δR2〉 = 0.0 ± 0.4 s−1. These results indicate that the extracted values of ka accurately report on the global folding process and that they are not significantly perturbed by any local conformational fluctuations within the folded or unfolded states. Similarly, the mean difference in ka between the local and global fits also is insignificant: 〈δka〉 = −0.1 ± 2 s−1. The larger standard deviation of δka arises because ka is not well determined in the individual fits for residues with low Δω. The large number of degrees of freedom involved in the global optimization enables sensitive and accurate determination of ka, even if Δω is small or if ka is close to the noise level of the relaxation dispersions. Global fitting was therefore applied to measure the folding and/or unfolding rate constant(s) at each set of conditions. The minimum value of Δω (218 s−1; observed for residue M70) and the maximum value of kex (99 s−1; determined by stopped flow) are obtained at 0.48 M GuHCl. These results verify that all residues are in the slow exchange regime at all denaturant concentrations and all temperatures. Furthermore, the values of Δω obtained from the relaxation dispersions agree extremely well with the chemical shift differences measured directly in the heteronuclear single quantum correlation (HSQC) spectrum, ΔωHSQC [〈Δω − ΔωHSQC〉 = −0.1 ± 0.8 ppm], again demonstrating that the method is robust and that the applied analysis is valid.
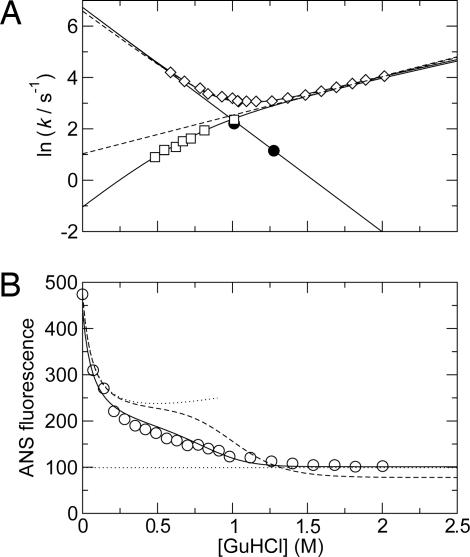
The resulting values of ka obtained from the CPMG relaxation dispersion experiments define an “inverted chevron” plot that complements the traditional chevron plot obtained by stopped-flow experiments, as shown in Fig. 3A.
Fig. 3.
Folding kinetics and equilibrium stability of ACBP as a function of [GuHCl] at pH 5.3 and 40°C. (A) Folding kinetics. Shown are the observed rates measured by stopped flow (◇) and the unfolding (□) and folding (•) rates measured by 15N CPMG relaxation dispersions. Solid lines represent fits of unfolding rates from relaxation dispersions to Eq. 3, folding rates from relaxation dispersions to Eq. 4, and the observed folding rate from stopped-flow data to Eq. 5. The dashed lines represent the best fit of the stopped-flow data to a two-state model and the unfolding rates extrapolated from the best fit of the stopped-flow data to the two-state model. See Table 1 for the optimized parameters. (B) Equilibrium stability measured by ANS fluorescence. The solid line represents the fit of the three-state model, and the dashed line represents the fit of the two-state model. The upper and lower dotted lines represent the baselines for N and D, respectively. The baseline for N was estimated as the fluorescence of ACBP/ANS in different concentrations of NaCl, and the baseline for D was estimated as the fluorescence of ANS in the absence of ACBP.
Equilibrium Unfolding and Stopped-Flow Kinetics.
For comparison with the folding rates determined from NMR relaxation dispersions, we also studied the folding process at 40°C by stopped-flow kinetics and equilibrium unfolding using various spectroscopic probes (Fig. 3; see also Fig. 5, which is published as supporting information on the PNAS web site). Initially, the rate constants (kobs) were fitted to an apparent two-state folding model. The resulting values of kf, mf, ku, and mu at 40°C are listed in Table 1. A two-state folding process was also fitted to the equilibrium unfolding data measured by Trp fluorescence, yielding ΔGeq = 14.7 ± 0.8 kJ·mol−1 and meq = 15.3 ± 0.6 kJ·mol−1, which are equal to the corresponding values obtained from the kinetic experiments. Folding kinetics was also measured at 35°C (data not shown). From the kinetic data obtained at 35°C and 40°C, together with previously published data on ACBP at 5°C and 25°C (23, 26), the Arrhenius energy of activation for ku at 0.55 M GuHCl was estimated to 225 ± 5 kJ·mol−1.
Table 1.
Observed folding kinetic parameters for ACBP
| Model | kDI | mDI | kID | mID | KNI | mNI |
|---|---|---|---|---|---|---|
| Two states | 721 ± 85 | −11.1 ± 0.5 | 2.8 ± 0.3 | 3.9 ± 0.1 | ||
| Three states | 838 ± 70 | −11.4 ± 0.2 | 4.1 ± 1.1 | 3.4 ± 0.4 | 0.10 ± 0.02 | 8.8 ± 1.4 |
Rates are given in s−1, and m values are given in kJ·mol−1·M−1.
Combined Analysis of NMR and Stopped-Flow Data.
Extrapolation of kf obtained from the two-state analysis of the stopped-flow data to the GuHCl concentrations covered by the CPMG data yields values in good agreement between the two methods (Fig. 3). In contrast, extrapolation of ku from stopped-flow data does not agree with the values obtained by NMR. The downward curvature in ka is clearly inconsistent with a two-state folding model and suggests the presence of an intermediate on the native side of the major transition state of the folding process (Fig. 3). For this reason, a linear three-state model was considered that involved the denatured state (D), the folded state (N), and an intermediate (I), with microscopic rate constants kij (Scheme 1). In Scheme 1, I is pictured as an on-path obligatory intermediate, but it should be noted that the following formalism is equally valid if I is an off-path minor state. In this model, the previously identified folding intermediate, Iu (7), and the unfolded state, U, are treated as a single denatured state, D. This simplification will scale the apparent folding rate constant kDI by the preequilibrium between U and Iu as kDI = kIu,N/(1 + KU,Iu) but will not influence the measured unfolding rate constants. Specifically, the unfolding rates determined by NMR will not be affected because only the flux from I to D is measured in the slow exchange regime. The NMR and stopped-flow data were fitted simultaneously to Eqs. 3–5 (see Methods). The excellent agreement of all kinetic data with the three-state model further indicates that the N to I transition also can be well approximated as a preequilibrium. The fitted parameters are shown in Table 1. The midpoint of the N to I transition occurs at 0.68 M GuHCl, which is well below the midpoint of the I to D transition at 0.93 M GuHCl. Therefore, the denaturant dependence of the N to I transition will have only a minor effect on the kID rate constant in the region where both kID and kDI significantly contribute to the observed rate in the stopped-flow experiments. Accordingly, the fitted values of kDI, mDI, kID and mID from the combined three-state fit are almost identical to the corresponding parameters km, mf, ku and mu from the two-state fit of the stopped-flow data. The intermediate state has a population of 10% in 0 M GuHCl and a maximal population of 47% at 0.78 M GuHCl.
Scheme 1.
For comparison, we carried out a parallel study with urea as a denaturant at 25°C (data not shown). The stopped-flow kinetic and equilibrium stability data are in agreement with a two-state model. The unfolding rates measured by NMR relaxation dispersion at low urea concentrations diverge from the values extrapolated from the stopped-flow data, just as in the case of GuHCl. These results indicate that the stabilization of I relative to N is the result of the denaturing properties of GuHCl and urea rather than of nonspecific electrostatic screening by GuHCl.
The influence of protein concentration on the measured rates was investigated by recording additional relaxation dispersions at 0.65 M GuHCl and protein concentrations of 0.25 mM and 0.75 mM. No change in the measured rate was observed, indicating that dimers or higher oligomers are not significantly populated during the unfolding process at these concentrations. These findings agree with previous NMR diffusion measurements (27).
Temperature Dependence of Unfolding Rates.
Differences in the residue-specific temperature dependencies of folding rates measured by spin relaxation dispersion have been shown to serve as a sensitive marker for the existence of a folding intermediate in the fyn Src homology 3 domain (19). In the present case, a comparison of the residue-specific unfolding rates at 35°C, 40°C, and 43°C reveals that each individual data set is not significantly different from the rest treated as a group. Similarly, the temperature dependence of the unfolding rate obtained from the global fit is not significantly different from the individual data sets treated as a group (data not shown). Thus, the temperature variation of ka measured by relaxation dispersion does not pinpoint the existence of an intermediate state in the present case, most likely because the chemical shifts of N and I are similar (see below). From the temperature dependence of ku,app obtained from the global fits, the Arrhenius energy of activation at 0.55 M GuHCl was estimated to 232.0 ± 0.4 kJ·mol−1, in excellent agreement with the stopped-flow data.
Near-UV CD and 8-Anilinonaphthalene-1-Sulfonic Acid (ANS) Binding.
To validate the three-state model, equilibrium unfolding titrations were followed by fluorescence of ANS and near-UV CD spectroscopy (Figs. 3B and 5B). ANS binds to the native state of ACBP as seen from an increase in the quantum yield and a distinct shift in the emission spectrum of ANS upon addition of ACBP (see Fig. 6, which is published as supporting information on the PNAS web site). Addition of 0.07–0.28 M GuHCl results in a large decrease in the fluorescence intensity of ANS (Fig. 6). This initial decrease in ANS fluorescence is due to quenching by Cl−, similar to what is observed for the Trp fluorescence in the native state of ACBP and several other proteins (28). The GuHCl-dependent baseline for the ANS fluorescence in the native state of ACBP was therefore determined from the ANS fluorescence at various concentrations of NaCl. Further addition of GuHCl up to a concentration of 2 M, gradually changes the ANS spectrum to become indistinguishable from the spectrum of free ANS (Fig. 6), which is independent of GuHCl concentration. Thus, the ANS fluorescence is independent of GuHCl in the D state. Assuming that the ANS fluorescence also is independent of GuHCl in the I state, the intensity of the ANS fluorescence in the I and D states were fit to the two- and three-state models, whereas ΔGND, ΔGNI, ΔGID, and the corresponding m values were kept fixed at the values calculated from Table 1. As shown in Fig. 3B, the three-state model fits the data significantly better than the two-state model (P < 0.0001, F test). The fitted fluorescence intensities for ANS in the I and D states are 101 ± 10 and 101 ± 5, respectively, which is the same as the intensity of 99 for ANS free in solution. Thus, neither I nor D binds ANS. Furthermore, it should be noted that the fluorescence from ANS bound to N is much lower than that from ANS bound to acid-unfolded ACBP (27) or from ANS bound to molten globule intermediates of other proteins (29).
The two- and three-state models also were fit to the near-UV CD data, using fixed parameters as in the case of the ANS fluorescence data and fitting only the signal intensities from N, I, and D, which were assumed to be independent of GuHCl. The F test indicates that the three-state model fits the data significantly better than the two-state model (P < 0.0001) (Fig. 5B), although the latter model appears to yield a satisfactory fit. The cumulative near-UV CD signal for the N, I, and D states are −741 ± 4, −586 ± 9, and 65 ± 3, respectively.
Chemical Shift Changes of the Folded State.
The cross-peak positions in the 1H–15N HSQC spectrum are only slightly sensitive to changes in GuHCl concentration (the 15N shifts changes by <0.4 ppm between 0 and 1 M GuHCl), and only two sets of cross peaks are observed, corresponding to states N + I and D. Furthermore, the 15N chemical shift differences measured in the HSQC spectra are very similar to those obtained from fitting Eq. 1 to the relaxation dispersions. Together, these observations suggest that I has chemical shifts similar to those of N (ΔωNI ≈ 0). Consequently, N and I are expected to be structurally similar.
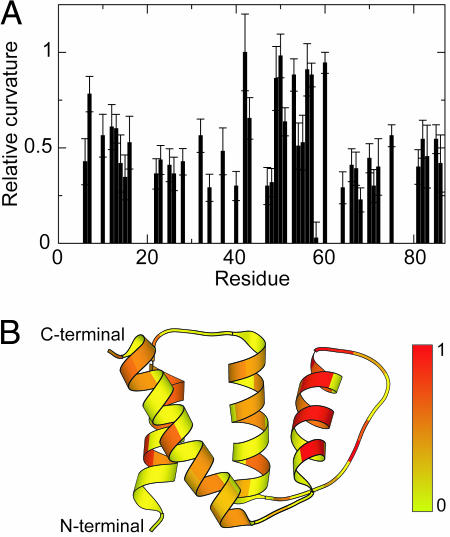
Any chemical shift differences between the states N and I should give rise to changes in the chemical shifts of the cross peaks from the folded state, as the equilibrium KNI changes with denaturant concentration. The subtle variation in chemical shift with GuHCl concentration is hard to interpret, because it depends both on changes in ionic strength and on structural differences between N and I. By contrast, 1HN chemical shifts vary linearly with temperature (30). Deviation from linearity of the temperature dependence serves as a sensitive indicator of minute changes in structural equilibria, as demonstrated for several proteins (31). The curvature depends in a complex manner on the chemical shift difference between the states, the temperature coefficients of the chemical shifts in each state, and ΔG between the exchanging states (32). In the present case of a global cooperative transition (where ΔG is the same for all residues), the magnitude of the curvature is a measure of the chemical shift difference between the two states and, hence, of their structural difference. The magnitude of the curvature can be assessed as the coefficient of the quadratic term in a second-order polynomial fit to the chemical shift changes. Fig. 4 summarizes these results for ACBP; graphs of the 1HN chemical shift changes versus temperature are shown in Fig. 7, which is published as supporting information on the PNAS web site. It is evident that the largest effects are observed on one side of helix 3 and for E42 in the overhand loop connecting helices 2 and 3. Residues in helix 1 also show curvatures above average. Upon addition of 0.2 M GuHCl, the curvatures decrease (data not shown), which is expected because increasing denaturant concentration (in the range 0–0.68 M GuHCl) acts to equalize the populations of the exchanging states N and I.
Fig. 4.
Relative curvature of the 1HN chemical shifts as a function of temperature in the absence of GuHCl. (A) Relative curvature plotted versus residue number. The curvature is normalized to the curvature of the chemical shifts for E42, which shows the largest deviation from linearity. The graph includes only those residues for which the chemical shift changes are significantly better represented by a second-order polynomial than a first-order polynomial (P < 0.05, F test). (B) The relative curvature mapped onto the structure of ACBP (Protein Data Bank ID code 1NTI). The data are color-coded from 0 (yellow) to 1 (red).
Discussion
The combined analysis of data from 15N CPMG relaxation dispersion and stopped-flow kinetic measurements reveals an intermediate in the unfolding process of ACBP. Neither of these techniques could detect the intermediate independently, but together the data are clearly inconsistent with a two-state model. We therefore propose a linear exchange pathway with an intermediate on the native side of the major energy barrier. The proposed model accounts for our observations in GuHCl and urea. From the present data we cannot rule out a triangular exchange mechanism with a direct but slow flux from N to D. The linear and triangular models can be distinguished based on the measured ku,app value only at significantly lower denaturant concentrations than those used here. However, at such low denaturant concentrations, the unfolding process is too slow to measure using the relaxation dispersion method. Therefore, we have taken the simpler linear model to represent the data.
Curvature in the unfolding arm of the inverted chevron plot also may result from a movement of the transition state for folding due to the Hammond effect (33, 34). However, if transition state movement was the source of the curvature in the unfolding kinetics, then a corresponding curvature should also be observed in the folding kinetics, which is not the case here, as concluded from the following arguments. The present experimental data for the folding arm of the chevron plot do not extend far into the linear region, which might mask some curvature in the folding kinetics. However, no curvature is observed in the chevron plots for ACBP obtained at 5°C, 25°C, and 35°C, where a more extensive part of the folding limb is sampled. Furthermore, virtually the same m values for folding are obtained at all these temperatures (mf covering the range 9–11 kJ·mol−1 between 5°C and 40°C), which confirms the linear denaturant dependence of the folding kinetics and rules out transition state movement as a source for the curvature in the inverted chevron plots.
Curvature in the unfolding kinetics could also result from a change in the relative height of two barriers surrounding a high-energy intermediate (9, 33). However, the minor changes in NMR peak positions upon addition of denaturant show that N and I must be highly similar in structure, which refutes the notion that N and I may have very different free energies. This conclusion is further emphasized by the nearly identical temperature dependencies of the unfolding rates measured by NMR and stopped-flow kinetics. Thus, KNI appears to be only slightly temperature-dependent, supporting the conclusion that the N and I states are similar in free energy and structure.
Lastly, curvature in the unfolding kinetics may result if the linear free energy relationship (Eq. 3) breaks down. The equilibrium stability that we measured in urea was 25 ± 1 kJ·mol−1 at 25°C, which is the same as the previously reported stability (25.2 ± 0.2 kJ·mol−1) in GuHCl at 25°C (22). Therefore, the linear extrapolation method appears to be valid for the current analysis.
The intermediate identified here shows that ACBP is more stable in water than what would be expected from standard measurements of equilibrium unfolding induced by GuHCl or urea. Importantly, the three-state model thus reconciles the apparent discrepancy between previous observations that the global stability measured by hydrogen exchange experiments at 25°C is ≈5.5 kJ·mol−1 higher than the stability obtained from equilibrium unfolding experiments (35). The fitted value of KIN obtained in the current study corresponds to ΔGNI = 5.7 kJ·mol−1 at 25°C, in excellent agreement with the hydrogen exchange results. Thus, the hydrogen exchange data strongly support our present conclusions. Although the folding equilibrium is well determined by hydrogen exchange, the kinetics is less reliable. Previous estimates of folding kinetics from hydrogen exchange experiments yield much lower rates than those found both in this and other studies of ACBP folding kinetics (35). These differences are most likely due to difficulties in measuring hydrogen exchange rates in the true EX1 limit.
We have identified a native-like intermediate in the folding process of ACBP. The three-state model is based on kinetic evidence, agrees with previous stability data from hydrogen exchange experiments, and further provides a significantly improved fit to the equilibrium stability curve measured by ANS binding but not by Trp fluorescence. Clearly, the difference in structure between N and I is subtle, as gauged from the chemical shifts and other spectroscopic probes. The temperature dependence of the 1HN chemical shifts suggests that the structural changes between N and I occur primarily in helix 3 and to some extent in helix 1, which is consistent both with the previous observation that the hydrogen bonds in helices 1 and 3 generally are less stable than those in the rest of ACBP (35) and with the 20% decrease in the near-UV CD signal of I with respect to N, which is mainly due to the two Trp residues in helix 3. Furthermore, mutations of residues in helix 3 do not substantially change the kinetics of the folding process (23), suggesting that this helix is formed late in the folding process. Conversely, the interaction between helix 3 and the remainder of the protein is likely perturbed early in the unfolding process, in agreement with the present results. Lastly, ACBP apparently binds ANS only in the N state, suggesting that the binding site for ANS is disrupted in I, and one can speculate that this involves helix 3 detaching from the remaining core. In addition, the lack of ANS binding to I indicates that this state is not molten globule-like.
The m value reflects the sum of changes in excluded volume and in interactions between the protein and denaturant, both of which are roughly proportional to the accessible surface area (ASA) (11). The m values in Table 1 indicate that the difference in ASA between N and I is 37% of the total difference between N and D [calculated as mNI/(mNI + mID + mDI)]. This difference may seem large in light of the minor chemical shift differences between N and I. However, the 15N chemical shift is mainly determined by the backbone conformation (36). Thus, if N and I do not differ substantially in secondary structure but mainly in the packing of helix 3 against the remaining core, then the two states can have similar 15N chemical shifts, although they differ in ASA. Detachment of the region that experiences the largest structural change between N and I (namely, residues 40–60) from the rest of the core would account for ≈15% of the difference in ASA between the folded state and an extended chain. However, the unfolded state of ACBP has a high degree of residual structure (21), implying that the relative change in ASA should be larger than that calculated by modeling D as an extended chain. Furthermore, if the I state exposes sites that interact more favorably with the denaturant than do either N or D, then the observed m value will be larger than expected from the change in ASA alone (11). The proposed structural difference between N and I could therefore lead to the observed value of mNI.
Including the present results, both native-like and unfolded-like intermediates have been observed in the folding process of ACBP. It should be noted that none of the intermediates were identified by traditional stopped-flow experiments but required the use of techniques that enable detection of transiently and weakly populated species.
The present report demonstrates the power of combining stopped-flow kinetic measurements with NMR relaxation dispersion methods for probing the folding and unfolding processes under the full range of conditions favoring the folded and unfolded states. The prospect of detecting transient intermediates on the native side of the folding barrier is directly relevant for probing the early phases of protein unfolding, which promises to increase our understanding of the molecular basis for a large number of folding-related diseases.
Methods
Protein Production and NMR Spectroscopy.
Unlabeled and 15N-labeled ACBP were expressed and purified as described in ref. 37. For the relaxation dispersion measurements, samples of 15N-labeled ACBP were prepared in 20 mM NaAc with 10% 2H2O at pH 5.3, at different concentrations of GuHCl. The protein concentration varied from 0.25 to 1 mM. All experiments were recorded on a Varian Inova 600. 15N CPMG relaxation-compensated experiments (38) were performed by using the constant time approach (39). Relaxation dispersion profiles were sampled with at least 12 CPMG field strengths (νCPMG) ranging from 33 to 1,000 Hz, where νCPMG is defined as 1/(4τCPMG) and 2τCPMG is the spacing between two successive 180° pulses in the CPMG pulse train. Each dispersion profile included two reference spectra recorded without any CPMG elements (i.e., with a relaxation delay of zero). Peak intensities were determined by fitting Gaussian line-shapes to each cross peak using the nmrpipe (40) command nlinLS or by summing the intensity in a 5- × 7-point window centered on the cross peak. Effective relaxation rates, R2eff, were obtained for all peaks at each CPMG field strength according to published procedures (39).
Relaxation dispersion profiles were fitted to the expression for a two-state process in the slow exchange regime, as described in ref. 18:
where R2 is the transverse relaxation rate of the folded (or unfolded) state in the absence of exchange, ka is the apparent rate-constant for unfolding (or folding) obtained by measuring R2eff of the folded (or unfolded) state, and Δω is the chemical-shift difference between the folded and unfolded states. Errors in the fitted parameters were estimated as the asymptotic standard errors.
The temperature dependence of the 1HN chemical shifts was measured on two samples (one without GuHCl and one with 0.2 M GuHCl) containing 0.8 mM ACBP in 20 mM NaAc, 10% 2H2O at pH 5.3, and 0.1 mM 2,2-dimethyl-2-silapentane-5-sulfonate as internal chemical shift standard. Eight 1H–15N HSQC spectra were recorded at temperatures ranging from 25°C to 46°C with an interval of 3°C. Proton chemical shifts of each amide cross peak were measured with an accuracy of ±1 part per billion. Both first- and second-order polynomials were fitted to the chemical shift versus temperature data for each 1HN resonance. Significant deviations from linearity were identified by F test (P < 0.05).
Stopped-Flow Kinetics.
Experiments were performed on 7.5 μM ACBP in 20 mM NaAc at pH 5.3 by using a Biologic SFM-3 with a dead-time of 10 ms. Unfolded ACBP was initially prepared in 3 M GuHCl. Kinetic experiments were performed by 10-fold dilution of (un)folded protein with appropriate concentrations of GuHCl. After excitation at 280 nm, Trp fluorescence was measured with a 305-nm long-pass filter.
Unfolding traces were fitted to monoexponential decays. Folding traces were fitted to biexponential decays because refolding involves a slow phase that is most likely due to cis–trans isomerization of peptidyl-prolyl peptide bonds (23); this phase was not considered further. The fast folding phase in the microsecond regime (7) is not observable by using the current experimental setup. However, the apparent folding rate kf will be modulated by the preequilibrium resulting in kf = kIu,N/(1 + KU,Iu), where U denotes the unfolded state, Iu denotes the collapsed folding intermediate, and N denotes the native state. kIu,N is the rate constant for the Iu-to-N transition, and KU,Iu is the equilibrium constant for the U-to-Iu transition. For simplicity, the preequilibrium between U and Iu is treated as a single denatured state, D.
Equilibrium Stability.
All experiments were carried out in 20 mM NaAc at pH 5.3 with various concentrations of GuHCl. The protein concentration was 5 μM for measurement of intrinsic Trp fluorescence and 40 μM for near-UV CD and ANS fluorescence measurements. Samples for ANS fluorescence measurements also included 200 μM ANS. Trp fluorescence was measured at 356 nm after excitation at 280 nm. ANS fluorescence was measured at 486 nm after excitation at 370 nm.
Analysis of Kinetic Folding Data.
Initially, folding data from stopped-flow kinetics and CPMG relaxation dispersions were fitted separately to two-state models (41). Simultaneous fitting of stopped-flow and NMR data were fit to a three-state model (Scheme 1). If kIN, kNI ≫ kDI, kID then the N-to-I transition is in equilibrium at all times and may be treated collectively as (N + I). Thus, at any time
such that the three-state model reduces to a pseudo two-state model for which Eq. 1 is still valid with ku,app = kIDKNI(1 + KNI)−1 in the unfolding reaction. Assuming that the logarithm of each microscopic rate constant depends linearly on the denaturant concentration (41), the unfolding rate measured by spin relaxation dispersion depends on the denaturant concentration according to:
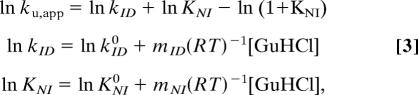 |
where the superscript 0 indicates the parameter in the absence of denaturant. The folding rates measured by spin relaxation dispersion are fit to
and the stopped-flow data are fit to
Supplementary Material
Acknowledgments
We thank Patrik Lundström for assistance with the relaxation experiments. This work was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, and the Knut and Alice Wallenberg Foundation (M.A.); the Carlsberg Foundation (F.M.P.); and the Villum Kann Rasmussen Foundation (K.T.).
Abbreviations
- ACBP
acyl-CoA binding protein
- ANS
8-anilinonaphthalene-1-sulphonic acid
- ASA
accessible surface area
- CPMG
Carr–Purcell–Meiboom–Gill
- GuHCl
guanidine hydrochloride
- HSQC
heteronuclear single quantum correlation
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wright P. E., Dyson H. J. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg A. L. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 3.Dobson C. M. Semin. Cell Dev. Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Uversky V. N., Fink A. L. Biochim. Biophys. Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Caughey B., Lansbury P. T. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 6.Glabe C. G. Trends Biochem. Sci. 2004;29:542–547. doi: 10.1016/j.tibs.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Teilum K., Maki K., Kragelund B. B., Poulsen F. M., Roder H. Proc. Natl. Acad. Sci. USA. 2002;99:9807–9812. doi: 10.1073/pnas.152321499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorski S. A., Capaldi A. P., Kleanthous C., Radford S. E. J. Mol. Biol. 2001;312:849–863. doi: 10.1006/jmbi.2001.5001. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez I. E., Kiefhaber T. J. Mol. Biol. 2003;325:367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 10.Friel C. T., Beddard G. S., Radford S. E. J. Mol. Biol. 2004;342:261–273. doi: 10.1016/j.jmb.2004.06.076. [DOI] [PubMed] [Google Scholar]
- 11.Schellman J. A. Biophys. J. 2003;85:108–125. doi: 10.1016/S0006-3495(03)74459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rupley J. A., Scheraga H. A. Biochemistry. 1963;13:421–431. doi: 10.1021/bi00903a005. [DOI] [PubMed] [Google Scholar]
- 13.Sivaraman T., Arrington C. B., Robertson A. D. Nat. Struct. Biol. 2001;8:331–333. doi: 10.1038/86208. [DOI] [PubMed] [Google Scholar]
- 14.Englander S. W. Annu. Rev. Biophys. Biomol. Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 15.Sridevi K., Udgaonkar J. B. Biochemistry. 2002;41:1568–1578. doi: 10.1021/bi011494v. [DOI] [PubMed] [Google Scholar]
- 16.Vugmeyster L., Kroenke C. D., Picart F., Palmer A. G., Raleigh D. P. J. Am. Chem. Soc. 2000;122:5387–5388. [Google Scholar]
- 17.Zeeb M., Balbach J. J. Am. Chem. Soc. 2005;127:13207–13212. doi: 10.1021/ja051141+. [DOI] [PubMed] [Google Scholar]
- 18.Tollinger M., Skrynnikov N. R., Mulder F. A., Forman-Kay J. D., Kay L. E. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 19.Korzhnev D. M., Salvatella X., Vendruscolo M., Di Nardo A. A., Davidson A. R., Dobson C. M., Kay L. E. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 20.Kristjansdottir S., Lindorff-Larsen K., Fieber W., Dobson C. M., Vendruscolo M., Poulsen F. M. J. Mol. Biol. 2005;347:1053–1062. doi: 10.1016/j.jmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Lindorff-Larsen K., Kristjansdottir S., Teilum K., Fieber W., Dobson C. M., Poulsen F. M., Vendruscolo M. J. Am. Chem. Soc. 2004;126:3291–3299. doi: 10.1021/ja039250g. [DOI] [PubMed] [Google Scholar]
- 22.Teilum K., Kragelund B. B., Poulsen F. M. J. Mol. Biol. 2002;324:349–357. doi: 10.1016/s0022-2836(02)01039-2. [DOI] [PubMed] [Google Scholar]
- 23.Kragelund B. B., Osmark P., Neergård T. B., Schiødt J., Kristiansen K., Knudsen J., Poulsen F. M. Nat. Struct. Biol. 1999;6:594–601. doi: 10.1038/9384. [DOI] [PubMed] [Google Scholar]
- 24.Teilum K., Kragelund B. B., Knudsen J., Poulsen F. M. J. Mol. Biol. 2000;301:1307–1314. doi: 10.1006/jmbi.2000.4003. [DOI] [PubMed] [Google Scholar]
- 25.Teilum K., Thormann T., Caterer N. R., Poulsen H. I., Jensen P. H., Knudsen J., Kragelund B. B., Poulsen F. M. Proteins. 2005;59:80–90. doi: 10.1002/prot.20340. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell K. L., Wildes D., Zarrine-Afsar A., De Los Rios M. A., Brown A. G., Friel C. T., Hedberg L., Horng J. C., Bona D., Miller E. J., et al. Protein Sci. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fieber W., Kragelund B. B., Meldal M., Poulsen F. M. Biochemistry. 2005;44:1375–1384. doi: 10.1021/bi0481949. [DOI] [PubMed] [Google Scholar]
- 28.Schmid F. In: Protein Folding Handbook: Part I. Buchner J., Kiefhaber T., editors. Weinheim, Germany: Wiley; 2005. pp. 22–44. [Google Scholar]
- 29.Semisotnov G. V., Rodionova N. A., Razgulyaev O. I., Uversky V. N., Gripas A. F., Gilmanshin R. I. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 30.Baxter N. J., Williamson M. P. J. Biomol. NMR. 1997;9:359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 31.Baxter N. J., Hosszu L. L., Waltho J. P., Williamson M. P. J. Mol. Biol. 1998;284:1625–1639. doi: 10.1006/jmbi.1998.2265. [DOI] [PubMed] [Google Scholar]
- 32.Williamson M. P. Proteins. 2003;53:731–739. doi: 10.1002/prot.10464. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. R. Proc. Natl. Acad. Sci. USA. 2000;97:14121–14126. doi: 10.1073/pnas.260502597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez I. E., Kiefhaber T. J. Mol. Biol. 2003;327:867–884. doi: 10.1016/s0022-2836(03)00171-2. [DOI] [PubMed] [Google Scholar]
- 35.Kragelund B. B., Heinemann B., Knudsen J., Poulsen F. M. Protein Sci. 1998;7:2237–2248. doi: 10.1002/pro.5560071101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart D. S., Case D. A. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]
- 37.Thomsen J. K., Kragelund B. B., Teilum K., Knudsen J., Poulsen F. M. J. Mol. Biol. 2002;318:805–814. doi: 10.1016/S0022-2836(02)00159-6. [DOI] [PubMed] [Google Scholar]
- 38.Loria J. P., Rance M., Palmer A. G. J. Am. Chem. Soc. 1999;121:2331–2332. [Google Scholar]
- 39.Mulder F. A., Skrynnikov N. R., Hon B., Dahlquist F. W., Kay L. E. J. Am. Chem. Soc. 2001;123:967–975. doi: 10.1021/ja003447g. [DOI] [PubMed] [Google Scholar]
- 40.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 41.Bachmann A., Kiefhaber T. In: Protein Folding Handbook: Part I. Buchner J., Kiefhaber T., editors. Weinheim, Germany: Wiley; 2005. pp. 379–410. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.