Abstract
γ-Secretase is an intramembrane-cleaving aspartyl protease required for the normal development of metazoans because it processes Notch within cellular membranes to release its signaling domain. More than two dozen additional substrates of diverse functions have been reported, including the Notch ligands Delta and Jagged, N- and E-cadherins, and a sodium channel subunit. The protease is causally implicated in Alzheimer’s disease because it releases the neurotoxic amyloid β-peptide (Aβ) from its precursor, APP. γ-Secretase occurs as a large complex containing presenilin (bearing the active site aspartates), nicastrin, Aph-1, and Pen-2. Because the complex contains at least 18 transmembrane domains, crystallographic approaches to its structure are difficult and remote. We recently purified the human complex essentially to homogeneity from stably expressing mammalian cells. Here, we use EM and single-particle image analysis on the purified enzyme, which produces physiological ratios of Aβ40 and Aβ42, to obtain structural information on an intramembrane protease. The 3D EM structure revealed a large, cylindrical interior chamber of ∼20–40 Å in length, consistent with a proteinaceous proteolytic site that is occluded from the hydrophobic environment of the lipid bilayer. Lectin tagging of the nicastrin ectodomain enabled proper orientation of the globular, ∼120-Å-long complex within the membrane and revealed ∼20-Å pores at the top and bottom that provide potential exit ports for cleavage products to the extra- and intracellular compartments. Our reconstructed 3D map provides a physical basis for hydrolysis of transmembrane substrates within a lipid bilayer and release of the products into distinct subcellular compartments.
Keywords: electron microscopy, image analysis, intramembrane protease
Inherited mutations in presenilin (PS), the active site component of γ-secretase, which was first recognized as the proteolytic activity responsible for generating amyloid β-peptide (Aβ) via cleavage of APP within cellular membranes, modulate the intramembrane cleavage of APP to increase levels of the highly amyloidogenic Aβ42 peptide in humans (1). APP, Notch, and other γ-substrates must undergo shedding of their large ectodomains by one of the α-secretases or by β-secretase before they can be processed within the membrane by γ-secretase (2). Beyond the intense interest in γ-secretase function for normal cell biology, the enzyme has emerged as a key therapeutic target for Alzheimer’s disease, and inhibitor design would be much facilitated by structural information about the proteolytic mechanism. PS is predicted to have eight or nine transmembrane domains (TMDs) (3–5), and it is catalytically inactive until it undergoes endoproteolysis between the two TMDs that bear the aspartates (nos. 6 and 7) to yield a heterodimer of the PS N- and C-terminal fragments (Fig. 1A). This activation requires the stepwise assembly of the four-protein complex. The seven-TMD Aph-1 protein apparently associates with the large, single-pass nicastrin (NCT) molecule early in the secretory pathway, followed by the binding of the uncleaved PS polypeptide and then the addition of the 2-TMD Pen-2, which allows endoproteolysis of PS to create the active enzyme (6, 7).
Fig. 1.
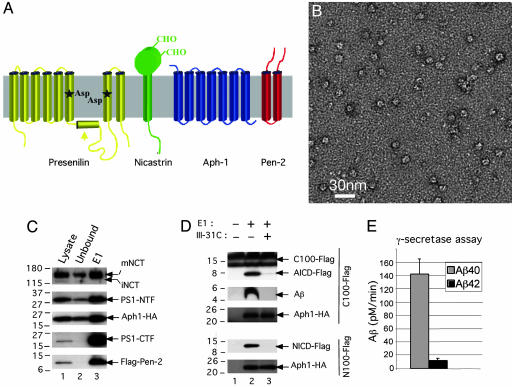
Characterization of purified γ-secretase. (A) Schematic drawing of the predicted transmembrane helices of the four component proteins in γ-secretase. (B) Electron micrograph of purified and negatively stained γ-secretase. (C) Detection by Western blot analysis of all four γ-secretase components recovered after the last step of the purification procedure. Twenty-five microliters each of the 1% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO)-solubilized membranes from γ-30 cells (starting lysate, lane 1), the material not binding to the M2 anti-Flag affinity resin (unbound, lane 2), and the fraction eluted from the M2 anti-Flag affinity resin (E1, lane 3) were loaded onto a 4–20% Tris-glycine SDS/PAGE gel and blotted with R302 (to NCT), Ab14 (to PS1-holo and -NTF), 13A11 (to PS1-CTF), 3F10 (to Aph-1-HA), and M2 (to Flag-Pen-2). (D) Specific proteolytic activity of the purified γ-secretase on both APP- and Notch-based substrates. The eluate fraction (as in C) was incubated in 0.1% phosphatidylcholine, 0.025% phosphatidylethanolamine with a 1 μM concentration of either C100Flag (APP-derived) or N100Flag (Notch-derived) substrates in the absence or presence of a 0.5 μM concentration of the active-site-directed γ-secretase inhibitor, III-31C, and the respective cleavage products (AICD-Flag and Aβ, or NICD-Flag) were detected as described in Materials and Methods. Aph-1 serves as an indicator of equal protein loading. (E) Purified γ-secretase cleavage of C100Flag generates a physiological ratio of Aβ42 and Aβ40 peptides. The specific eluate (as in C) was diluted in 0.2% CHAPSO-Hepes, pH 7.5, and incubated at 37°C for 4 h with 1 μM C100Flag substrate in 0.1% phosphatidylcholine, 0.025% phosphatidylethanolamine, and Aβ40 and Aβ42 were measured by ELISA.
Results
Purified γ-Secretase Was Active and Produced the Physiological Ratio of Aβ42/Aβ40.
To examine the 3D structure of γ-secretase, we purified intact, proteolytically active γ-secretase complexes from Chinese hamster ovary cells that stably overexpress human PS1, Aph-1, and Pen-2 in the presence of high levels of endogenous NCT (designated γ-30 cells) (8). Western blots of the sample recovered after the last purification step (Fig. 1C, lane 3) (silver stains published in ref. 8), combined with in vitro activity assays performed by using both APP- and Notch-based recombinant substrates (C100Flag and N100Flag) (Fig. 1D), confirmed that the purified, four-component γ-secretase complex was active and was sensitive to III-31C, (IC50 = 20 nM), a well characterized transition-state-analog γ-secretase inhibitor. Importantly, the C100Flag cleavage products, Aβ40 and Aβ42, were quantified by ELISA (Fig. 1E), revealing a physiological Aβ42/Aβ40 cleavage ratio (0.11) indistinguishable from that in normal human cerebrospinal fluid and plasma (10) and the media of cultured human neural cells (11). Moreover, the purified, active complexes contained no detectable immature NCT (Fig. 1C, top panel, compare lanes 1 and 3), signifying that the final preparation contained no significant level of immature complexes. This conclusion is further supported by nondenaturing blue native PAGE, which reveals only a single band of ∼300 kDa that is immunoreactive for each of the γ-secretase components (PS1-NTF, PS1-CTF, Aph-1, NCT, and Pen-2) (see Fig. 4A in ref. 12). However, we cannot exclude the possibility that a portion of the purified complexes is proteolytically inactive.
Fig. 4.
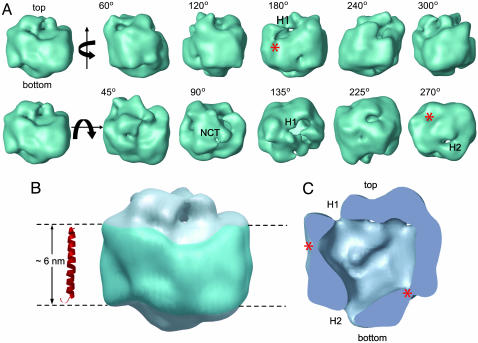
3D structure of the γ-secretase complex. (A) Surface rendering of the 3D reconstruction. The first row displays side views generated by rotating the map around a vertical axis, and the second row shows tilted views by rotating around a horizontal axis. The rotation angles are shown within each view. Two openings at the top and bottom are labeled H1 and H2, respectively, where visible. The top density is labeled NCT because the lectin labeling (Fig. 2) showed that the NCT ectodomain is located at this surface. (B) The potential transmembrane segment with the belt-like structure is outlined in blue by two parallel dashed lines, 60 Å apart. For size comparison, a typical transmembrane α-helix, taken from the rhodopsin structure (Protein Data Bank ID code 1GZM), is shown to the left of the structure. (C) A cut-open view of the γ-secretase complex from the side, revealing a large central chamber and one opening (H1) at the top and one at the bottom (H2). Two weak-density lateral regions are labeled with asterisks.
3D Reconstruction of γ-Secretase Structure from EM Images.
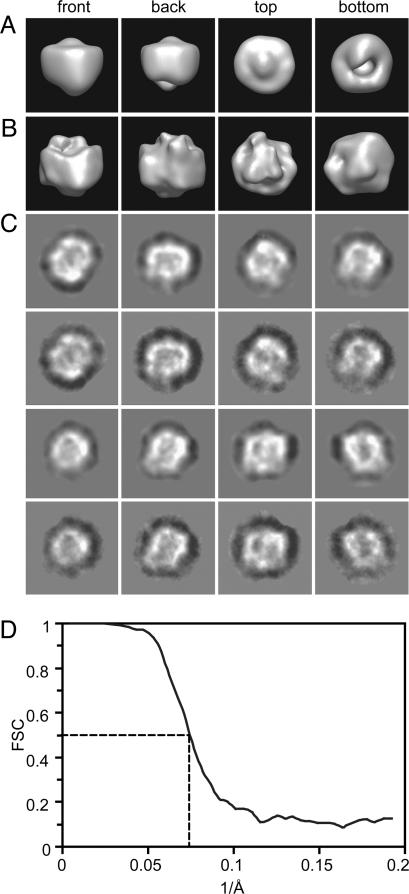
EM of the negatively stained complexes purified from γ-30 cells in a final concentration of 0.1% digitonin revealed fields of round, relatively homogeneous particles that measured 80–100 Å in diameter (Fig. 1B). This size was consistent with a particle of ∼300 kDa in mass, a value that has been reported by using biochemical sizing methods, including blue native PAGE (7, 8, 12). The random conical tilt method (13) was used to generate a series of initial 3D models, one from each class. The resultant models were highly similar, as judged by their general appearance and by the cross-correlation coefficients (≥0.9) (data not shown). These models were aligned and merged to produce a starting model (Fig. 2A). A total of 12,000 individual particle images were then used for model refinement. The orientations of the enzyme particles on the supporting carbon film were found to distribute evenly in angular space (data not shown), likely because of the round, globular shape of the active protease. The refined 3D reconstruction is shown in Fig. 2B. A comparison of eight selected reprojections of the refined 3D map (Fig. 2C, rows 1 and 3) with their corresponding class averages (Fig. 2C, rows 2 and 4) revealed essentially the same structural features. The most striking finding was a large, central low-density region measuring ∼40 Å in the longest dimension that was observed in the center of many of the class averages. The resolution of this map, as estimated by the Fourier shell correlation of two maps calculated separately from two halves of the data set, was ∼15 Å (Fig. 2D) (14, 15).
Fig. 2.
3D reconstruction from particle images of the γ-secretase complex. (A) Views of the starting model derived from the random conical tilt method. In this method, the tilt angle needed for calculating the 3D map is determined experimentally. (B) Corresponding views of the refined 3D map. (C) A gallery of selected reprojections of the reconstructed 3D map (rows 1 and 3) and their corresponding class averages (rows 2 and 4). (D) The Fourier shell correlation (FSC) curve suggests a resolution of 15 Å.
Lectin Labeling Identified the Location of the NCT Ectodomain.
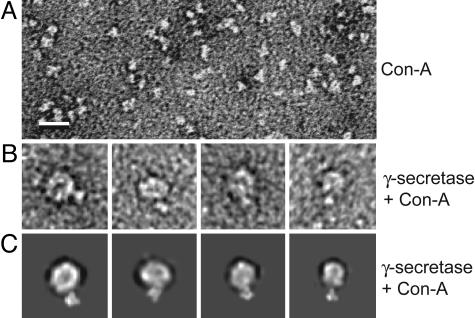
To determine the basic orientation of the complex in the membrane and ascertain whether the top surface in our 3D map (as shown in the first panels of Fig. 2 A and B) faces the extracellular or the cytoplasmic space, we localized the extracellular domain of the enzyme complex by taking advantage of the fact that multiple N-linked oligosaccharide groups occur exclusively in the NCT ectodomain, and that the plant lectin, Con A, can bind their mannose and glucose residues highly specifically. Con A is known to be an asymmetric tetramer (104 kDa) formed by orthogonal association of two homodimers, with four carbohydrate-binding sites exposed at the tip of each monomer (16). Pure Con A was visible in negatively stained electron micrographs as roughly triangular particles of 30–60 Å in the longest dimension, depending on their orientation relative to the incident electron beam (Fig. 3A); this appearance was similar to that reported for Con A (16). After incubating an aliquot of the purified γ-secretase complexes with Con A, we selected 20 γ-secretase particles with a clearly protuberant Con A attachment for further image analysis (Fig. 3B). Importantly, this protuberance was never observed in γ-secretase complexes that had not undergone Con A labeling. The density attributed to Con A was then manually masked from the image, and the resulting raw γ-secretase particle images were classified with the reprojections of our 3D reconstruction in a multireference alignment procedure. This correlative analysis established the assignment of the extracellular glycosylation sites to the top density in the 3D reconstruction. Fig. 3B shows the raw images of four such particles, and Fig. 3C shows the corresponding reprojections of the model when the Con A structure (Protein Data Bank ID code 1AZD) was low-pass filtered and manually placed on the top of the map. We note that the locations of the Con A in the raw images are generally consistent with those in the reprojections, but they are not identical, as expected, because of the multiple Con A binding sites available on the NCT ectodomain at the top of the γ-secretase complex (16 potential sites). Our lectin-binding analysis therefore revealed that the reconstructed 3D structure of γ-secretase shown in Fig. 2B (first panel from the left) is oriented with the top density at the extracellular surface and the long axis in the membrane bilayer, so that the smooth and continuous belt-like density (see Fig. 2B and Fig. 4, described below) faces the hydrophobic lipid environment. This orientation is consistent with the knowledge that all four component proteins (PS1, Aph-1, NCT, and Pen-2) are integral membrane proteins that can be found together in a proteolytically active complex on the plasma membrane (17, 18).
Fig. 3.
Lectin labeling of the γ-secretase complex. (A) A small area of an electron micrograph showing the negatively stained Con A particles, which are known to be 104-kDa asymmetric homotetramers 30–60 Å in size. (B) Four selected raw particle images of the γ-secretase–Con A complex. (Scale bar: 100 Å.) (C) The corresponding reprojections of the 3D reconstruction with the atomic structure of Con A low-pass-filtered and placed at the top when the map is oriented in the front view (i.e., as in the first panels in Fig. 2 A and B).
Structural Description of the γ-Secretase Map.
The 3D map of the γ-secretase complex was rendered into surface views at a threshold including 80% of the expected molecular mass of the complex (Fig. 4A and Movie 1, which is published as supporting information on the PNAS web site). The basic structural features and the overall size were not sensitive to the particular threshold used. Purified, active γ-secretase was an elongated globular structure, with the longest dimension being ∼120 Å (in the membrane) and the other two dimensions perpendicular to the long axis being ∼70–80 Å each. A major feature of the 3D map was a belt-like density ∼60 Å high circumferentially around the particle that is very likely to be the transmembrane portion of γ-secretase (Fig. 4B). A typical transmembrane helix, such as the one from rhodopsin structure (Protein Data Bank ID code 1GZM) shown in red, fitted well with the height of the belt-like density (Fig. 4B). Similar features are often observed in the EM structures of the membrane-embedded portions of membrane protein complexes, such as the proton pumping Vo-subcomplex of the V-ATPase (14) and the voltage-dependent tetrameric K+ channel in complex with a Fab fragment (15). A cut-open view is displayed in Fig. 4C. The most dramatic feature of the structure was the presence of a low-density central chamber. The shape of the chamber was irregular, with a diameter of 20–40 Å. The extracellular density at the top of the map (Fig. 4A, second row) can be attributed to part of the large (669 aa) ectodomain of NCT because it comprises a long extracellular polypeptide (∼70 kDa) onto which N-linked oligosaccharide groups are attached. At the display threshold, we observed two openings to the interior chamber: one of ∼20 Å size (designated H1) facing up toward the exterior of the cell and the other ∼10 Å in size (designated H2) facing down toward the cytoplasm (Fig. 4A). We also observed two weak-density regions (asterisks in Fig. 4 A and C) on the attributed intramembrane surface.
Discussion
The most enigmatic feature of γ-secretase and other intramembrane proteases is their location within the hydrophobic environment of the membrane and yet their requirement for water molecules to accomplish peptide bond hydrolysis. How γ-secretase releases its two cleavage products into distinct subcellular compartments has also been mysterious. Our 3D EM structure of the purified, proteolytically active, overexpressed human enzyme, which contains only mature complexes and produces physiologically correct ratios of human APP cleavage products, provides plausible explanations for these questions. The observed interior chamber and apical and basal pores would allow the entry of water molecules and their sequestration from the lipid surrounded by the proteinaceous microenvironment provided by the many TMDs. The two thin-density regions in the transmembrane portion (labeled with asterisks in Fig. 4C) may represent sites that could be opened up transiently to enable entrance of the α-helically conformed substrates, i.e., the TMDs of various single transmembrane proteins. After a substrate (e.g., C99 of APP or the analogous NΔE fragment of Notch) enters the proteinaceous central chamber and undergoes hydrolysis by the 2-aspartate-containing catalytic site in PS, the cleavage product that includes the extracellular half of the substrate’s TMD (e.g., Aβ or Np3) could be released through the H1 channel to the extracellular surface, and the product that includes the cytoplasmic half of the substrate’s TMD plus the intracellular domain (e.g., AICD or NICD) could be released through the H2 channel to the cytoplasm (Fig. 4).
If the catalytic site is located inside the proteinaceous chamber of our 3D structure, the architectural design of γ-secretase resembles to some extent that of the well characterized cytoplasmic proteasome (19, 20). We note that the γ-secretase may employ a compartmentalization strategy similar to that of the proteasome to prevent uncontrolled proteolysis, given its very broad substrate specificity (21). Substrate unfolding and gate opening of the eukaryotic proteasomes are tightly regulated by ATPases associated with various cellular activities (AAA)-type proteins (20). In the case of γ-secretase, there is now strong evidence that an initial substrate-docking site is near to but physically distinct from the catalytic site (22). The 3D EM structure reveals no apparent opening in the transmembrane portion of the complex through which a docked TMD substrate could translocate into the central chamber. We postulate that there may be a gate-opening mechanism at the two low-density lateral regions of the intramembrane barrel that can transiently modify the conformation of some of the TMDs. A lateral gate-opening mechanism was recently proposed for the protein-conducting channel of Sec61 that enables the translocation of TMDs into the lipid bilayer (23). In the case of γ-secretase, a related mechanism could account for the opposite direction of TMD translocation: from the lipid bilayer into the central proteinaceous chamber.
Using the natural affinity of lectins for the glycosylated ectodomain of NCT allowed its localization within the complex and revealed that it partially covers the top of the chamber, limiting the size of the H1 opening to ∼20 Å. This position of the NCT ectodomain suggests a role as a type of flexible lid that could regulate the entry of water molecules into the central chamber and the exit of hydrophilic ectodomain products. This localization is also quite consistent with the recent discovery that NCT acts as a type of receptor that binds the extracellular amino terminus of a substrate to facilitate its subsequent endoproteolysis by γ-secretase within the membrane (24).
Attempts to record cryo-electron microscopic images of γ-secretase are underway, but much more purified enzyme will be needed. However, faithful structures of a large number of protein complexes have been determined at medium resolution from images of negatively stained particles such as those we have analyzed (14, 15, 25–27). This structural analysis of an intramembrane-cleaving protease reveals a globular architecture of the physiologically active γ-secretase complex, with the long-axis in the plane of the membrane, a sizable interior chamber, and discrete openings to the extracellular and cytoplasmic spaces. We believe that the overall architecture and the interior chamber of the complex are well determined from the negatively stained samples, but the precise sizes of the two openings cannot yet be specified because they could be affected by the local stain salt affinity and the relatively limited resolution. On the basis of these initial views, further structural and biochemical studies examining expressed, purified γ-secretase complexes that bear natural or artificial mutations in the subunit proteins will now build on this information to steadily refine the EM structure and reveal details of its unusual mechanism.
Materials and Methods
Purification of γ-Secretase.
The multistep procedure for the high-grade purification of human γ-secretase from the γ-30 cells was performed as described in ref. 8, except that the III-31C affinity purification step was omitted.
In Vitro γ-Secretase Assays.
By using the recombinant APP-based substrate C100Flag and the recombinant Notch-based substrate N100Flag, γ-secretase assays were performed as reported in refs. 28 and 29. Briefly, the proteolytic reaction mixtures contained C100Flag or N100Flag substrate at concentrations of 1 μM, purified γ-secretase solubilized in 0.2% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO)-Hepes (pH 7.5, at 10-fold dilution from stock), 0.025% phosphatidylethanolamine (PE), and 0.10% phosphatidylcholine (PC). All reactions were stopped with 0.5% SDS, and the samples were assayed for Aβ40 and Aβ42 by ELISA as described in ref. 30 or electrophoresed on 4–20% Tris-glycine gels and transferred to poly(vinylidene difluoride) membranes to detect AICD-Flag with anti-Flag M2 antibody (1:1,000; Sigma) and NICD with Notch Ab1744 antibody (1:1,000; Cell Signaling Technology), which is selective for the N terminus of NICD. Aliquots of the same samples were blotted onto nitrocellulose membranes to detect Aβ with the antibody 6E10. The γ-secretase inhibitor III-31C was prepared as described in ref. 28 and added to the reaction mixtures from a DMSO stock (final DMSO concentration of 1%, which alone did not affect γ-secretase activity).
Western Blotting and Antibodies.
For Western blot analysis of PS1-NTF, PS1-CTF, Aph1α2-HA, Flag-Pen-2, and NCT, the purified γ-secretase complexes were run on 4–20% Tris-glycine PAGE gels, transferred to poly(vinylidene difluoride), and probed with Ab14 (for PS1-NTF, 1:2,000; gift of S. Gandy, Jefferson Medical College, Philadelphia), 13A11 (for PS1-CTF, 5 μg/ml; gift of Elan Pharmaceuticals, South San Francisco, CA), 3F10 (for Aph1α2-HA, 50 ng/ml; Roche), anti-Flag M2 (for Flag-Pen-2, 1:1,000; Sigma), or R302 antibodies (for NCT, 1:4,000; gift of D. Miller and P. Savam, Institute for Basic Research in Developmental Disorders, Staten Island, NY).
Lectin Labeling.
The lyophilized Con A was purchased from Sigma. A stock solution of 5 mg/ml was prepared in a buffer (10 mM Hepes, pH 7.0) containing 50 mM NaCl, 1 mM CaSO4, and 1 mM MnCl2. The mixture of Con A and γ-secretase at the molar ratio of 3:1 was incubated on ice for 30 min before making EM grids.
EM.
The purified γ-secretase complexes were applied to glow-discharged carbon-coated copper grids, and excess solution was blotted before staining with uranyl acetate (2%) for 30 sec. The stain was blotted, and the grids were left to air dry. Images were recorded under low dose condition (10 e/Å2) in a JEOL JEM 1200EX electron microscope operated at 120 kV with a magnification of ×50,000 and a defocus of 1.5–2.0 μm. The best-stained regions were selected with a charge-coupled device camera before recording the images on Kodak SO-163 negative film. Micrographs were inspected with an optical diffraction apparatus, and only micrographs without drift and astigmatism were further processed. The micrographs were digitized with a Nikon Super Coolscan at a step size of 12.7 μm, corresponding to a pixel size of 2.54 Å at the sample level. The size of digitonin micelles, as observed in EM grids prepared from a digitonin solution at the same concentration and buffer condition as that of the purified γ-secretase preparation, was ∼40–50 Å. Thus, the detergent micelles were clearly distinguishable from the much larger γ-secretase particles.
Image Analysis and 3D Reconstruction.
Image analysis was carried out with spider (31) and eman (32). By using spider, the defocus and amplitude contrast for each EM image were found, and these parameters were used for correcting the effect of the contrast transfer function. Raw particle images were subjected to reference-free classification through multivariate statistic analysis. For asymmetrical particles, it is essential to establish a reliable starting model. We used three approaches for this purpose: a Gaussian ellipsoid with added noise model; a starting model calculated from selected class averages based on cross-common line technique; and a model from the random conical tilt method. These methods resulted in similar but not identical structures. In the random conical tilt method, the models from different views were remarkably similar with high correlations, therefore these models were merged to serve as the starting model for projection-matching-based refinement. The convergence of the refinement process was monitored by examining the Fourier shell correlation between the successive 3D reconstructions. A total of 12,000 particles were used to calculate the final map. The resolution of the map was estimated by the Fourier shell correlation of two maps calculated separately from two halves of the data set. The map was low-pass filtered to the estimated resolution and rendered into surface views with chimera (33).
Supplementary Material
Acknowledgments
This work was supported by Brookhaven National Laboratory Laboratory-Directed Research and Development Fund 05-111, Department of Energy Grant KP1102010 (to H.L.), Swiss National Science Foundation Grant 81FR-68842 (to P.C.F.), and National Institutes of Health Grant AG1537 (to D.J.S. and M.S.W.).
Abbreviations
- Aβ
amyloid β-peptide
- PS
presenilin
- TMD
transmembrane domain
- NCT
nicastrin
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Aguzzi A., Haass C. Science. 2003;302:814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 2.Struhl G., Adachi A. Mol. Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 3.Li X., Greenwald I. Proc. Natl. Acad. Sci. USA. 1998;95:7109–7114. doi: 10.1073/pnas.95.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henricson A., Kall L., Sonnhammer E. L. FEBS J. 2005;272:2727–2733. doi: 10.1111/j.1742-4658.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 5.Laudon H., Hansson E. M., Melen K., Bergman A., Farmery M. R., Winblad B., Lendahl U., von Heijne G., Naslund J. J. Biol. Chem. 2005;280:35352–35360. doi: 10.1074/jbc.M507217200. [DOI] [PubMed] [Google Scholar]
- 6.Takasugi N., Tomita T., Hayashi I., Tsuruoka M., Niimura M., Takahashi Y., Thinakaran G., Iwatsubo T. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 7.LaVoie M. J., Fraering P. C., Ostaszewski B. L., Ye W., Kimberly W. T., Wolfe M. S., Selkoe D. J. J. Biol. Chem. 2003;278:37213–37222. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- 8.Fraering P. C., Ye W., Strub J. M., Dolios G., LaVoie M. J., Ostaszewski B. L., van Dorsselaer A., Wang R., Selkoe D. J., Wolfe M. S. Biochemistry. 2004;43:9774–9789. doi: 10.1021/bi0494976. [DOI] [PubMed] [Google Scholar]
- 9.Esler W. P., Das C., Campbell W. A., Kimberly W. T., Kornilova A. Y., Diehl T. S., Ye W., Ostaszewski B. L., Xia W., Selkoe D. J., Wolfe M. S. Nat. Cell Biol. 2002;4:E110–E111. doi: 10.1038/ncb0502-e110b. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner D., Eckman C., Jensen M., Song X., Citron M., Suzuki N., Bird T. D., Hardy J., Hutton M., Kukull W., et al. Nat. Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 11.Asami-Odaka A., Ishibashi Y., Kikuchi T., Kitada C., Suzuki N. Biochemistry. 1995;34:10272–10278. doi: 10.1021/bi00032a022. [DOI] [PubMed] [Google Scholar]
- 12.Fraering P. C., LaVoie M. J., Ye W., Ostaszewski B. L., Kimberly W. T., Selkoe D. J., Wolfe M. S. Biochemistry. 2004;43:323–333. doi: 10.1021/bi035748j. [DOI] [PubMed] [Google Scholar]
- 13.Radermacher M., Wagenknecht T., Verschoor A., Frank J. J. Microsc. (Oxford) 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 14.Venzke D., Domgall I., Kocher T., Fethiere J., Fischer S., Bottcher B. J. Mol. Biol. 2005;349:659–669. doi: 10.1016/j.jmb.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Q. X., Wang D. N., MacKinnon R. Nature. 2004;430:806–810. doi: 10.1038/nature02735. [DOI] [PubMed] [Google Scholar]
- 16.Bouckaert J., Hamelryck T., Wyns L., Loris R. Curr. Opin. Struct. Biol. 1999;9:572–577. doi: 10.1016/s0959-440x(99)00007-x. [DOI] [PubMed] [Google Scholar]
- 17.Tarassishin L., Yin Y. I., Bassit B., Li Y. M. Proc. Natl. Acad. Sci. USA. 2004;101:17050–17055. doi: 10.1073/pnas.0408007101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chyung J. H., Raper D. M., Selkoe D. J. J. Biol. Chem. 2005;280:4383–4392. doi: 10.1074/jbc.M409272200. [DOI] [PubMed] [Google Scholar]
- 19.Baumeister W., Walz J., Zuhl F., Seemuller E. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 20.Pickart C. M., Cohen R. E. Nat. Rev. Mol. Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 21.Voges D., Zwickl P., Baumeister W. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 22.Kornilova A. Y., Bihel F., Das C., Wolfe M. S. Proc. Natl. Acad. Sci. USA. 2005;102:3230–3235. doi: 10.1073/pnas.0407640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 24.Shah S., Lee S., Tabuchi K., Hao Y., Yu C., LaPlant Q., Ball H., Dann C. E., III, Südhof T., Yu G. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Andel F., III, Ladurner A. G., Inouye C., Tjian R., Nogales E. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- 26.Golas M. M., Sander B., Will C. L., Luhrmann R., Stark H. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 27.Viadiu H., Stemmann O., Kirschner M. W., Walz T. Nat. Struct. Mol. Biol. 2005;12:552–553. doi: 10.1038/nsmb935. [DOI] [PubMed] [Google Scholar]
- 28.Esler W. P., Kimberly W. T., Ostaszewski B. L., Ye W., Diehl T. S., Selkoe D. J., Wolfe M. S. Proc. Natl. Acad. Sci. USA. 2002;99:2720–2725. doi: 10.1073/pnas.052436599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimberly W. T., Esler W. P., Ye W., Ostaszewski B. L., Gao J., Diehl T., Selkoe D. J., Wolfe M. S. Biochemistry. 2003;42:137–144. doi: 10.1021/bi026888g. [DOI] [PubMed] [Google Scholar]
- 30.Xia W., Zhang J., Ostaszewski B. L., Kimberly W. T., Seubert P., Koo E. H., Shen J., Selkoe D. J. Biochemistry. 1998;37:16465–16471. doi: 10.1021/bi9816195. [DOI] [PubMed] [Google Scholar]
- 31.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 32.Ludtke S. J., Baldwin P. R., Chiu W. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.