Abstract
Embryonic stem (ES) cells hold great promise for the future of medicine. To elucidate the molecular mechanisms that control ES cell self-renewal and differentiation, a comprehensive knowledge of the molecules involved in these processes is required. Here we describe an effective approach for genomewide identification of functionally active genes in ES cells. This approach combines genetic screens based on cDNA libraries with microarray detection methods to permit high-throughput functional analyses. We implement this strategy to identify genes whose overexpression can maintain phenotypic properties of undifferentiated mouse ES cells under differentiation-inducing conditions, specifically in the absence of leukemia inhibitory factor. The identified genes encode a variety of regulatory proteins whose function in ES cells was previously unknown. Moreover, our approach is capable of detecting genes whose overexpression promote differentiation or cell death. Overall, our studies establish a methodology for highly sensitive identification of genes that confer particular phenotypes on ES cells.
Keywords: cDNA library, differentiation, microarray, phenotype, self-renewal
Mouse and human ES cells can be propagated extensively in culture as homogeneous self-renewing populations (1). These cells are widely used for studies of developmental processes and thought to provide a system for design of novel transplantation therapies (2, 3). In vitro, murine ES cells are maintained in the presence of leukemia inhibitory factor (LIF), a growth factor activating the gp130/Stat3 signaling pathway (4). Undifferentiated ES cells rapidly proliferate, form tight colonies with a characteristic morphology, possess high alkaline phosphatase activity levels, and preserve their multilineage differentiation capacities. Upon removal of LIF, ES cells undergo differentiation associated with decreased proliferation rates and morphological changes. Previous studies uncovered various aspects of ES self-renewal and differentiation (5–11). In addition, genomewide analyses have identified specific sets of transcripts and splicing variants generated in ES and other stem cell populations (12–14). However, the current understanding of molecular mechanisms regulating self-renewal and differentiation is far from complete (15). For example, only two genes, the transcription factors Nanog and Myc, have been shown to be sufficient for maintaining undifferentiated ES cells in the absence of LIF (5, 6, 16).
Gain-of-function genetic screens are a powerful method to identify genes sufficient to confer a particular cellular phenotype (17). Such screens have a long history in studies of microorganisms. Gain-of-function screens generally begin with an introduction of cDNA expression libraries into cells, followed by specific selective regimens. In mammalian cells, these screens usually require iterative rounds of selection and yield only one or few functional gene products that are identified by clone sequencing. To facilitate the isolation of small numbers of gene products amenable to direct sequencing, harsh selection conditions are used to minimize background noise. As a result, most gain-of-function screens identify only the “most potent” phenotype-conferring genes. For example, the only currently described gain-of-function screen in mouse ES cells identified a single gene encoding the transcription factor Nanog (5). Moreover, the clone sequencing method is incapable of negative detection, complicating the identification of gene products that promote differentiation or apoptosis (18). These limitations prevent the comprehensive identification of functional genes in mammalian systems.
In this study, we applied the microarray technology as a method of large-scale parallel analysis to conduct comprehensive gain-of-function screens in ES cells (Fig. 1A). Our approach was designed to simultaneously monitor the activity of all gene products in a cDNA library as they function to mediate a given phenotype. We implemented this approach to identify genes whose overexpression is sufficient to maintain undifferentiated mouse ES cells in the face of differentiation-inducing conditions, specifically in the absence of LIF. We also show that our approach is capable of negative detection and can identify gene products that promote differentiation or cell death.
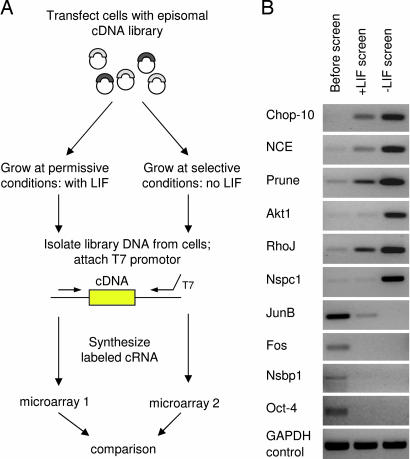
Fig. 1.
Microarray-assisted gain-of-function genetic screen in ES cells. (A) Schematic diagram for comprehensive functional analysis. The ES cells were transfected with a cDNA library, selected and propagated at permissive or selective conditions in the presence or absence of LIF, respectively. The cDNA insert populations before and after the screen were labeled and analyzed with microarrays. (B) Confirmation of microarray results. Changes in cDNA representation detected with microarrays were confirmed by using PCR with gene-specific primers and libraries before and after the screen as templates. GAPDH primers were used as a control.
Results
Microarray-Assisted Gain-of-Function Screens.
We introduced an episomal cDNA library constructed from ES cells into the E14/T mouse ES cell line (Fig. 1A; for complete experimental details, see Materials and Methods; see also Supporting Materials and Methods, which is published as supporting information on the PNAS web site). This cell line constitutively expresses the polyoma virus large T protein and can stably maintain episomal vectors carrying the polyoma origin of replication (4, 19). Transfected cells were divided into two pools and propagated for 18 days in the presence or absence of LIF: permissive and selective conditions, respectively. The decreased proliferation rate associated with differentiation served as a selection criterion in our genetic screens. We reasoned that ES cells harboring gene products that maintain rapid proliferation rates would progressively increase their relative representation in the cell population propagated without LIF. Similarly, we expected that cells harboring differentiation-inducing gene products would undergo progressive representational decreases during culture in the presence of LIF. To simultaneously identify all gene products that confer these phenotypes, we used microarrays. Episomal libraries were isolated from transfected ES cells after selection and used to prepare material for hybridization to microarrays. For analysis of obtained microarray data, we applied a 5-fold threshold to ensure identification of genes that undergo significant representational changes during selection. This threshold is higher than the 2-fold value generally applied in the microarray experiments and considered biologically significant. It is also justified statistically: Only ≈1% of probes were identified as “increased” by using this value. We identified >560 probes (representing 499 genes) whose intensity increased at least 5-fold on microarrays hybridized with material from the “−LIF” conditions, relative to “+LIF” conditions (Table 2, which is published as supporting information on the PNAS web site). Probes representing Nanog and Oct4, whose overexpression is known to promote self-renewal and differentiation (5, 7), respectively, showed expected changes in their signal intensity (Tables 3 and 4, which are published as supporting information on the PNAS web site; microarray data). Intensity values for housekeeping control genes such as Gapdh did not show significant changes. Analysis of replicate experiments indicated reproducibility of screens conducted at the same conditions (Fig. 4, which is published as supporting information on the PNAS web site). Correlation coefficients of all microarray values obtained at the same (both permissive) or different (permissive and selective) conditions were 0.96 and 0.67, respectively. We used PCR with gene-specific primers designed toward 3′ of coding regions to confirm microarray-detected representational changes above the selected 5-fold threshold. All of the tested genes were confirmed, and the examples are shown in Fig. 1B.
Identification of Genes that Rescue ES Cell Phenotype in the Absence of LIF.
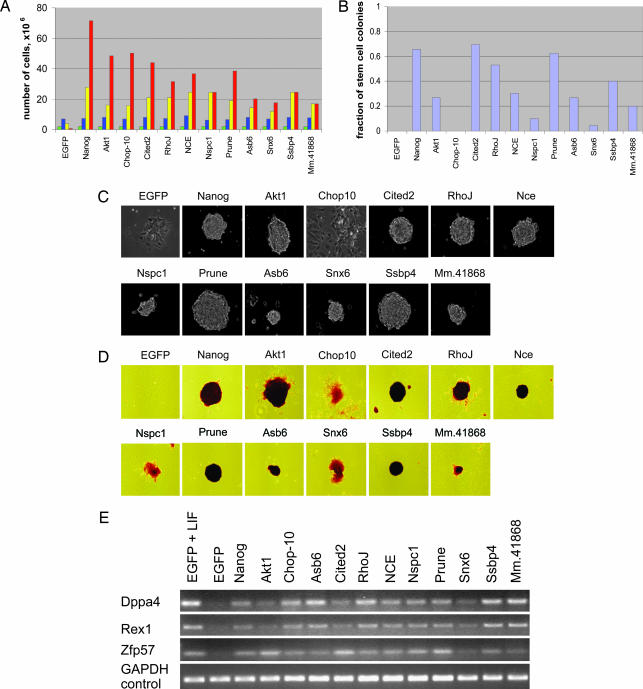
From the set of probes whose intensity values increased in the absence of LIF, we selected 25 genes for individual confirmation. No previous knowledge was used in selection of these genes. Full-length cDNAs derived from these genes were introduced individually into the episomal vector, and each was tested for its ability to maintain growth of ES cells in the absence of LIF. We further tested whether the identified genes can rescue other phenotypic properties of undifferentiated ES cells such as colony formation at a clonal density, characteristic morphology, alkaline phosphatase activity, and the expression of pluripotency markers. Cells expressing Nanog and EGFP served as positive and negative controls. In 11 of 25 tested cases (≈40%), the identified genes were sufficient to at least partially “rescue” ES cell proliferation rates in the absence of LIF (Fig. 2 and Table 1). Overexpression of these genes maintained at least a 10-fold higher number of cells after 9 days without LIF, in comparison with the EGFP control. In comparison, these genes do not render a significant growth advantage in the presence of LIF. The identified genes encode an array of regulatory proteins and show different levels of rescue activity in the various assays. For example, the transcription factor Cited2 and signaling protein Prune were sufficient to maintain all tested phenotypic properties at levels observed with the positive Nanog control. On the other hand, the transcription regulator Chop10 maintained robust cell proliferation rates; however, a large fraction of transfected cells displayed differentiated morphologies. The weakly characterized Polycomb factor Nspc1 and intracellular transport regulator Snx6 partially rescued cell growth; however, their overexpression was not sufficient to allow colony formation at a clonal density. The identified genes also were capable of promoting the continued expression of the ES cell pluripotency markers Rex1, Dppa4, and Zfp57 (20, 21). Expression levels of these markers decrease in the absence of LIF.
Fig. 2.
Confirmation of function for identified gene products. (A) Cumulative number of cells was determined for genes that rescue ES cell proliferation in the absence of LIF. After transfection, ES cells were propagated in the absence of LIF, passaged, and counted every 3 days. Nanog and EGFP served as positive and negative controls, respectively. Colored bars indicate the following: green, day 0; blue, day 3; yellow, day 6; red, day 9. (B) Fraction of stem cell colonies formed by transfected cells at a clonal density after 7 days in the absence of LIF. (C and D) Morphology (C) and alkaline phosphatase staining (D) of colonies formed at a clonal density after 7 days in the absence of LIF. (E) The functionally identified genes rescue expression of ES cell molecular markers after 4 days in the absence of LIF, as determined by RT-PCR. Cells transfected with EGFP and Nanog served as a negative and positive control, respectively.
Table 1.
Genes that rescue ES cell phenotypic properties
| Genes | Rescued ES properties | ||
|---|---|---|---|
| Rapid growth | Morphology | Alkaline phosphatase | |
| Akt1/PKB, Mm.6645, protein kinase | + | + | + |
| Chop-10, Mm.110220, DNA damage-inducible transcript 3 (Ddit3) | + | − | +/− |
| Cited2, Mm.272321, Cbp/p300-interacting transactivator 2 | + | + | + |
| RhoJ, Mm.27467, Ras homolog gene family, member J | + | + | + |
| NCE, Mm.337238, NEDD8-conjugating enzyme | + | + | +/− |
| Nspc1, Mm.12261, nervous system polycomb 1 | + | +/− | +/− |
| Prune, Mm.14155, Prune homolog (Drosophila) | + | + | + |
| Asb6, Mm.27656, ankyrin repeat and SOCS box-containing protein 6 | +/− | +/− | +/− |
| Snx6, Mm.28240, sortin nexin 6 | + | +/− | +/− |
| Ssbp4, Mm.6667, single-stranded DNA-binding protein 4 | + | + | + |
| Mm.41868, hypothetical protein | +/− | +/− | +/− |
To compare the efficiency and sensitivity of our microarray-based approach with the traditional individual clone sequencing methods, we randomly picked and sequenced 50 clones from the cDNA library isolated from ES cells cultured without LIF. None of the 11 functionally confirmed genes was found among these sequenced clones. These observations clearly demonstrate that our microarray-based approach can identify functional genes at a level of sensitivity not possible by using traditional clone-by-clone sequencing approaches.
To test the functional specificity of the identified genes in the ES cell context, we analyzed the activities of Bmi1 and Cited1. These genes are closely related homologues of Nspc1 and Cited2, respectively. Neither Bmi1 nor Cited1 was capable of rescuing the properties of undifferentiated ES cells in the absence of LIF. In fact, the overexpression of Cited1 and Bmi1 induced an opposite phenotypic effect, differentiation, both with and without LIF (Fig. 5, which is published as supporting information on the PNAS web site). These data suggest that the observed ES cell functional activity is specific for individual members of multigene families.
Transcriptional Regulation of the Identified Genes.
It is reasonable to expect that at least some of the genes that confer LIF independence are regulated by LIF signaling at the transcriptional level. To test this hypothesis, we measured changes in their expression levels in response to withdrawal of LIF during a 5-day time period (Fig. 6, which is published as supporting information on the PNAS web site). At the used conditions, expression levels of Socs-3, a known downstream target of the LIF/Stat3 signaling pathway, rapidly decreased (22). In comparison, the expression levels of the functionally identified genes remained unchanged. These data indicate that the identified genes are not activated by LIF at the transcriptional level. Transcription of Nanog also was shown to be independent on LIF (5).
Identification of Genes that Induce Differentiation.
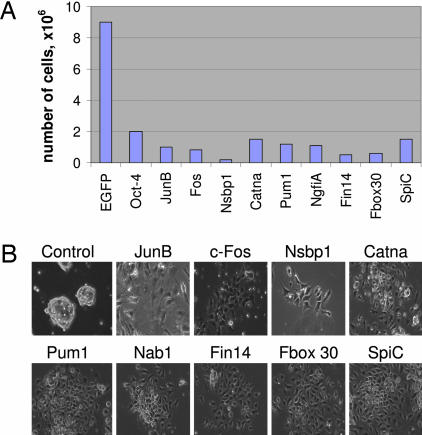
The microarray analysis also has identified >3,000 probes (representing 2,953 genes) whose intensity decreased >5-fold in the library harboring ES cells propagated at the permissive, LIF-containing conditions. From this set, we have selected 20 genes for functional confirmation. Our functional analyses found that nine of the selected genes induced the expected phenotypic effect, differentiation, when overexpressed in ES cells (Fig. 3 and Table 4). Cells transfected with these genes decreased their proliferation rate and underwent typical morphological changes associated with differentiation. The identified genes encode various regulatory proteins, including transcription factors, RNA-binding factors, and signaling molecules. Among the transcription factors, we found the well known protooncogenes JunB and c-Fos, transcription repressor Nab1, and ETS family member SpiC. We also identified RNA-binding proteins, the ATP-dependent helicase Ddx3, and translation regulator Pumilio 1. Other identified genes included Catenin α 1, nucleosome-binding protein Nsbp1, and ubiquitination regulator Fbxo 30. These results demonstrate that our microarray-assisted approach also is capable of negative detection and can identify genes that induce differentiation.
Fig. 3.
Confirmation of function for genes promoting differentiation or cell death. (A) Number of cells was determined after transfected ES cells were propagated in the presence of LIF for 6 days. The EGFP construct served as a control. (B) Cell morphology of transfected cells.
Sources of Noise.
Although PCR analyses using gene-specific primers confirmed microarray-detected changes (Fig. 1B), the rate of functional confirmation for individual genes currently achieved in our experiments is ≈40% (Fig. 2). The following observations suggest that this experimental “noise” may be, at least partially, due to differences between cDNAs representing identified genes in the library used in the screen and cDNAs whose function was tested individually in our confirmation experiments. For example, the microarray data showed a significant increase in representation of the IFN Response Factor 1 (IRF1) gene, which was further confirmed by PCR with specific primers. A 5′ truncated cDNA corresponding to an amino terminus-deficient version of this protein was identified by sequencing of clones isolated from cells after the screen. Surprisingly, overexpression of a full-length IRF1-encoding cDNA induced cell death, opposite to the expected phenotype, whereas cells carrying the 5′ truncated construct proliferated normally. Similarly, truncated transcripts whose functions strongly differ from the full-length genes were detected in gain-of-function screen studies previously published by other groups (23). As another example, constitutive expression of a Nanog cDNA with a truncated 3′ UTR induced higher rates of proliferation than the full-length version. A possible explanation is the presence of repetitive B2 elements in the 3′ UTR that can alter transcription from our vector (5).
Because of incomplete cDNA synthesis reactions, cDNA libraries usually contain a significant number of clones representing truncated transcripts. Approximately 50% of the clones in the particular library used in our studies are full-length, and a similar percentage was detected among clones sequenced after selection. Alternative splicing and alternative polyadenylation affect >50% of mammalian genes, including those expressed in ES cells (14, 24). Because microarray probes generally cover the 3′ portions of mRNAs and, therefore, cannot effectively distinguish between full-length, truncated, or alternative isoforms, these phenomena may contribute to false positives observed in our studies. Additional possible sources of false positives also may include, for example, artificial fluctuations in the clone representation during propagation of transfected cells in culture. However, to preserve the clone representation, the number of transfected cells used in our studies was 100-fold higher than the number of independent recombinants present in the starting cDNA library.
We expect that employing full-length and completely characterized cDNA libraries will significantly improve the confirmation rate in our approach, so that each microarray-detected change in cDNA representation could be considered as a significant indication of biological function. However, even in their current format, our studies provide a significant advancement for genetic analysis in mammalian cells, as shown by a large number of functionally identified genes.
Discussion
The development of large-scale robust methods for functional analyses is necessary to reduce the rapidly increasing gap between the vast amount of gene expression data and the knowledge of biological function. Such methods have been widely applied in studies of microorganisms, yeast, and Escherichia coli (25–28). In these studies, we developed a combination of cDNA library screens and microarray analyses to identify functional genes in mouse ES cells. Although previously applied in E. coli (28), this methodology is used here in a mammalian system. Other large-scale gain-of-function techniques developed for mammalian cells include transfections of arrayed cDNA collections and transfected-cell microarrays (29, 30). Although valuable and capable of providing genomewide functional information, these approaches require state-of-the-art procedures and special equipment such as robotic printers and analyzers. In comparison, our approach is cost-effective and easily adaptable because it can be applied with any commercially available microarrays and in any proliferating cell type after establishing moderate selective conditions. We expect this approach will provide information complementary to the previously described large-scale loss-of-function approaches based on the RNA interference technology (31, 32).
Using the developed approach, we were able to identify an array of genes that rescue the ES cell phenotypic properties in the absence of LIF and whose function in ES cells was previously unknown. The identified genes encode a wide range of regulatory proteins. We hypothesize that these genes inhibit differentiation processes induced at the withdrawal of LIF and they exert their activity through a variety of molecular pathways. For example, one of the identified genes encodes the Akt1 kinase, a known downstream component of the phosphatidylinositol 3-kinase (PI3-K) pathway (33). Inhibition of this pathway was shown to induce growth arrest and differentiation of ES cells in the presence of LIF (34, 35). Then, it is reasonable to speculate that the PI3-K pathway regulates genes that function downstream of the LIF/Stat3 pathway. The rescue activity of another gene, the Nedd8-conjugation enzyme NCE, suggests involvement of ubiquitin-mediated protein degradation in ES cell regulation. Previous studies have shown that NCE regulates levels of the cell cycle inhibitor p27 (36). Another identified gene, Prune, was shown to undergo amplification in human sarcomas and possess a cAMP phosphodiesterase activity (37). It also was shown to interact with nm23-H1, a tumor suppressor gene encoding a nucleoside diphosphate kinase. An additional gene with the rescue activity encodes the transcription factor Cited2 that was shown previously to enhance proliferation of embryonic fibroblasts (38). Interestingly, its close homologue, Cited1, induced an opposite effect, differentiation, which implies specificity of the identified functional activity across multigene families. We also monitored changes in transcriptional levels of the genes with the rescue activity to test whether any of them is directly regulated by LIF signaling. We did not detect significant changes in their expression as a result of LIF withdrawal. These results indicate that the identified genes are not downstream transcriptional targets of the LIF/Stat3 signaling pathway. A similar lack of correlation between transcriptional regulation and functional activity was observed in genomewide studies of yeast deletion mutants (27).
The set of genes inducing differentiation in ES cells included two well known protooncogenes, JunB and c-Fos, that are known to enhance proliferation and inhibit differentiation in other cell types (39). In this set, we also identified SpiC, a member of the ETS domain family of transcription factors activated in erythroleukemias (40). These data indicate that the functional activity of the identified genes depends on a specific context, which is an overall set of molecules available in a particular type of cells. Therefore, gene functional roles must be experimentally determined in each cell type of interest that further underlines the requirement for large-scale functional analyses such as those presented in these studies.
What is the role of the identified genes in vivo? Previous studies indicate that a deficiency in either the Cited2 orAkt1 gene is not detrimental to ES cell precursors in the mouse epiblast (41, 42). A possible explanation lies in the fundamental difference between gain-of-function and loss-of-function analyses. Gain-of-function analysis identifies genes that are sufficient to confer a specific phenotype, whereas loss-of-function analysis identifies genes that are necessary. The former type of analysis cannot determine whether genes are essential, whereas the latter is affected by functional redundancy in a studied system. For example, cyclin genes (e.g., cyclin E), whose functional roles were firmly established in biochemical and gain-of-function experiments, were shown to be dispensable for mouse development (43). An additional possible explanation is that regulatory mechanisms in cultured ES cells do not accurately reflect regulatory pathways present in the early mouse embryo. For example, although LIF is required to maintain undifferentiated ES cells in vitro, mouse mutants that lack the LIF gene are viable (44). Further loss-of-function experiments are required to test whether the genes identified in our studies are essential for maintenance of the ES cell phenotype.
Ultimately, a complete list of genetic perturbations, gain and loss of function, and their phenotypic consequences will be necessary to understand complex molecular mechanisms regulating ES cell self-renewal and differentiation. Our studies establish an efficient approach to comprehensively determine gain-of-function genetic modifications that confer particular fates on ES cells. Using lineage-specific reporter lines, this approach also can be used to identify gene products inducing differentiation toward a particular cell type and, thus, is potentially useful for development of stem cell therapies.
Materials and Methods
ES Cell Culture.
The E14/T ES cell line (gift of Austin Smith, University of Edinburgh) was used for most of the experiments. This cell line constitutively expresses the polyoma virus large T antigen and, therefore, can maintain plasmid vectors (episomes) carrying the polyoma virus origin of replication such as pPyCAGIP (4, 19). Cells were grown without feeder cells as described elsewhere, in the presence of 15% FBS and LIF (4).
cDNA Library.
The cDNA library in pPyCAGIP (gift of Austin Smith) prepared from undifferentiated ES cells cultured on embryonic fibroblasts has been described in ref. 5. We measured the fraction of full-length cDNAs. Sequences of 11 of 20 randomly chosen clones included known initiation codons as compared with annotations at the National Center for Biotechnology Information database.
Gain-of-Function Screens in ES Cells.
For the library screen, 5 × 107 E14/T cells were electroporated with 50 μg of the library DNA. The transfected cells were selected with puromycin (2 μg/ml) for 6 days in regular growth conditions. After selection, the cells were divided into two pools that were separately propagated in the presence or absence of LIF for 18 days. At the end of the screen, the library cDNA was extracted from cells, and cDNA inserts were amplified by using PCR with primers complimentary to the regions just outside of the cloning site; the reverse primer was attached to the T7 promoter sequence. The products of the reaction were used for in vitro biotinylation transcription reaction, and the biotinylated cRNA was analyzed by using Mouse430 microarrays (Affymetrix), according to the standard procedure.
Analysis of Data from Genomewide Genetic Screens.
To identify genes that increase their representation in the total cDNA population in the absence of LIF, we compared microarray probe intensity values at −LIF versus +LIF conditions. We identified 560 probes that were recognized by the Affymetrix system as “present” and “increased” >5-fold (arbitrary threshold) at the −LIF conditions. To identify these genes that induce differentiation or cell death and, therefore, decrease their representation, we compared microarray probe intensity values between the library propagated at +LIF conditions and the initial library. We identified 3,524 probes that were present in the initial library and “decreased” >5-fold at the +LIF conditions. We applied PCR with gene-specific primers to confirm the microarray data. The primers were designed to be in the 3′ ends of coding regions corresponding to the design of microarray probes that are selected from 600 bases most proximal to the 3′ end of the mRNA (Affymetrix) according to the manufacturer’s instructions.
Experimental Confirmation of Gene Function.
For experimental confirmation, we selected genes whose full-length cDNA clones were available at the Mammalian Gene Collection (Open Biosystems, Huntsville, AL). The cDNA fragments were cloned between the XhoI and NotI sites of the pPyCAGIP vector. The resulting constructs were individually tested in the E14/T cells, under conditions similar to those used in screens. To monitor proliferation, transfected cells were counted every 3 days. To determine an ability of identified genes to support stem cell colony formation at clonal density, transfected cells were seeded 1,000 per cm2 in the absence of LIF and presence of puromycin. The fraction of stem cell colonies was determined in 7 days by using an alkaline phosphatase staining kit (Sigma).
Analysis of Gene Expression After LIF Withdrawal.
ES cells were cultured in growth medium without LIF: 2 × 106, 1 × 106, 7 × 105, 5 × 105, and 3 × 105 cells per 10-cm plate for 1, 2, 3, 4, and 5 days without replating. At these conditions, initial low-cell densities were used to avoid culture overgrowth. Total RNA was collected and isolated with TRIzol (Invitrogen) and analyzed by using Mouse430 microarrays (Affymetrix), according to the standard procedure. PCR for 20–25 cycles with gene-specific primers and AmpliTaq-Gold enzyme (Applied Biosystems) was used to confirm expression profiles.
Transcriptional Analysis of ES Cell Molecular Markers.
Transfected cells were selected in the presence of LIF and puromycin and then plated in the absence of LIF and presence of puromycin for 4 days. Total RNA was collected, and gene expression was analyzed as described above.
Supporting Information.
An extensive description of materials and methods is included in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Natalia Ivanova (Princeton University), Max Gassmann (University of Zurich), and Austin Smith and Ian Chambers (University of Edinburgh) for providing materials for this work. This work was supported by funds from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. M.P. was supported by the Burroughs Wellcome Fund fellowship in Biological Dynamics.
Abbreviation
- LIF
leukemia inhibitory factor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Smith A. G. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 2.Hochedlinger K., Jaenisch R. N. Engl. J. Med. 2003;349:275–286. doi: 10.1056/NEJMra035397. [DOI] [PubMed] [Google Scholar]
- 3.Lengerke C., Daley G. Q. Exp. Hematol. 2005;33:971–979. doi: 10.1016/j.exphem.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Niwa H., Burdon T., Chambers I., Smith A. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 6.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 7.Niwa H., Miyazaki J., Smith A. G. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 8.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 9.Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., et al. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane T., Dylla S. J., Muijtjens M., Weissman I. L. Proc. Natl. Acad. Sci. USA. 2005;102:3312–3317. doi: 10.1073/pnas.0500167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anneren C., Cowan C. A., Melton D. A. J. Biol. Chem. 2004;279:31590–31598. doi: 10.1074/jbc.M403547200. [DOI] [PubMed] [Google Scholar]
- 12.Ivanova N. B., Dimos J. T., Schaniel C., Hackney J. A., Moore K. A., Lemischka I. R. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 13.Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R. C., Melton D. A. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 14.Pritsker M., Doniger T. T., Kramer L. C., Westcot S. E., Lemischka I. R. Proc. Natl. Acad. Sci. USA. 2005;102:14290–14295. doi: 10.1073/pnas.0502132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boiani M., Scholer H. R. Nat. Rev. Mol. Cell. Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 16.Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. Development (Cambridge, U.K.) 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 17.Stark G. R., Gudkov A. V. Hum. Mol. Genet. 1999;8:1925–1938. doi: 10.1093/hmg/8.10.1925. [DOI] [PubMed] [Google Scholar]
- 18.Singhi A. D., Kondratov R. V., Neznanov N., Chernov M. V., Gudkov A. V. Proc. Natl. Acad. Sci. USA. 2004;101:9327–9332. doi: 10.1073/pnas.0403080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gassmann M., Donoho G., Berg P. Proc. Natl. Acad. Sci. USA. 1995;92:1292–1296. doi: 10.1073/pnas.92.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bortvin A., Eggan K., Skaletsky H., Akutsu H., Berry D. L., Yanagimachi R., Page D. C., Jaenisch R. Development (Cambridge, U.K.) 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- 21.Akagi T., Usuda M., Matsuda T., Ko M. S., Niwa H., Asano M., Koide H., Yokota T. Biochem. Biophys. Res. Commun. 2005;331:23–30. doi: 10.1016/j.bbrc.2005.03.118. [DOI] [PubMed] [Google Scholar]
- 22.Duval D., Reinhardt B., Kedinger C., Boeuf H. FASEB J. 2000;14:1577–1584. doi: 10.1096/fj.14.11.1577. [DOI] [PubMed] [Google Scholar]
- 23.Erez N., Milyavsky M., Goldfinger N., Peles E., Gudkov A. V., Rotter V. Oncogene. 2002;21:6713–6721. doi: 10.1038/sj.onc.1205867. [DOI] [PubMed] [Google Scholar]
- 24.Modrek B., Lee C. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 25.Smith V., Botstein D., Brown P. O. Proc. Natl. Acad. Sci. USA. 1995;92:6479–6483. doi: 10.1073/pnas.92.14.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badarinarayana V., Estep P. W., 3rd, Shendure J., Edwards J., Tavazoie S., Lam F., Church G. M. Nat. Biotechnol. 2001;19:1060–1065. doi: 10.1038/nbt1101-1060. [DOI] [PubMed] [Google Scholar]
- 27.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 28.Gill R. T., Wildt S., Yang Y. T., Ziesman S., Stephanopoulos G. Proc. Natl. Acad. Sci. USA. 2002;99:7033–7038. doi: 10.1073/pnas.102154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziauddin J., Sabatini D. M. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q., Raya A., DeJesus P., Chao S.-H., Quon K. C., Caldwell J. S., Chanda S. K., Izpisua-Belmonte J. C., Schultz P. G. Proc. Natl. Acad. Sci. USA. 2004;101:3456–3461. doi: 10.1073/pnas.0308562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddison P. J., Silva J. M., Conklin D. S., Schlabach M., Li M., Aruleba S., Balija V., O’Shaughnessy A., Gnoj L., Scobie K., et al. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 32.Berns K., Hijmans E. M., Mullenders J., Brummelkamp T. R., Velds A., Heimerikx M., Kerkhoven R. M., Madiredjo M., Nijkamp W., Weigelt B., et al. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe S., Umehara H., Murayama K., Okabe M., Kimura T., Nakano T. 2006 Jan 9; doi: 10.1038/sj.onc.1209307. Oncogene, 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 34.Jirmanova L., Afanassieff M., Gobert-Gosse S., Markossian S., Savatier P. Oncogene. 2002;21:5515–5528. doi: 10.1038/sj.onc.1205728. [DOI] [PubMed] [Google Scholar]
- 35.Paling N. R., Wheadon H., Bone H. K., Welham M. J. J. Biol. Chem. 2004;279:48063–48070. doi: 10.1074/jbc.M406467200. [DOI] [PubMed] [Google Scholar]
- 36.Podust V. N., Brownell J. E., Gladysheva T. B., Luo R.-S., Wang C., Coggins M. B., Pierce J. W., Lightcap E. S., Chau V. Proc. Natl. Acad. Sci. USA. 2000;97:4579–4584. doi: 10.1073/pnas.090465597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Angelo A., Garzia L., Andre A., Carotenuto P., Aglio V., Guardiola O., Arrigoni G., Cossu A., Palmieri G., Aravind L., et al. Cancer Cell. 2004;5:137–149. doi: 10.1016/s1535-6108(04)00021-2. [DOI] [PubMed] [Google Scholar]
- 38.Kranc K. R., Bamforth S. D., Braganca J., Norbury C., van Lohuizen M., Bhattacharya S. Mol. Cell. Biol. 2003;23:7658–7666. doi: 10.1128/MCB.23.21.7658-7666.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z. Q., Liang J., Schellander K., Wagner E. F., Grigoriadis A. E. Cancer Res. 1995;55:6244–6251. [PubMed] [Google Scholar]
- 40.Bemark M., Martensson A., Liberg D., Leanderson T. J. Biol. Chem. 1999;274:10259–10267. doi: 10.1074/jbc.274.15.10259. [DOI] [PubMed] [Google Scholar]
- 41.Bamforth S. D., Braganca J., Eloranta J. J., Murdoch J. N., Marques F. I., Kranc K. R., Farza H., Henderson D. J., Hurst H. C., Bhattacharya S. Nat. Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- 42.Cho H., Thorvaldsen J. L., Chu Q., Feng F., Birnbaum M. J. J. Biol. Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 43.Geng Y., Yu Q., Sicinska E., Das M., Schneider J. E., Bhattacharya S., Rideout W. M., Bronson R. T., Gardner H., Sicinski P. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 44.Stewart C. L., Kaspar P., Brunet L. J., Bhatt H., Gadi I., Kontgen F., Abbondanzo S. J. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.