Abstract
The enteric nervous system (ENS) is composed of neurons and glial cells, organized as interconnected ganglia within the gut wall, which controls persistalsis of the gut wall and secretions from its glands. The Ret receptor tyrosine kinase is expressed throughout enteric neurogenesis and is required for normal ENS development; humans with mutations in the RET locus have Hirschsprung disease (HSCR, an absence of ganglia in the colon), and mice lacking Ret have total intestinal aganglionosis. The Ret mutant mouse provides a tool for identifying genes implicated in development of the ENS. By using RNA from WT and Ret mutant (aganglionic) gut tissue and DNA microarrays, we have conducted a differential screen for ENS-expressed genes and have identified hundreds of candidate ENS-expressed genes. Forty-seven genes were selected for further analysis, representing diverse functional classes. We show that all of the analyzed genes are expressed in the ENS and that the screen was sensitive enough to identify genes marking only subpopulations of ENS cells. Our screen, therefore, was reliable and sensitive and has identified many previously undescribed genes for studying ENS development. Moreover, two of the genes identified in our screen Arhgef3 and Ctnnal1, have human homologues that map to previously identified HSCR susceptibility loci, thus representing excellent candidates for HSCR genes. This comprehensive profile of ENS gene expression refines our understanding of ENS development and serves as a resource for future developmental, biochemical, and human genetic studies.
Keywords: microarray, Ret, Sox2, Arhgef3, Ctnnal
The enteric nervous system (ENS) is composed of a vast number of neurons and glial cells, which form interconnected ganglia that control the contractility of the smooth muscle of the gut wall and the secretory activity of its glands (1). The ENS is derived from vagal and sacral enteric neural crest cells (ENCs), which invade the foregut and hindgut, respectively (2, 3). Once situated within the gut, ENCs migrate along the developing bowel, proliferate, and differentiate to form many different neuronal subtypes and make synaptic connections (4, 5). Migration of ENCs within the gut requires signaling between these cells and the gut environment and depends on dynamic changes in cell shape and adhesive properties. How cell shape and adhesion are controlled to allow regulated and directed migration of ENCs currently is unknown. Furthermore, how cell fate decisions are controlled to specify the correct number and subtypes of neurons and glial cells within ENS ganglia or how correct synaptic circuits are established also is unknown.
Genetic studies in mouse and human have identified several genes whose function is required for normal ENS development (6, 7). For example, the Ret receptor tyrosine kinase is expressed in ENCs during migration into the gut and persists during later ENS development (8). Humans carrying mutations in RET develop congenital megacolon [Hirschsprung disease (HSCR) OMIM 142623; ref. 7], which is characterized by the absence of enteric ganglia from the colon (colonic aganglionosis), whereas Ret-deficient mice (Retk−/k−) have total intestinal aganglionosis (9, 10). Ret signaling has been implicated in regulating migration, proliferation, differentiation, and axonogenesis within the ENS (11–13). How Ret signaling regulates these diverse cellular responses required for normal ENS development is not understood.
HSCR occurs relatively frequently in the human population (1:5,000 live births). Mutations in RET and the Endothelin receptor type B (EDNRB) account for 50% and 5% of familial HSCR cases, respectively. Mutations in a number of other genes account for a further small percentage of HSCR cases, including the Ret ligands GDNF and NTN, the EDNRB ligand EDN3, the EDN3-converting enzyme ECE1, SOX10, PHOX2B, and ZFHX1B (7). Additional susceptibility loci have been identified, although the corresponding genes have yet to be determined (7).
The key to understanding the molecular and cellular processes required for normal ENS development, and the corresponding defects that lead to HSCR, is an accurate profile of the cellular and molecular makeup of the ENS. In this study, we have applied DNA microarray techniques to profile gene expression in the developing ENS. Our screen has identified a large cohort of candidate ENS marker genes within the mammalian gut, and we have verified the ENS expression of a representative subset of genes. The identified genes are expressed during ENS migration, proliferation, differentiation, and axonogenesis and provide a valuable resource for further understanding of the cellular events critical for enteric neurogenesis. Finally, our studies have revealed candidate genes for previously characterized HSCR susceptibility loci.
Results
Microarray Screen for ENS Expressed Genes.
To identify genes expressed during mammalian ENS development we took advantage of the mouse Retk− mutation, which in homozygosity results in total intestinal aganglionosis (9, 10). We reasoned that transcripts more abundantly expressed in Ret+/+ versus Retk−/k− intestines should represent ENS-expressed genes. We therefore isolated RNA from embryonic day 15.5 (E15.5) Ret+/+ and Retk−/k− intestines, because sufficient amounts of RNA could be isolated at this time point and because genes known to be key ENS development regulators, such as Ret, Grfa1, and Sox10, are expressed at this stage (14).
To enable a broad and unbiased comparison of gene expression between Ret+/+ and Retk−/k− intestines, we examined gene expression profiles by using Affymetrix microarrays. Of the ≈22,000 probe sets represented on the array, 372 were expressed more abundantly in Ret+/+ versus Retk−/k− intestinal samples, ranging from 89-fold higher to 1.28-fold higher in normal versus aganglionic gut tissue (Table 4, which is published as supporting information on the PNAS web site). The 372 probe sets correspond to 329 distinct genes and represent candidate ENS-expressed genes. Consistent with this idea, we find several ENS-expressed genes amongst this list of differentially expressed genes, including Ret (1.9-fold), Phox2b (6.1-fold), Sox10 (12.5-fold), and Gfra1 (2.8 fold) (14).
Analysis of Gene Expression of Candidate ENS Expressed Genes.
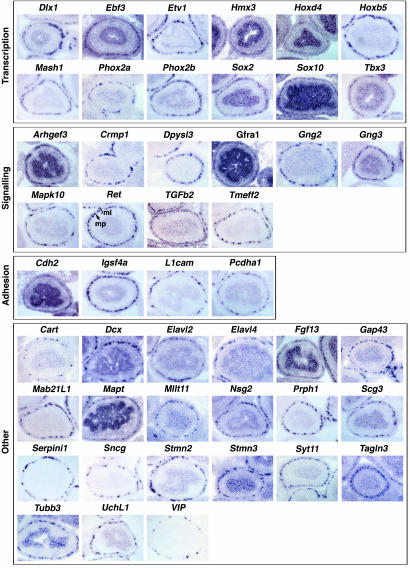
To assess the validity of the list of candidate ENS markers identified by our microarray screen, we examined the expression of 47 genes whose differential representation in Ret+/+ versus Retk−/k− intestines ranged from 89-fold to 1.8-fold (Table 1). The genes chosen for analysis were selected to represent diverse functional classes and were either previously uncharacterized or only partially characterized (such as Crmp1 and Hmx3), or known ENS marker genes (such as Ret, Phox2a, Phox2b, Sox10, and Gfra1) that serve as a reference for the many previously uncharacterized gene expression patterns described. The expression of our selected genes was examined by RNA in situ hybridization on E15.5 embryo sections (Fig. 1), because this method provided valuable spatial information. Ret mRNA shows the characteristic distribution in ENS ganglia comprising the myenteric plexus (mp; Figs. 1 and 3, Ret).
Table 1.
Genes selected for validation by in situ hybridization from the full list of genes on the MOE430A chip more abundantly expressed in Ret+/+ versus Retk−/k− intestine
| Gene symbol | Gene name | Fold change WT/MUT | Unigene |
|---|---|---|---|
| Mapk10 | Mitogen-activated protein kinase 10 | 89.88 | Mm.39253 |
| Sncg | Synuclein, γ | 59.42 | Mm.282800 |
| Mab21i1 | Mab-21-like 1 | 51.19 | Mm.252244 |
| Prph1 | Peripherin | 33.04 | Mm.2477 |
| Ascl1 | Achaete-scute complex homolog-like 1 | 30.99 | Mm.136217 |
| Stmn3 | Stathmin-like 3 | 29.01 | Mm.2319 |
| Tubb3 | Tubulin, β3 | 28.96 | Mm.40068 |
| Dlx1 | Distal-less homeobox 1 | 25.29 | Mm.4543 |
| Serpini1 | Serine (or cysteine) peptidase inhibitor, clade I, member 1 | 20.19 | Mm.41560 |
| Elavl4 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | 18.01 | Mm.3970 |
| Pcdha | Protocadherin α | 17.28 | Mm.308500 |
| Phox2a | Paired-like homeobox 2a | 16.83 | Mm.358574 |
| Vip | Vasoactive intestinal polypeptide | 16.17 | Mm.98916 |
| Mapt | Microtubule-associated protein τ | 16.05 | Mm.1287 |
| Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | 15.16 | Mm.318042 |
| Stmn2 | Stathmin-like 2 | 14.37 | Mm.29580 |
| Sox10 | SRY-box containing gene 10 | 12.53 | Mm.276739 |
| Sox2 | SRY-box containing gene 2 | 10.97 | Mm.4541 |
| Uchl1 | Ubiquitin carboxyl-terminal hydrolase PGP9.5 | 10.77 | Mm.29807 |
| Dcx | Doublecortin | 10.72 | Mm.12871 |
| Tagln3 | Transgelin 3 | 9.968 | Mm.24183 |
| Ebf3 | Early B cell factor 3 | 9.837 | Mm.258708 |
| Cart | Cocaine and amphetamine regulated transcript | 9.657 | Mm.75498 |
| Nsg2 | Neuron-specific gene family member 2 | 8.049 | Mm.3304 |
| Gap43 | Growth-associated protein 43 | 7.615 | Mm.1222 |
| Hmx3 | H6 homeobox 3 | 7.291 | Mm.323562 |
| Syt11 | Synaptotagmin XI | 7.061 | Mm.379376 |
| Scg3 | Secretogranin III | 6.567 | Mm.2386 |
| Arhgef3 | Rho guanine nucleotide exchange factor 3 | 6.191 | Mm.248606 |
| Phox2b | Paired-like homeobox 2b | 6.098 | Mm.62505 |
| Gng3 | Guanine nucleotide binding protein (G protein), γ3 subunit | 5.623 | Mm.329700 |
| Hoxb5 | Homeobox B5 | 5.298 | Mm.207 |
| Hoxd4 | Homeobox D4 | 5.219 | Mm.1214 |
| Dpysl3 | Dihydropyrimidinase-like 3 | 5.181 | Mm.8180 |
| Tmeff2 | Transmembrane protein with EGF-like and two follistatin-like domains 2 | 4.936 | Mm.245154 |
| Fgf13 | Fibroblast growth factor 13 | 4.827 | Mm.7995 |
| Crmp1 | Collapsin response mediator protein 1 | 4.386 | Mm.290995 |
| Gng2 | Guanine nucleotide-binding protein (G protein), γ2 subunit | 3.988 | Mm.41737 |
| Mllt11 | Myeloid/lymphoid or mixed-lineage leukemia (tri-thorax homolog Drosophila) translocated to 11 | 4.222 | Mm.331208 |
| Igsf4a | Immunoglobulin superfamily, member 4A | 3.42 | Mm.234832 |
| Cdh2 | Cadherin 2 | 3.279 | Mm.257437 |
| L1cam | L1 cell adhesion molecule | 2.807 | Mm.260568 |
| Gfra1 | Glial cell line-derived neurotrophic factor family receptor α1 | 2.772 | Mm.88367 |
| Tgfb2 | Transforming growth factor, β2 | 2.326 | Mm.18213 |
| Ret | Ret proto-oncogene | 1.913 | Mm.57199 |
| Etv1 | Ets variant gene 1 | 1.885 | Mm.4866 |
| Tbx3 | T-box 3 | 1.812 | Mm.219139 |
Genes from the full list of genes on the MOE430A chip more abundantly expressed in Ret+/+ versus Retk−/k− intestine (Table 3) were selected for validation by RNA in situ hybridization on tissue sections (Fig. 1). In this table, we identify those genes by (i) gene symbol, as assigned by the National Center for Biotechnology Information (NCBI), (ii) gene name as assigned by NCBI; (iii) fold change in Ret+/+ versus Retk−/k− intestine; and (iv) Unigene number. Previously undescribed ENS marker genes are in bold.
Fig. 1.
Expression profile of selected genes in the E15.5 mouse gut. RNA in situ hybridization of the genes identified in Table 1 is on transverse cryosections of E15.5 mouse small intestine. At E15.5, the enteric nervous system is organized into the myenteric plexus (mp), situated between the developing muscle layers (ml). Comparison of gene expression of known ENS markers reveals that all genes analyzed have equivalent expression within the myenteric region (mp in Ret).
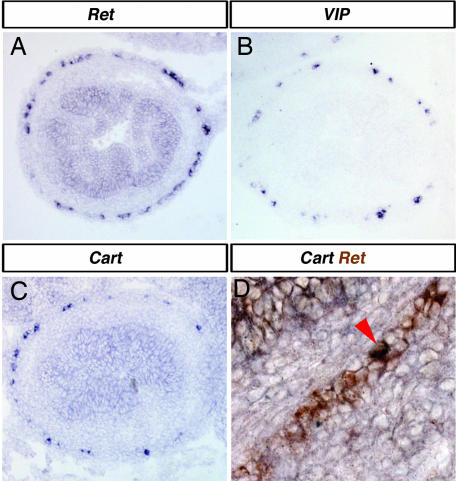
Fig. 3.
VIP and Cart are expressed in a subpopulation of Ret-expressing enteric neurons. Comparisons of gene expression profiles of Ret (A) to VIP (B), and Cart (C) by RNA in situ hybridization suggest that VIP and Cart are expressed in a smaller proportion of the ENS than Ret. A direct comparison of Ret expression (brown in D) and Cart expression reveals that Cart expression is found in only a small proportion of Ret-expressing cells (coexpression of Cart and Ret is seen as black; arrowhead in D).
A number of transcription factors (TFs) were expressed in the ENS in patterns indistinguishable from the established ENS-expressed TF markers Mash1, Phox2a, Phox2b, and Sox10 (Fig. 1, Transcription). Our analysis confirmed the expression of the homeobox-containing TFs Dlx1, Hmx3 (or Nkx5.1), and Hoxb5 (15–17) and identified previously undescribed TFs expressed in the ENS, including the early b cell factor Ebf3 (or O/E-2), the ets family TF Etv1 (or Er81), the homeobox-containing TF Hoxd4, the SOX family TF Sox2, and the T-box TF Tbx3.
Several of the identified genes representing components of signaling pathways, such as secreted ligands, receptors, and intracellular-signaling mediators, are expressed in the ENS in a profile indistinguishable from Ret and Gfra1 (Fig. 1, Signaling). We have identified as previously undescribed ENS markers the ligand Tgfb2 and transmembrane Tmeff2. Our screen has also identified previously undescribed ENS expression of intracellular mediators of signaling pathways, including the Rho guanine nucleotide exchange factor Arghef3, the mitogen-activated protein kinase family member Mapk10 (or Jnk3), and the G protein-γ subunits Gng2 and Gng3. Consistent with previous reports (18), we observed ENS expression of the collapsin response mediator family members Crmp1 and Dpysl3 (or Crmp4). Some of the genes that we demonstrate to be expressed in the ENS have been implicated in cell adhesion and may also participate in intracellular signaling events (ref. 19; Fig. 1, Adhesion). Our analysis confirms ENS expression of L1cam (20) and identifies previously undescribed ENS expression of the cadherin family gene Cdh2 (or Ncad), the protocadherin Pcdha1, and the Ig superfamily family member Igsf4a(or SgIGSF/SynCam).
The remaining genes examined for ENS expression by in situ hybridization fall into a broad range of functional categories (Fig. 1, Other). Microtubule-associated proteins, which may function in both cell migration and axon outgrowth during ENS development, include Dcx, Mapt (or tau), Stmn2 (or SCG10), Stmn3, Tagln3 (or NP25), and Tubb3. We have characterized the expression of several genes whose product can be detected by using common antibodies, including Gap43, Prph1, UchL1 (PGP9.5), Elavl2, and Elavl4 (or HuD and HuB) and confirmed the previously described ENS expression of membrane-associated Fgf13 (or FHF2) (21). In addition, we show previously undescribed expression of genes encoding for synapse-associated proteins in the ENS, such as Serpini1, Sncg, and Syt11. We have confirmed the ENS expression of the neuropeptide VIP and demonstrate that the neuropeptide Cart is expressed in the developing ENS. Interestingly, our screen also identified a number of genes that we show are previously undescribed ENS marker genes whose molecular function is unknown, including Mab21l1, Mllt11 (or AF1q), Nsg2, and Scg3.
Expression of Candidate ENS Marker Genes Is Absent in Retk−/k− Intestines.
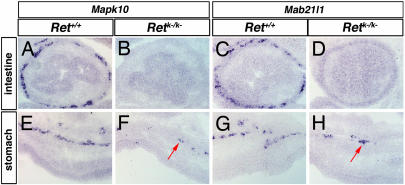
To further verify the ENS expression of the 47 genes described in Fig. 1, we examined their expression in Ret+/+ versus Retk−/k− intestines. mRNA for two representative examples, Mapk10 and Mab21l/1, was not detected in Retk−/k− intestine (Fig 2 B and D), indicating that their expression is restricted to the ENS. Consistent with this fact is the expression of Mapk10 and Mab21l1 in the small number of enteric neurons that colonize the stomach in a Ret-independent manner (Fig. 2 F and H; ref. 10). Equivalent results were observed for all 47 independently verified genes (data not shown).
Fig. 2.
Mapk10 and Mab21l1 are expressed in Ret+/+ but not Retk−/k− intestines. Representative comparison of gene expression in Ret+/+ versus Retk−/k− embryos by RNA in situ hybridization: Mapk10 (A, B, E, and F) and Mab21l1 (C, D, G, and H). Both genes are expressed in the myenteric layer of the Ret+/+ intestine (A and C), but expression is lost in the Retk−/k− intestine (B and D). We see expression throughout the myenteric plexus of the Ret+/+ stomach for both Mapk10 (E) and Mab21l1 (G) but only a small number of Mapk10- or Mab21l1-expressing cells in the equivalent region of the Retk−/k− stomach (arrows in F and H).
Sensitivity of Screen Allows Identification of Genes Expressed in Subsets of ENS.
The ENS comprises <5% of the total cell population of the E15.5 intestine (22). Consequently, the major difference between Ret+/+ and Retk−/k− intestines at this stage is the loss of this 5% of cells in Retk−/k− intestines. Despite this difference, our microarray approach is clearly capable of detecting differences in RNA composition. Moreover, two of the genes identified by using this approach, Cart and VIP, appear to be expressed in a fraction of ENS cells (compare Fig. 3A, B, and C). Comparison of the Cart and Ret expression domains by double in situ hybridization shows that Cart is expressed only in a subset of the Ret-expressing ENS cells (Fig. 3D). Nevertheless, the microarray screen was able to detect the differential expression of Cart.
Broad Temporal Expression of ENS-Expressed Genes.
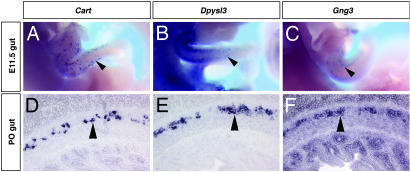
To further characterize the expression of the ENS-expressed genes, we examined the expression of a subset of these genes at a variety of embryonic and postnatal stages. We find that, like Ret, these ENS markers identified in our screen are expressed during early and late ENS development and elsewhere during neural development (Fig. 4; data not shown; ref. 8). For example, Cart is expressed in a subset of ENS neurons at E15.5 can be observed within the ENS from E11.5 when ENS precursors are migrating rostrocaudally through the gut (Fig. 4A) and persists until at least postnatal day (P) 0 (Fig. 4D). Dpysl3 and Gng3 also are expressed within the migrating ENS (Fig. 4 B and E) and persist through at least P0 (Fig. 4 C and F).
Fig. 4.
Analysis of Cart, Dpysl3, and Gng3 gene expression from E10 to P0. RNA in situ hybridization on E11.5 whole embryos (A–C) and on transverse sections of P0 intestines (D–F). Cart is expressed in a subset of enteric neurons at E11.5 (arrowhead in A) and at P0 (arrowhead in D). In the ENS, Dpysl3 expression can be seen at E11.5 (arrowhead in B) and P0 (arrowhead in E). Gng3 is expressed in the ENS at E11.5 (arrowhead in C) and P0 (arrowhead in F).
Human Homologues of ENS-Expressed Genes Map to HSCR Susceptibility Loci.
One of the confounding factors in genetic studies of HSCR is that it is thought to be a multifactorial disease; whereas HSCR is hereditary, inheritance is not Mendelian, with risk influenced by a number of parameters, notably gender (7). More than one-half of all cases of HSCR can be attributed to mutations in specific genes (7). Although the mutations responsible for the remaining HSCR cases have yet to be determined, human geneticists have identified several susceptibility loci, including 3p21, 19q21 (23), 9q31 (24), and 16q23 (25).
To explore whether any of the ENS expressed genes identified in our screen are candidate genes at the known susceptibility loci, we have identified human homologues of these genes and determined their chromosomal position (Tables 2 and 3 and data not shown). The markers used to define the RET-dependent modifier at 3p21, D3S2408, and D3S1766 are mapped to 3p14 in Ensembl genome build, NCBI35. Interestingly, ARHGEF3, the human homologue of Arhgef3, also maps to the region defined by these markers. We also have identified a gene whose human homologue maps to 9q31; catenin (cadherin associated protein) α-like 1 (Ctnnal1) is expressed more abundantly in Ret+/+ than Retk−/k− intestines (1.42-fold; Table 4) and human CTNNAL1 maps to 9q31.
Table 2.
Human homologues of ENS marker genes map to HSCR susceptibility loci
| Susceptability loci for HSCR | Genes mapping to HSCR loci |
|---|---|
| 3p21 | ARHGEF3 |
| 9q31 | CTNNAL1 |
| 16q23 | — |
| 19q21 | — |
The human homologues of the genes expressed more abundantly in Ret+/+ versus Retk−/k− intestine (Table 4), and their chromosomal position on the current Ensembl genome build (NCBI35) was determined. The markers used to identify the susceptibility loci at 3p21 (D3S2408 and D3S1766) (23) are mapped to 3p14 in NCBI35. The human homologue of Arhgef3, ARHGEF3, maps between these markers at 3p14. The human homologue of Ctnnal1 (also identified in our screen, Table 4), CTNNAL1, maps to the 9q31, another HSCR susceptibility loci (24). No currently identifiable human homologues of the genes identified in our screen map to susceptibility loci at 16q23 or 19q21.
Table 3.
Human homologues of ENS marker genes map to the X chromosome
| Genes that map to the X chromosome | Position |
|---|---|
| PCSK1N | Xp11.23 |
| PRKX | Xp22.33 |
| BEX2 | Xq22.1 |
| GPRASP1 | Xq22.1 |
| DCX | Xq23 |
| DIAPH2 | Xq21.3 |
| FGF13 | Xq26.3 |
| L1CAM | Xq28 |
Eight genes have human homologues on the X chromosome. These include proprotein convertase subtilisin/kexin type 1 inhibitor (PCSK1N); protein kinase, X-linked (PRKX); brain expressed X-linked 2 (BEX2); G protein-coupled receptor-associated sorting protein 1 (GPRASP1); doublecortin (DCX); diaphanous homolog 2 (DIAPH2); fibroblast growth factor 13 (FGF13); and L1 cell adhesion molecule (L1CAM).
The sex bias for HSCR is four males to one female (7), suggesting that other susceptibility loci may exist on the X chromosome. We have identified eight ENS expressed genes whose human homologues are found on the X chromosome (Table 3).
Discussion
Our microarray gene expression profiling has identified hundreds of genes more abundantly expressed in Ret+/+ than Retk−/k− intestines, which are candidate ENS-expressed genes. We have independently verified the results of our microarray screen by using RNA in situ hybridization for 47 selected genes and demonstrated that all of these genes are expressed in the ENS. The fidelity of this screen gives us a high level of confidence that the total list of candidate genes will be highly enriched for ENS-expressed genes.
At E15.5, the ENS contains cells that are proliferative, progressively differentiating and terminally differentiated. The genes identified in our screen therefore reflect these various cell states. We have identified markers of early differentiating neurons (such as Gap43 and Elavl2/4), early differentiating glial cells (such as Fabp7; Table 4), terminally differentiated neurons (such as VIP), axon outgrowth (such as Stmn2/3, Dcx), and synaptogenesis (such as Syt11 and Snap25; Table 4). Examination of the entire list of ENS-expressed genes refines our understanding of ENS development. Moreover, as we will discuss, this screen has identified previously undescribed genes, implicated previously undescribed processes, and is likely to inform and direct future studies.
Sox2 as a Potential Marker of Proliferative and Stem Cell Populations.
Within the CNS, highly proliferative neural progenitors and neural stem cells can be identified by their expression of Sox2. Moreover, Sox2 functions to maintain cells in a neural progenitor state (26). Our microarray experiments have identified Sox2 as a previously undescribed ENS-expressed gene. The identification of Sox2 is consistent with the fact that even at E15.5, ENS neurons are highly proliferative; during ENS development, unlike the CNS, cells expressing pan-neural markers such as Hu or Gap43 and the glial marker S100b, continue to proliferate while progressively differentiating (27, 28).
Significantly, the identification of Sox2 may represent expression within a population of ENS stem cells present in the gut. Sox2 expression is detectable in the ENS from E11.5 to at least P4 (data not shown). Proliferative ENS progenitors can be isolated from fetal and postnatal gut that, when grown in culture, will self-renew and give rise to neurons and glial cells (29). It is possible that, as in the CNS, Sox2 is expressed within the neural stem cell population that can be expanded in these culture conditions. If so, resources that are used to enrich for CNS neural stem cells, such as the Sox2-EGFP mouse (30), could be applied in future studies of the ENS stem cell population.
Cart: An Early Differentiation Marker.
Despite the fact that terminally differentiated neurons represent only a subpopulation of the ENS at E15.5 (31), our experiments detected VIP and CART expression, underscoring the sensitivity of this screen. In addition, we can detect the expression of Cart within a subpopulation of the developing ENS as early as E11.5 (Fig. 4). This finding establishes Cart as one of the earliest markers of a phenotypically distinct, terminally differentiated cell type in the ENS that could facilitate future studies of differential cell fate choice.
Semaphorin Signaling in the ENS.
The identification of two mediators of semaphorin signaling in our analysis (Crmp1 and Dpysl3) and the semaphorin receptor neuropilin1 (Nrp1; Table 4) suggests a role for semaphorin signaling within ENS development. Moreover, the repulsive semaphorin Sema3a is expressed in the chick gut (32). Semaphorins are transmembrane or secreted molecules that bind to plexin and/or neuropilin receptors to mediate a variety of cellular responses, including cell migration and axon guidance. Crmp1 and Dpysl3 are required within the plexin/neuropilin-expressing cell to mediate Sema3a-induced growth cone collapse (33) and are also proposed to mediate neurite extension controlled by the neurotrophins NGF and NT-3 (34). Semaphorin signaling also plays a role in neuronal migration (35), indicating that Crmp1, Dpysl3, and Nrp1 may mediate early cellular migration responses in addition to later axon extension responses.
A Potential Role for FGF Signaling in ENS Development.
In some instances, the ENS-expressed genes we have identified can be grouped into functional modules. For example, we have confirmed the ENS expression of Dlx1 and identified Etv1 and Tbx3 as previously undescribed ENS markers. Dlx, Ets, and Tbx transcription factors are regulated by FGFs in other developmental contexts (36, 37). The presence of FGF-responsive genes within the ENS highlights a possible role for FGF signaling in ENS development. Such a role is supported by expression of the FGF receptor FGFR2 in postnatal ENS ganglia (20). Also, FGF2 (basic FGF) is an essential component of the culture media used to stimulate growth of both ENS progenitor cells and neural stem cells in vitro and maintains cells in a continuously expanding state (29, 38). Finally, the mice mutant for the FGF receptor antagonist Sprouty2 exhibits ENS hyperganglionosis (39). Taken together, these findings suggest roles for FGFs in ENS development, which remain to be explored. One of the genes identified in our screen, Fgf13, shares strong sequence and structural similarity to FGFs but is not secreted and does not activate FGF receptors (40). Therefore, FGF13 is unlikely to mediate the FGF responses we propose. More likely, the relevant FGFs are expressed in the mesenchyme and, therefore, would not be identified in this screen for ENS markers.
c-Jun N-Terminal Kinase Pathway in ENS Development.
As a receptor tyrosine kinase, Ret can activate various signaling pathways, such as the RAS/extracellular signal-regulated kinase, phosphatidylinositol 3-kinase/AKT, p38 mitogen-activated protein kinase, and c-Jun N-terminal kinase pathways (41). These pathways are activated through phosphorylation of Tyr-1062 of Ret (42), a residue required for normal Ret function in vivo (43). Which of these pathways downstream of Ret are acting during the many phases of normal ENS development is unknown. Interestingly, our microarray screen has identified Mapk10 (Jnk3) expression within the ENS. Mapk10 activation is associated with responses to inflammation and cellular stresses but also with cytoskeletal changes associated with neuronal growth; Mapk10 binds and phosphorylates Stmn2 (Scg10) (44), that, in turn, can regulate the microtubule-destabilizing activity of Stnm2 (45). Because we show that Ret, Mapk10, and Stmn2 are all expressed in the ENS at this stage and we know that GDNF stimulation leads to axonal outgrowth (12), we can hypothesize that these molecules may function together to link an extracellular signal (GDNF) to the rearrangement of the neuronal cytoskeleton required for axonal outgrowth.
Implications for Human Diseases of the ENS.
We have explored the possibility that human homologues of the ENS marker genes identified in our screen map to HSCR susceptibility loci or to the X chromosome. ARHGEF3 maps to the RET-dependent susceptibility locus identified at 3p21 (23) and encodes a Rho guanine nucleotide exchange factor (RhoGEF). CTTNAL1 maps to the RET-dependent susceptibility locus at 9q31 (24) and is known to interact with RhoGEFs (46). RhoGEFs regulate the activity of Rho family GTPase to mediate cellular responses such as adhesion, migration, and axon growth (47). The interaction of CTNNAL1 with RhoGEFs has been proposed to modulate Rho pathway signaling by providing a scaffold for RhoGEFs (46). Dramatically, therefore, the two genes that map to susceptibility loci also are potentially interacting in the regulation of cell movement. The chromosomal positions of ARHGEF3 and CTNNAL1, in addition to the likely functional involvement of RhoGEFs in mediating the cellular events underway during normal ENS development, make these two genes excellent candidates for further tests to identify the modifier genes at 3p21 and 9q31. Finally, the fact that Rho has been shown to mediate GNDF/Ret signaling downstream of phosphatidylinositol 3-kinase in neuroblastoma cell lines (48) is a further indication that ARHGEF3 and CTNNAL1 are worthy of further study as a candidate genes to account for RET-dependent HSCR susceptibility at these loci.
We also have identified a number of human homologues of ENS-expressed genes that map to the X chromosome and, therefore, are candidates to account for the greater susceptibility of males to HSCR, including DCX and L1CAM. Both Dcx and L1cam have proposed roles in regulating cell migration and axonal outgrowth (49, 50), making these attractive candidates for susceptibility factors on the X chromosome. Furthermore, a role for L1CAM in HSCR is consistent with the observation of HSCR in a patient with L1CAM mutations (51). The localization of L1CAM to Xq28 also makes this gene a good candidate for a HSCR-related neuropathic motility disorder, chronic idiopathic intestinal pseudoobstruction, which has been mapped to this region (52).
Genes up-regulated in Retk−/k− versus Ret+/+ intestines have not been explored here (Table 5, which is published as supporting information on the PNAS web site), because they did not fulfill the remit of classifying ENS expressed genes. However, such genes are worthy of further study in light of evidence that aganglionic smooth muscle is a suboptimal substrate for growth of cells (53). This finding has significant implications for cell replacement therapy approaches being explored as treatment options for HSCR patients. Characterization of genes more abundant in Retk−/k− versus Ret+/+ intestines may uncover the changes to anganglionic tissue that affect cell growth, thereby facilitating improved therapeutic methods.
In summary, our experiments have profiled gene expression within the developing ENS and have identified ENS-expressed genes with high fidelity. The sensitivity and reliability of our screen gives us confidence that the complete data set will be highly enriched for ENS-expressed genes. Although our study does not represent a saturation screen, the genes identified form a large cohort of ENS-expressed genes, and this resource can be continually mined in silico to inform ongoing developmental and biochemical studies of the mammalian ENS. Finally, our findings indicate that cell-specific gene expression profiling is an efficient means of identifying candidate genes for human disease susceptibility loci.
Materials and Methods
Isolation of Ret+/+ and Retk−/k− Intestinal Total RNA.
Mice carrying the Retk− mutation and their genotyping have been described in refs. 9 and 10. Litters from intercrosses of Ret+/k− animals were harvested at E15.5. Embryos were dissected, with intestines (gut minus esophagus and stomach) taken for RNA isolation and tails taken for genotyping. Individual intestines were either flash-frozen and stored at −80°C or stored in RNAlater (Qiagen, Valencia, CA). Upon genotyping, pools of ≈10 intestines of a given genotype, either Ret+/+ or Retk−/k−, were combined to form single samples. Three samples of each genotype were generated. Tissue was homogenized with the QIAshredder kit (Qiagen), and total RNA was isolated by using the RNeasy kit (Qiagen).
Microarray Experiments and Data Analysis.
Three 8-mg total RNA samples for each genotype were used to prepare material for hybridization of six GeneChip Mouse Expression Set MOE430A arrays (Affymetrix) according to the manufacturer’s protocol. Hybridization, scanning, and generation of raw expression data were performed by a facility at the Columbia University Institute of Cancer Genetics. Data analysis was performed by using genespring 7 software (Agilent Technologies, Palo Alto, CA). For data normalization details and data output, see Tables 4 and 5. Identification and chromosomal position of human homologues of the genes from Table 1 used the Ensembl BioMart data mining tool, www.ensembl.org/Homo_sapiens/martview, by using Ensebl 36 Homo sapians genes (NCBI35) and Mus musculus genes (NCBIM34). Fine-map position of genes and markers was determined by querying NCBI35.
RNA in Situ Hybridization.
Section or whole-mount RNA in situ hybridization was performed as described in refs. 11 and 54. For double-section RNA in situ hybridization, the digoxigenin-labeled (Roche) probe was detected with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (BCIP), the precipitate was fixed with 4% paraformaldehyde, and alkaline phosphatase inactivated by 30 min in 0.1 M glycine, pH 2.2. After the second antibody incubation, the fluorescein-labeled probe (Roche) was detected with iodonitrotetrazdium violet/alkaline phosphatase. Antisense probes were generated as described in ref. 54. All probe information is in Table 6, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Benson Lu and Frank Costantini for facilitating microarray hybridizations and members of our laboratory and Malcolm Logan for critical comments. This work was funded by National Institutes of Health Project Grant CA23767.
Abbreviations
- En
embryonic day n
- ENC
enteric neural crest cell
- ENS
enteric nervous system
- HSCR
Hirschsprung disease
- Pn
postnatal day n
- RhoGEF
Rho guanine nucleotide exchange factor
- TF
transcription factor.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Gershon M. D., Kirchgessner A. L., Wade P. R. Functional Anatomy of the Enteric Nervous System. New York: Raven; 1994. [Google Scholar]
- 2.Burns A. J., Douarin N. M. Development (Cambridge, U.K.) 1998;125:4335–4347. doi: 10.1242/dev.125.21.4335. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R. B., Stewart A. L., Young H. M. Cell Tissue Res. 2006;323:11–25. doi: 10.1007/s00441-005-0047-6. [DOI] [PubMed] [Google Scholar]
- 4.Gershon M. D. Curr. Opin. Neurobiol. 1997;7:101–109. doi: 10.1016/s0959-4388(97)80127-4. [DOI] [PubMed] [Google Scholar]
- 5.Sang Q., Young H. M. Cell Tissue Res. 1996;284:39–53. doi: 10.1007/s004410050565. [DOI] [PubMed] [Google Scholar]
- 6.Newgreen D., Young H. M. Pediatr. Dev. Pathol. 2002;5:224–247. doi: 10.1007/s10024-001-0142-y. [DOI] [PubMed] [Google Scholar]
- 7.Brooks A. S., Oostra B. A., Hofstra R. M. Clin. Genet. 2005;67:6–14. doi: 10.1111/j.1399-0004.2004.00319.x. [DOI] [PubMed] [Google Scholar]
- 8.Pachnis V., Mankoo B., Costantini F. Development (Cambridge, U.K.) 1993;119:1005–1017. doi: 10.1242/dev.119.4.1005. [DOI] [PubMed] [Google Scholar]
- 9.Schuchardt A., D’Agati V., Larsson-Blomberg L., Costantini F., Pachnis V. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- 10.Durbec P. L., Larsson-Blomberg L. B., Schuchardt A., Costantini F., Pachnis V. Development (Cambridge, U.K.) 1996;122:349–358. doi: 10.1242/dev.122.1.349. [DOI] [PubMed] [Google Scholar]
- 11.Taraviras S., Marcos-Gutierrez C. V., Durbec P., Jani H., Grigoriou M., Sukumaran M., Wang L. C., Hynes M., Raisman G., Pachnis V. Development (Cambridge, U.K.) 1999;126:2785–2797. doi: 10.1242/dev.126.12.2785. [DOI] [PubMed] [Google Scholar]
- 12.Young H. M., Hearn C. J., Farlie P. G., Canty A. J., Thomas P. Q., Newgreen D. F. Dev. Biol. 2001;229:503–516. doi: 10.1006/dbio.2000.0100. [DOI] [PubMed] [Google Scholar]
- 13.Natarajan D., Marcos-Gutierrez C., Pachnis V., de Graaff E. Development (Cambridge, U.K.) 2002;129:5151–5160. doi: 10.1242/dev.129.22.5151. [DOI] [PubMed] [Google Scholar]
- 14.Newgreen D., Young H. M. Pediatr. Dev. Pathol. 2002;5:329–349. doi: 10.1007/s10024-002-0002-4. [DOI] [PubMed] [Google Scholar]
- 15.Qiu M., Bulfone A., Martinez S., Meneses J. J., Shimamura K., Pedersen R. A., Rubenstein J. L. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- 16.Pitera J. E., Smith V. V., Thorogood P., Milla P. J. Gastroenterology. 1999;117:1339–1351. doi: 10.1016/s0016-5085(99)70284-2. [DOI] [PubMed] [Google Scholar]
- 17.Bober E., Baum C., Braun T., Arnold H. H. Dev. Biol. 1994;162:288–303. doi: 10.1006/dbio.1994.1086. [DOI] [PubMed] [Google Scholar]
- 18.Inagaki H., Kato Y., Hamajima N., Nonaka M., Sasaki M., Eimoto T. Histochem. Cell Biol. 2000;113:37–41. doi: 10.1007/s004180050005. [DOI] [PubMed] [Google Scholar]
- 19.Viollet C., Doherty P. Cell Tissue Res. 1997;290:451–455. doi: 10.1007/s004410050952. [DOI] [PubMed] [Google Scholar]
- 20.Yoneda A., Wang Y., O’Briain D. S., Puri P. Pediatr. Surg. Int. 2001;17:299–303. doi: 10.1007/s003830100598. [DOI] [PubMed] [Google Scholar]
- 21.Hartung H., Feldman B., Lovec H., Coulier F., Birnbaum D., Goldfarb M. Mech. Dev. 1997;64:31–39. doi: 10.1016/s0925-4773(97)00042-7. [DOI] [PubMed] [Google Scholar]
- 22.Iwashita T., Kruger G. M., Pardal R., Kiel M. J., Morrison S. J. Science. 2003;301:972–976. doi: 10.1126/science.1085649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel S. B., Salomon R., Pelet A., Angrist M., Amiel J., Fornage M., Attie-Bitach T., Olson J. M., Hofstra R., Buys C., et al. Nat. Genet. 2002;31:89–93. doi: 10.1038/ng868. [DOI] [PubMed] [Google Scholar]
- 24.Bolk S., Pelet A., Hofstra R. M. W., Angrist M., Salomon R., Croaker D., Buys C. H. M., Lyonnet S., Chakravarti A. Proc. Natl. Acad. Sci. USA. 2000;97:268–273. doi: 10.1073/pnas.97.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrasquillo M. M., McCallion A. S., Puffenberger E. G., Kashuk C. S., Nouri N., Chakravarti A. Nat. Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- 26.Episkopou V. Trends Neurosci. 2005;28:219–221. doi: 10.1016/j.tins.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Baetge G., Gershon M. D. Dev. Biol. 1989;132:189–211. doi: 10.1016/0012-1606(89)90217-0. [DOI] [PubMed] [Google Scholar]
- 28.Young H. M., Turner K. N., Bergner A. J. Cell Tissue Res. 2005;320:1–9. doi: 10.1007/s00441-004-1057-5. [DOI] [PubMed] [Google Scholar]
- 29.Bondurand N., Natarajan D., Thapar N., Atkins C., Pachnis V. Development (Cambridge, U.K.) 2003;130:6387–6400. doi: 10.1242/dev.00857. [DOI] [PubMed] [Google Scholar]
- 30.Ellis P., Fagan B. M., Magness S. T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M., Pevny L. Dev. Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 31.Pham T. D., Gershon M. D., Rothman T. P. J. Comp. Neurol. 1991;314:789–798. doi: 10.1002/cne.903140411. [DOI] [PubMed] [Google Scholar]
- 32.Shepherd I. T., Raper J. A. Dev. Biol. 1999;212:42–53. doi: 10.1006/dbio.1999.9294. [DOI] [PubMed] [Google Scholar]
- 33.Liu B. P., Strittmatter S. M. Curr. Opin. Cell Biol. 2001;13:619–626. doi: 10.1016/s0955-0674(00)00260-x. [DOI] [PubMed] [Google Scholar]
- 34.Quach T. T., Duchemin A. M., Rogemond V., Aguera M., Honnorat J., Belin M. F., Kolattukudy P. E. Mol. Cell Neurosci. 2004;25:433–443. doi: 10.1016/j.mcn.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Gammill L. S., Gonzalez C., Gu C., Bronner-Fraser M. Development (Cambridge, U.K.) 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- 36.Firnberg N., Neubuser A. Dev. Biol. 2002;247:237–250. doi: 10.1006/dbio.2002.0696. [DOI] [PubMed] [Google Scholar]
- 37.Thomas B. L., Liu J. K., Rubenstein J. L., Sharpe P. T. Development (Cambridge, U.K.) 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 38.Conti L., Pollard S. M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q. L., Cattaneo E., et al. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taketomi T., Yoshiga D., Taniguchi K., Kobayashi T., Nonami A., Kato R., Sasaki M., Sasaki A., Ishibashi H., Moriyama M., et al. Nat. Neurosci. 2005;8:855–857. doi: 10.1038/nn1485. [DOI] [PubMed] [Google Scholar]
- 40.Goldfarb M. Cytokine Growth Factor Rev. 2005;16:215–220. doi: 10.1016/j.cytogfr.2005.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichihara M., Murakumo Y., Takahashi M. Cancer Lett. 2004;204:197–211. doi: 10.1016/S0304-3835(03)00456-7. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi H., Ichihara M., Iwashita T., Murakami H., Shimono Y., Kawai K., Kurokawa K., Murakumo Y., Imai T., Funahashi H., et al. Oncogene. 2000;19:4469–4475. doi: 10.1038/sj.onc.1203799. [DOI] [PubMed] [Google Scholar]
- 43.Wong A., Bogni S., Kotka P., de Graaff E., D’Agati V., Costantini F., Pachnis V. Mol. Cell. Biol. 2005;25:9661–9673. doi: 10.1128/MCB.25.21.9661-9673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neidhart S., Antonsson B., Gillieron C., Vilbois F., Grenningloh G., Arkinstall S. FEBS Lett. 2001;508:259–264. doi: 10.1016/s0014-5793(01)03090-3. [DOI] [PubMed] [Google Scholar]
- 45.Antonsson B., Kassel D. B., Di Paolo G., Lutjens R., Riederer B. M., Grenningloh G. J. Biol. Chem. 1998;273:8439–8446. doi: 10.1074/jbc.273.14.8439. [DOI] [PubMed] [Google Scholar]
- 46.Park B., Nguyen N. T., Dutt P., Merdek K. D., Bashar M., Sterpetti P., Tosolini A., Testa J. R., Toksoz D. J. Biol. Chem. 2002;277:45361–45370. doi: 10.1074/jbc.M202447200. [DOI] [PubMed] [Google Scholar]
- 47.Bloch-Gallego E., Causeret F., Ezan F., Backer S., Hidalgo-Sanchez M. Brain Res. Rev. 2005;49:253–266. doi: 10.1016/j.brainresrev.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Murakami H., Iwashita T., Asai N., Iwata Y., Narumiya S., Takahashi M. Oncogene. 1999;18:1975–1982. doi: 10.1038/sj.onc.1202514. [DOI] [PubMed] [Google Scholar]
- 49.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M. C., Friocourt G., McDonnell N., Reiner O., Kahn A., et al. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 50.Kenwrick S., Watkins A., De Angelis E. Hum. Mol. Genet. 2000;9:879–886. doi: 10.1093/hmg/9.6.879. [DOI] [PubMed] [Google Scholar]
- 51.Parisi M. A., Kapur R. P., Neilson I., Hofstra R. M., Holloway L. W., Michaelis R. C., Leppig K. A. Am. J. Med. Genet. 2002;108:51–56. doi: 10.1002/ajmg.10185. [DOI] [PubMed] [Google Scholar]
- 52.Auricchio A., Brancolini V., Casari G., Milla P. J., Smith V. V., Devoto M., Ballabio A. Am. J. Hum. Genet. 1996;58:743–748. [PMC free article] [PubMed] [Google Scholar]
- 53.Langer J. C., Betti P. A., Blennerhassett M. G. Cell Tissue Res. 1994;276:181–186. doi: 10.1007/BF00354798. [DOI] [PubMed] [Google Scholar]
- 54.Riddle R. D., Johnson R. L., Laufer E., Tabin C. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.