Abstract
A key regulatory gene in metamorphosing (holometabolous) insect life histories is the transcription factor broad (br), which specifies pupal development. To determine the role of br in a direct-developing (hemimetabolous) insect that lacks a pupal stage, we cloned br from the milkweed bug, Oncopeltus fasciatus (Of’br). We find that, unlike metamorphosing insects, in which br expression is restricted to the larval–pupal transition, Of’br mRNA is expressed during embryonic development and is maintained at each nymphal molt but then disappears at the molt to the adult. Induction of a supernumerary nymphal stage with a juvenile hormone (JH) mimic prevented the disappearance of br mRNA. In contrast, induction of a precocious adult molt by application of precocene II to third-stage nymphs caused a loss of br mRNA at the precocious adult molt. Thus, JH is necessary to maintain br expression during the nymphal stages. Injection of Of’br dsRNA into either early third- or fourth-stage nymphs caused a repetition of stage-specific pigmentation patterns and prevented the normal anisometric growth of the wing pads without affecting isometric growth or molting. Therefore, br is necessary for the mutable (heteromorphic) changes that occur during hemimetabolous development. Our results suggest that metamorphosis in insects arose as expression of br, which conveys competence for change, became restricted to one postembryonic instar. After this shift in br expression, the progressive changes that occur within the nymphal series in basal insects became compressed to the one short period of morphogenesis seen in the larva-to-pupa transition of holometabolous insects.
Keywords: evolution of metamorphosis, heteromorphosis, Oncopeltus, juvenile hormone, allometry
Life history strategies are highly plastic within animal phyla; some groups develop directly, whereas related taxa pass through a metamorphosis. Regulation of stage-specific differences may be under either environmental or hormonal control, but relatively little is known of the molecular switches involved or how changes in the timing of these switches can lead to evolutionary change (1). In insects, metamorphosis arose once from a direct-developing ancestor ≈300 million years ago (2). A key regulatory gene in metamorphosing (holometabolous) insect life histories is the transcription factor broad (br) (3–7). In both moths and flies, epidermal expression of br is restricted to the larval–pupal transition (3, 5–7), and its expression at this time is required for activation of pupal-specific gene expression, as well as suppression of larval- and adult-specific gene expression (3, 7). Accordingly, Drosophila null mutants never enter the pupal stage; instead, they remain in a prolonged larval state (8). In addition, gynander larvae mosaic for br null and br+ tissue produced mosaic larval and pupal tissue, respectively, at the larval–pupal transition (9). Loss of br also prevents the larval–pupal transition in the silkmoth; tissues that were transformed with a vector driving br RNA interference were unable to produce adult structures, and transformed larval organs were not destroyed at metamorphosis (10).
The restriction of br expression at the larval–pupal transition of holometabolous insects occurs through the action of two hormones: the steroid 20-hydroxyecdysone (20E) and the sesquiterpenoid juvenile hormone (JH). Peaks of 20E trigger molts between stages, whereas the presence or absence of JH determines the type of cuticle that is produced (11) and whether br is expressed (5–7). During larval life, the presence of JH suppresses metamorphosis and br expression (5, 7). As JH titers disappear in the last larval stage, a small peak of 20E triggers “pupal commitment” as it induces br (3–7). Although JH levels again rise at the pupal molt, when they suppress precocious adult development, JH does not suppress br at this stage. In fact, topical application of JH during the adult molt, which normally occurs in the absence of JH and br, causes the reinduction of br and the production of a second pupal cuticle (7).
To determine the role of br in a nonholometabolous, direct-developing insect, we have isolated br from the milkweed bug, Oncopeltus fasciatus. Here, we show that, as in metamorphosing insects, br is required for morphogenesis and that its expression is regulated by JH at molts. In this insect, however, br is expressed at each nymphal molt, and its expression is required for progressive changes in proportions and pattern from instar to instar. Our results suggest that metamorphosis in insects arose as expression of this factor, which conveys competence for change, became restricted to one postembryonic instar.
Results
Using PCR with a set of primers to the N-terminal Broad–Tramtrack–Bric-a-brac (BTB) domain of br, we isolated a 600-bp br fragment from cDNA of O. fasciatus (Of’br) (GenBank accession no. DQ176004) and used RT-PCR to study its expression. Oncopeltus progresses through five nymphal instars (defined as the form observed during each successive intermolt stage or stadium) and then molts to the adult with functional wings and genitalia. br mRNA first appears during embryogenesis and is present in the first two nymphal stages (data not shown). Fig. 1 shows br mRNA levels at daily intervals through the last three nymphal stages. br mRNA is present through the third and fourth instars but is up-regulated during the molt when the next nymphal cuticle is made (Fig. 1 A and B). By contrast, no br mRNA is present during the latter part of the fifth nymphal instar when the adult cuticle is made (Fig. 1C).
Fig. 1.
RT-PCR analysis of Of’br mRNA during nymphal life. (A–C) br expression at 24-h intervals from the onset of the third, fourth, and fifth instars. Data are typical of two to three determinations for each instar. (D) br mRNA in fifth-instar nymphs after treatment with 2 μg of the JH mimic pyriproxifen within 12 h after ecdysis (typical of three determinations). The treated animals ecdysed to supernumerary sixth-instar nymphs after 4 days. (E) br mRNA expression in a fourth-instar nymph given 2 μg of precocene II early in the third instar to cause precocious metamorphosis at the end of the fourth stadium. The average duration of the fourth instar increased to 7.7 ± 0.1 days, and little, if any, br was detected at the end of the instar (typical of three determinations). Each cDNA sample also was separately tested for Of18S ribosomal RNA expression.
Because br expression was correlated with nymphal molts, but not with the adult molt, we asked whether gain or loss of a nymphal molt is accompanied by a corresponding gain or loss of br mRNA expression. JH is present throughout the nymphal stages of hemimetabolous insects but disappears in the final instar to allow adult differentiation (12, 13). JH treatment of Oncopeltus at the onset of the fifth instar prevents adult differentiation, resulting in a supernumerary sixth-stage nymph (14). In this situation, the duration of the fifth stadium was shortened, and br was up-regulated on the last day of the instar (Fig. 1D), a pattern similar to the earlier nymphal instars (compare with Fig. 1 A and B). To determine whether br up-regulation occurs only at nymphal molts, we used precocene to destroy the corpora allata, the source of JH, in fourth-instar nymphs (15, 16). After precocene treatment, fourth-stage nymphs molted to precocious adults, and their br expression resembled that of a fifth-instar nymph. That is, the duration of the instar expanded, and only trace levels of br mRNA were detected during the molt to the precocious adult (compare Fig. 1 E and C). Thus, br expression apparently requires the presence of JH, which is normally present only during a nymphal molt.
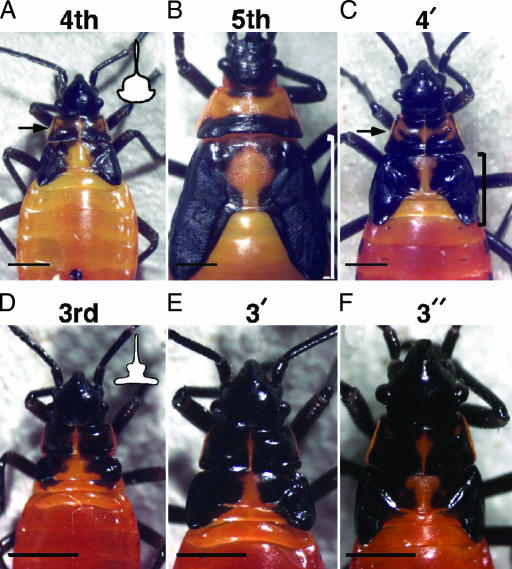
Because both JH and Of’br are absent during the adult molt, we wanted to determine whether the removal of Of’br from a nymphal molt was sufficient to redirect the molt to adult differentiation. Consequently, we injected Of’br dsRNA into staged nymphs to knock down Of’br expression. This technique has been used effectively to remove gene function from Oncopeltus nymphs (17). Of’br dsRNA injection into the fifth instar (n = 17) or latter half of the fourth instar (n = 14) had no significant effect on the subsequent molt. The former group molted into normal adults, whereas the latter molted to normal fifth-stage nymphs and then to adults. However, when Of’br dsRNA was injected during the first half of the fourth instar, 36 of 39 nymphs molted to a fifth instar of normal size but retained the pigmentation pattern characteristic of the fourth-instar nymph (the 4′ nymph; Fig. 2C). In addition, their wing pads were significantly smaller than those of control fifth-instar nymphs (compare bracket length in Fig. 2 B and C) and had the proportions of the fourth-instar wing pad (compare Fig. 2 C and A). To see whether the repeat of instar-specific characteristics was unique to the penultimate (fourth) nymphal stage, we also injected third-instar nymphs with dsRNA within the first 24 h after ecdysis. Eighteen of 34 nymphs molted to larger versions of the third instar (3′ nymph; Fig. 2E). The wing pads of these 3′ nymphs were smaller than those of control fourths, although the overall body size of the two groups was similar. The remaining nymphs that molted to normal fourth instars subsequently formed 4′ nymphs during their next molt. In contrast, all of the day-3 third-instar nymphs given Of’br dsRNA molted to normal fourth instars and then to 4′ nymphs (n = 11). As a negative control, we injected dsRNA made from the cricket Acheta domesticus br gene (Acd’br) into either third-instar (n = 12) or fourth-instar (n = 32) nymphs of various ages. All of these animals molted to normal fourth- or fifth-instar nymphs, respectively.
Fig. 2.
br dsRNA prevents changes in nymphal thoracic pigmentation. (A) Normal fourth-instar nymph. The prothorax is characterized by two melanized squares containing swirls of orange, unmelanized cuticle (arrow). (Inset) The shape outlined on the dorsal thorax resembles a handbell. (B) Normal fifth-instar nymph. The anterior two-thirds of the prothorax is entirely orange, whereas the posterior one-third is melanized. The wing pads (in brackets) are longer than in the fourth instar. (C) Effects of Of’br dsRNA given during the first half of the fourth instar on characteristics of the next stage nymph (a 4′ nymph). The prothoracic melanin in the fifth stadium consisted of two black squares with orange swirls (arrow). In addition, the wing pads were significantly smaller (brackets) than those of a normal fifth instar. The fourth-instar abdominal melanin pattern also was repeated in this 4′ nymph (data not shown). (D) Normal third-instar nymph. The two prothoracic tergal squares either were completely melanized (65%; n = 29) or had small spots of orange in the center of each square (35%). The third instar is also distinguished by the shape outlined in melanin on the dorsal thorax, which resembles the profile of a candle and candlestick (Inset). (E) Effect of injection of Of’br dsRNA in the third instar. The resultant 3′ nymph resembled the third instar; the prothoracic squares were filled or nearly filled, and the shape on the dorsal thorax more closely resembled the outline of a candlestick. (F) Effect of two doses of concentrated (8–9 μg/μl) Of’br dsRNA at the onsets of the third and 3′ instars. The resultant 3″ nymph was the size of the fifth instar but retained the third-instar-type prothoracic patterns and wing pad morphology. (Scale bars, 1 mm.)
We used real-time PCR to show that Of’br dsRNA knocked down Of’br transcript levels. In control nymphs, br transcripts on day 4 of the fourth instar measured 4.1- and 0.9-fg relative mRNA in two different control samples. In fourth-instar nymphs that were fated to form 4′ nymphs because of Of’br dsRNA treatment in the late third instar, br transcripts were undetectable on either day 4 (n = 2) or day 5 (n = 1).
These Of’br dsRNA knock-down experiments suggest that Of’br is required for the changes in cuticle character, or heteromorphoses, that occur between nymphal stages. If this explanation is correct, then loss of Of’br in consecutive instars should result in further repetition of the initial nymphal pattern. To test this prediction, we generated 3′ nymphs by Of’br dsRNA injection at the onset of the third instar and then gave a second dose of Of’br dsRNA at the onset of the 3′ instar. Five of eight 3′ nymphs that were injected at the onset of the 3′ instar molted to a fifth stadium but still retained the pigmentation pattern characteristic of a third instar (Fig. 2F). In addition, the wing pads of these 3″ nymphs were extremely reduced (Fig. 2F) compared with the normal fifth-instar pads or those of the 4′ nymphs (Fig. 2F).
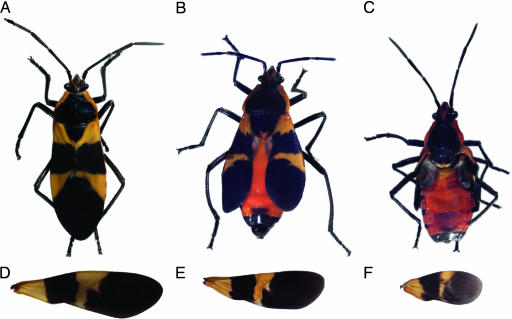
Although the premature knock-down of Of’br during nymphal life blocked the morphological transition from one nymphal instar to the next, it did not alter the number of nymphal instars or the transition to the adult. For example, nymphs that went through a progression series such as 3–4-4′ or 3–3′–4 (instead of 3–4-5) subsequently underwent an adult molt. Many of these individuals (46 of 59) died without completely shedding the last nymphal cuticle. Of the animals that were able to emerge (n = 13), all had typical adult pigmentation and body morphology (compare Fig. 3B and C with A), except that their wings were significantly reduced and often blistered and folded. Those receiving injections of Of’br dsRNA in the third instar were more affected than those injected during the fourth instar (Fig. 3 E and F compared with D). Although the disappearance of Of’br is associated with the nymphal-to-adult molt, these data suggest that the removal of Of’br alone is not sufficient to cause the formation of the adult. Rather, the function of br in these more basal insects appears to reside in orchestrating transitions within the nymphal series itself.
Fig. 3.
Wings of normal adults and those given br RNA interference as nymphs. (A) A normal, uninjected adult Oncopeltus has wings that project past the posterior end of the abdomen. (B) A typical adult formed from a 4′ nymph produced by injection of Of’br dsRNA into an early fourth-instar nymph. The wings were significantly reduced and were often held out to the side. Eighty-three percent (n = 47) of the 4′ nymphs died during the next molt as pharate adults. Of these, 8 of 39 had ruptured dorsal thoracic (notal) cuticles. (C) A typical adult produced after injection of Of’br dsRNA into a third-instar nymph that subsequently underwent molts to a 3′ and then a 4′ nymph. Adult pigmentation and morphology was normal, but the wings were smaller and held out to a greater extent than those of adults produced from a normal fifth-instar or 4′-instar nymph. (D–F) Typical wings of a normal adult (D), an adult that followed a 4–4′ nymphal series (E), and an adult that followed a 3–3′–4 nymphal series (F).
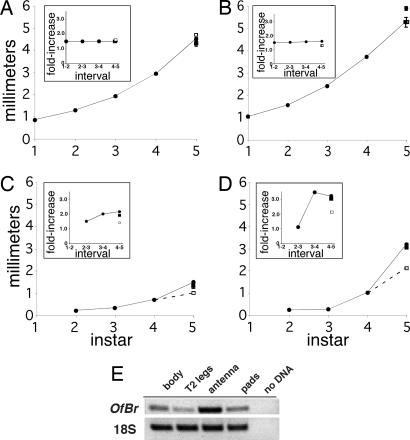
In contrast to holometabolous insects, where differential growth during postembryonic life is typically restricted to metamorphosis (18), differential growth in hemimetabolous insects occurs progressively through the nymphal stages and is largely restricted to the wing pads. When we followed the growth of the legs and antennae after injection of Of’br dsRNA into early fourth-instar nymphs, we found that these appendages, which normally grow at a constant rate (1.5-fold in each molt), were unaffected by Of’br knock-down (Fig. 4A and B Insets). In contrast to the legs and antennae, the wing pads grow during the fourth instar by a factor of 2.0 and 3.0 at the base and diagonal, respectively (Fig. 4 C and D Insets). Knock-down of Of’br at the onset of the fourth instar caused a decrease in this growth, so that the respective growth ratios were 1.5 and 2.0. Therefore, Of’br has no effect on the isometric growth of the legs and antennae but is critical for the anisometric growth of the wings. In its absence, wing growth becomes more isometric, like the remainder of the body. We were able to detect Of’br expression in all nymphal tissues (Fig. 4E).
Fig. 4.
Effects of Of’br dsRNA on growth of the antenna, leg, and wing. (A and B) Both antennae (A) and legs (B) of uninjected nymphs (filled circles) grew steadily throughout the nymphal instars and increased by a factor of 1.5 at each molt (Insets). “Interval” in Insets indicates the growth that occurs between the two indicated stadia. Injection into early fourth-instar nymphs of dsRNA made from Acd’br (filled squares) or Of’br (open squares) had no effect on the growth trajectory of legs or antennae. (C and D) Effect of Of’br dsRNA on the width of the wing pad base (C) and the wing pad diagonal (D). Standard error is shown where the width of the bars is greater than the data points. Between 23 and 31 measurements were made for each Of’br dsRNA mean; between 5 and 7 measurements were performed for each Acd’br dsRNA mean. (E) Ethidium gel showing RT-PCR of Of’br mRNA in day-4 fourth-instar body, T2 legs, antennae, and wing pads. Data are typical of two determinations.
Discussion
These studies show that in the direct-developing insect Oncopeltus, br is expressed throughout nymphal life with high levels at the times of the molt and then disappears during the formation of the adult. Premature removal of br by use of dsRNA prevented the heteromorphic transitions that normally occur during nymphal molts without interfering with the molt itself. Thus, an ancestral role of br is to confer mutability that provides for differential growth between nymphal instars.
After knock-down of br expression in the nymph, wing pad growth continued but became more isometric, and its proportions from the previous stage were repeated. This aspect of br function appears to be retained during metamorphosis of the fly, because the wings of a br allele (lacking the Z2 isoform of br) are defective in their morphology and are shortened and “broad” (19–21), a phenotype similar to that seen in the wings of Oncopeltus that lack Of’br during the third or fourth nymphal stages (Fig. 3 E and F). In contrast to the progressive role that Of’br plays through successive nymphal molts, the function of br in the Drosophila wing disk is restricted to the final larval instar as the wing disk translates patterning information to produce the pupal wing (22). Therefore, the ancestral function of br, to support progressive anisometric growth of the developing wing pad over a number of instars, has been restricted to the premetamorphic period in the last larval instar of holometabolous insects.
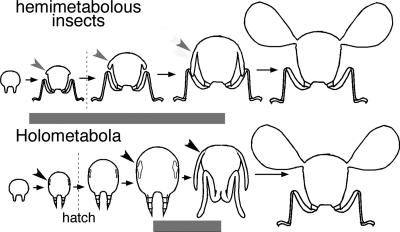
The homology of the pupal stage to a developmental stage of hemimetabolous insects has been a recurring issue among naturalists (23). During the latter half of the 20th century, the prevailing theory considered the pupal state to be derived from the final nymphal stage (24). An older idea considered pupal development to be more akin to the events that occur during embryonic development of direct developers (25, 26), which has recently been expanded into the pronymph hypothesis (27). Our data support this older idea. In both crickets and milkweed bugs, br mRNA is present during the latter half of embryonic development (28). In these hemimetabolous embryos, this period is characterized by differential growth as the embryo progresses from the phylotypic germ band stage to a miniature version of the adult (27, 29). In contrast, br mRNA is not present in the epidermis of holometabolous embryos (30), and the growth during the corresponding phase of embryogenesis is more isometric (31). This isometric growth then persists through postembryonic development until br reappears at the larval–pupal transition to help direct the differential growth needed to generate the adult form. Because br is required for postembryonic differential growth in the hemimetabolous insect Oncopeltus, we suggest that metamorphosis emerged in insects as br expression and its regulation of differential growth became transposed from late embryonic development to the penultimate postembryonic molt (Fig. 5).
Fig. 5.
Model depicting how changes in br expression follow the evolution of metamorphosis in insects. Each image is a cross section of the second thoracic segment. Gray arrowheads denote wing pads in nymphs, and black arrowheads indicate wing imaginal discs. The time of hatching is indicated by a vertical dashed line. In hemimetabolous insects, br (horizontal bar) is present throughout the nymphal stages. This expression correlates with a bout of differential growth that occurs late in embryogenesis, as well as the progressive differential growth of the wings that occurs during the nymphal stages. In holometabolous insects, the latter half of embryonic development is largely isometric, and this isometric growth phase persists until metamorphosis, when br permits differential growth of imaginal structures.
br may confer mutability to insect life history stages through its BTB/POZ domain, a motif implicated in the establishment and maintenance of complex differentiated states (32). Many BTB-containing proteins regulate complex states through chromatin deacetylation, thereby affecting the access of subsequent transcription factors to response elements (32–34). In the context of nymphal changes in Oncopeltus, the loss of Br may prevent the access of transcription factors to response elements that are needed for change from one stage to the next.
Materials and Methods
Cloning.
The br gene encodes a complex locus in which a BTB-containing “core” domain is fused to one of four C2H2 zinc fingers (21).
Acd’br.
A segment of Acd’br was cloned by PCR from genomic DNA by using degenerate primers to the BTB domain (35). RNA from embryos at the onset of katatrepsis, or dorsal closure, was used to determine the 5′ and 3′ ends of the cDNA, respectively, by using the SMART RACE cDNA Amplification Kit (Clontech). Analysis of the PCR and RACE products predicts a 507-aa protein (GenBank accession no. DQ176003). The 55-aa zinc finger region contains two C2H2 zinc fingers that show highest identity to the Z1 fingers found in Anopheles and Aedes Broad (63% in each case). The Acd’br BTB domain has greatest identity with that of Aedes (94%) and Anopheles (92%).
Of’br.
A segment of Of’br was also cloned by PCR from genomic DNA by using the primers described for Acheta. The SMART RACE cDNA Amplification Kit was again used to acquire the 5′ and 3′ ends of the transcript from mRNA isolated at midembryogenesis. The predicted 118-aa Oncopeltus BTB domain showed high identity with that of other insects: 90% with Drosophila melanogaster and the lepidopteran species, Bombyx mori and Manduca sexta, and 92% with the cricket A. domesticus.
RNA Isolation.
Individuals were staged from ecdysis, and two individuals were homogenized at 24-h intervals thereafter. After an initial extraction with TRIzol (Invitrogen), a second phenol-chloroform extraction was performed. This homogenate was DNase-treated and extracted again with phenol-chloroform.
RT-PCR.
RNA was subjected to further DNase treatment just before cDNA synthesis, as described in the product literature for Promega RNase-free DNase. cDNA was made from 1 μg of total RNA and random hexamers with the cDNA Synthesis Kit (Fermentas, Hanover, MD). To detect Br BTB and core expression in Oncopeltus cDNA, we used the primers Of’br12 (5′-CACCGAAGGCAGAAATGTTG-3′) and Of’br15 (5′-ACGATTAAACGACGGCCAAG-3′). The PCRs were run at 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s for 33 or 34 cycles. To normalize the reactions, we amplified each cDNA reaction with primers designed to amplify 18S ribosomal RNA [Of18S-B (5′-ATGGAACAGGACCTTGGTTC-3′) and Of18S-D (5′-GTATCTGATCGCCTTCGAAC-3′)] at the same conditions as above for Of’br12 and Of’br15, except that the temperature was 64°C and only 17 or 18 cycles were necessary. (When 33 cycles were used to detect Of’br, 17 cycles were used to detect Of18S mRNA. Likewise, when 34 cycles were used to detect Of’br, 18 cycles were used to detect 18S mRNA.) This measure was taken to control for variation in Taq activity.
No genomic DNA contamination was found in sham cDNA reactions made without reverse transcriptase when we screened ≈30% of the RNA samples. Furthermore, “no-DNA” controls were included with each PCR to ensure that the observed amplification was due to cDNA. Each time point was tested twice, from separate pools of RNA. The PCR products were run out on 2% gels and stained with ethidium bromide. Photographs of the gels were scanned and inverted by using photoshop (Adobe Systems, San Jose, CA).
dsRNA Injections.
Separate RNA strands were made from plasmids containing the fragments of Of’br or Acd’br by using the MEGAscript kit (Ambion, Austin, TX), and the sense and antisense strands were annealed as described by Hughes and Kaufman (36). Nymphal dsRNA injections were performed with a Hamilton syringe with a 32-gauge needle, as described by Liu and Kaufman (37) for parental dsRNA. Nymphs were injected into the ventral abdomen until they became distended (D. Angelini, personal communication). Unless otherwise indicated, nymphs were injected with 4–10 μg of dsRNA in a volume of 2–5 μl made from one of the following: (i) 125 bp of the 5′ UTR plus 600 bp of 5′ coding region of Of’br BTB domain and partial core sequence, (ii) the 151-bp region of the Of’br BTB domain, (iii) 218 bp of the Of’br 5′ UTR plus the first 107 bp of the BTB domain, (iv) an equimolar mixture of (ii) and (iii), (v) 151 bp of the Acd’br BTB domain, or (vi) 124 bp of the 5′ UTR plus ≈250 bp of the Acd’br BTB domain. We could not detect any difference in activity among the different pieces of Of’br used for RNA interference experiments, and we then used them interchangeably.
Real-Time PCR.
The primers Of’br10 (5′-CCTCTTCGTCTCCTGATATG-3′) and Of’br15 (5′-ACGATTAAACGACGGCCAAG-3′) were used in a Smart Cycler (Cepheid, Sunnyvale, CA) with LightCycler-FastStart DNA Master SYBR Green I (Roche Applied Science, Indianapolis) under the following conditions: 95°C for 600 s and then 95°C for 15 s, 64°C for 6 s, and 72°C for 6 s repeated 45 times with 3 mM Mg and a 0.5 μM concentration of each primer. One-twentieth of a cDNA reaction made from 1 μg of total RNA was used in each PCR. 18S ribosomal RNA was used to normalize the reaction with the primers Of18S-B and Of18S-D, except that for this reaction, the annealing temperature was 65°C and the Mg concentration was 4 mM. After normalization, amounts were calculated by using a standard curve made from known concentrations of Of’br plasmid DNA.
Morphometrics.
Ecdysed nymphal cuticles were hardened in 70% ethanol overnight, mounted in Fluoromount (Southern Biotechnology Associates) and photographed at ×4 magnification on a Nikon Optiphot microscope using a digital camera (Sony, Tokyo) and Apple video software (Macintosh). Cuticle dimensions were measured by using nih image software. To measure the wing pad diagonal, the most anterior corner of the wing pad and opposing distalmost point of the wing pad were used.
Drug and Hormone Treatment.
Two micrograms of Precocene II (Sigma) was applied to the tergum in 0.2 μl of HPLC-grade acetone (Aldrich) with a 10-μl Hamilton syringe to nymphs within 6 h of ecdysis to the third instar. For JH treatment, 2 μg of pyriproxifen (a JH mimic; a gift from H. R. Oouchi, Sumitomo Chemical, Osaka) was applied in 2 μl of HPLC-grade acetone to nymphs within 8 h of ecdysis to the fifth instar.
Acknowledgments
We thank Prof. Thomas Kaufman (Indiana University, Bloomington) and members of his laboratory, particularly David Angelini and Paul Liu, for the Oncopeltus stock and for encouragement and for sharing methods and materials; Hans Kelstrup for assistance with the precocene experiment; and Dr. Jason Hodin for insightful comments on the manuscript. This work was supported by National Science Foundation Grant IBN-9904959 (to J.W.T. and L.M.R.) and National Institutes of Health Grant GM60122 (to L.M.R.).
Abbreviations
- br
broad
- Of’br
br from Oncopeltus fasciatus
- JH
juvenile hormone
- BTB
Broad–Tramtrack–Bric-a-brac
- Acd’br
Acd’brbr from Acheta domesticus.
Footnotes
References
- 1.Heyland A., Hodin J., Reitzel A. M. BioEssays. 2004;27:64–75. doi: 10.1002/bies.20136. [DOI] [PubMed] [Google Scholar]
- 2.Kukalova-Peck J. In: The Insects of Australia: A Textbook for Students and Research Workers. 2nd Ed. Commonwealth Scientific and Industrial Research Organization, editor. Ithaca, NY: Cornell Univ. Press; 1991. pp. 141–179. [Google Scholar]
- 3.Karim F. D., Guild G. M., Thummel C. S. Development (Cambridge, U.K.) 1993;118:977–988. doi: 10.1242/dev.118.3.977. [DOI] [PubMed] [Google Scholar]
- 4.Bayer C., von Kalm L., Fristrom J. In: Metamorphosis: Postembryonic Reprogramming of Gene Expression in Amphibian and Insect Cells. Gilbert L., Atkinson B., Tata J., editors. San Diego: Academic; 1996. pp. 321–361. [Google Scholar]
- 5.Zhou B., Hiruma K., Shinoda T., Riddiford L. M. Dev. Biol. 1998;203:233–244. doi: 10.1006/dbio.1998.9059. [DOI] [PubMed] [Google Scholar]
- 6.Zhou B., Riddiford L. M. Dev. Biol. 2001;231:125–137. doi: 10.1006/dbio.2000.0143. [DOI] [PubMed] [Google Scholar]
- 7.Zhou X., Riddiford L. M. Development (Cambridge, U.K.) 2002;129:2259–2269. doi: 10.1242/dev.129.9.2259. [DOI] [PubMed] [Google Scholar]
- 8.Kiss I., Szabad J., Major J. Mol. Gen. Genet. 1978;164:77–83. [Google Scholar]
- 9.Kiss I. Nature. 1976;262:136–138. doi: 10.1038/262136a0. [DOI] [PubMed] [Google Scholar]
- 10.Uhlirova M., Foy B. D., Beaty B. J., Olson K. E., Riddiford L. M., Jindra M. Proc. Natl. Acad. Sci. USA. 2003;100:15607–15612. doi: 10.1073/pnas.2136837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddiford L. M. Arch. Insect Biochem. Physiol. 1996;32:271–286. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<271::AID-ARCH2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 12.Bruning E., Saxer A., Lanzrein B. Int. J. Invert. Reprod. Dev. 1985;8:269–278. [Google Scholar]
- 13.Riddiford L. M. In: Advances in Insect Physiology. Evans P. D., editor. Vol. 24. London: Academic; 1994. pp. 213–274. [Google Scholar]
- 14.Lawrence P. A. Dev. Biol. 1969;19:12–40. doi: 10.1016/0012-1606(69)90068-2. [DOI] [PubMed] [Google Scholar]
- 15.Bowers W. S., Ohta T., Cleere J.-S., Marsella P. A. Science. 1976;193:542–547. doi: 10.1126/science.986685. [DOI] [PubMed] [Google Scholar]
- 16.Bowers W. S. Gen. Comp. Endocrinol. 1979;37:156–166. doi: 10.1016/0016-6480(79)90103-5. [DOI] [PubMed] [Google Scholar]
- 17.Angelini D. R., Kaufman T. C. Dev. Biol. 2004;286:57–88. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Williams C. In: Insect Biology in the Future. Locke M., Smith D. S., editors. New York: Academic; 1980. pp. 369–383. [Google Scholar]
- 19.Waddington C. H. Proc. Natl. Acad. Sci. USA. 1939;25:299–307. doi: 10.1073/pnas.25.7.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waddington C. H. J. Genet. 1940;41:75–139. [Google Scholar]
- 21.DiBello P. R., Withers D. A., Bayer C. A., Fristrom J. W., Guild G. M. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emery I. F., Bedian V., Guild G. M. Development (Cambridge, U.K.) 1994;120:3275–3287. doi: 10.1242/dev.120.11.3275. [DOI] [PubMed] [Google Scholar]
- 23.Sehnal F., Svacha P., Zrzavy J. In: Metamorphosis. Gilbert L. I., Tata J. R., Atkinson B. G., editors. San Diego: Academic; 1996. pp. 3–58. [Google Scholar]
- 24.Hinton H. E. Proc. R. Entomol. Soc. London; 1963. pp. 77–85. [Google Scholar]
- 25.Lubbock J. On the Origin and Metamorphoses of Insects. London: Macmillian; 1883. [Google Scholar]
- 26.Berlese A. Redia. 1913;9:121–136. [Google Scholar]
- 27.Truman J. W., Riddiford L. M. Nature. 1999;401:447–452. doi: 10.1038/46737. [DOI] [PubMed] [Google Scholar]
- 28.Erezyilmaz D. Ph.D. thesis. Seattle: University of Washington; 2004. p. 94. [Google Scholar]
- 29.Erezyilmaz D. F., Riddiford L. M., Truman J. W. Dev. Genes Evol. 2004;214:313–323. doi: 10.1007/s00427-004-0408-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhou B. Ph.D. thesis. Seattle: Univ. of Washington; 2000. [Google Scholar]
- 31.Wheeler D. E., Nijhout F. H. Amer. Nat. 1996;148:40–56. [Google Scholar]
- 32.Albagli-Curiel O. Oncogene. 2003;22:507–516. doi: 10.1038/sj.onc.1206152. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad K. F., Engel C. K., Prive G. G. Proc. Natl. Acad. Sci. USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad K. F., Melnicik A., Lax S., Bouchard D., Liu J., Kiang C. L., Mayer S., Takahashi S., Licht J. D., Prive G. G. Mol. Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 35.Zollman S., Godt D., Prive G. G., Couderc J.-L., Laski F. A. Proc. Natl. Acad. Sci. USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes C. A., Kaufman T. C. Development (Cambridge, U.K.) 2000;127:3683–3694. doi: 10.1242/dev.127.17.3683. [DOI] [PubMed] [Google Scholar]
- 37.Liu P. Z., Kaufman T. C. Development (Cambridge, U.K.) 2004;131:1515–1527. doi: 10.1242/dev.01046. [DOI] [PubMed] [Google Scholar]