Abstract
Haplotypes are a powerful tool for identifying the genetic basis of common complex diseases. Disease-association mapping requires molecular methods for haplotyping biallelic SNP variation and highly complex polymorphisms. We developed a method for phasing HLA-A, HLA-B, and HLA-DRB1 alleles on chromosome 6 in unrelated individuals. This method uses the highly polymorphic HLA-B locus to discriminate the two HLA haplotypes in heterozygous individuals and its ideal location 1.4 Mbp telomeric to HLA-DRB1 and 1.2 Mbp centromeric to HLA-A to capture 2-Mbp-long genomic DNA. Genomic DNA representing a single HLA-B-captured haplotype is genotyped for HLA-A and HLA-DRB1 alleles and linkage to HLA-B is established. Proof of principle was established in a large blinded study of phase-known samples. Availability of an efficient method for MHC haplotype phase determination will facilitate the mapping of causative MHC-resident genes in many human diseases and has the potential to be broadened to other polymorphic gene complexes.
Keywords: HLA haplotypes
Haplotypes are a powerful tool for locating disease-susceptibility genes and understanding functional properties of haplotype-linked variation (1, 2). By querying a few known markers that display strong linkage disequilibrium with the causal variant, maximal information can be gleaned without the need to genotype all variations in the region of interest. The availability of a haplotype map of the human genome now makes it feasible to perform genomewide genetic association studies (3). Tools for haplotype determination have included pedigree analysis and statistical methods for haplotype estimation (4, 5). Although family study provides definitive phase information, relatives may not be available. Statistical methods provide inferred haplotype estimates based on linkage disequilibrium, but their accuracy for the study of rare variations or regions of weak linkage disequilibrium may limit their application (6).
Laboratory methods for phase determination have been developed including single-molecule PCR, sperm typing, allele-specific PCR, pyrosequencing, bacterial artificial chromosome cloning, polymerase colony, construction of somatic cell hybrids, atomic force microscopy, rolling circle amplification, and mass spectrometry (7–23). Although these methods can define SNP haplotypes, not all are suitable for haplotyping the multilocus MHC region because of its extensive polymorphism and uneven distribution of coding variation (www.ebi.ac.uk/imgt/hla) and the physical distance between each HLA locus.
The MHC region has been an intense area of interest, encoding an unprecedented number of genes that confer susceptibility to autoimmunity, infection, and cancer and play a fundamental role in transplantation (24–26). Its extensive polymorphism and linkage disequilibrium have provided a model system for understanding genetic organization and human evolution (27, 28). To address an unmet need for an MHC haplotyping method for unrelated individuals, we developed an approach for phasing the classical three-locus HLA-A, HLA-B, HLA-DRB1 haplotype in genomic DNA from heterozygous individuals. The two HLA haplotypes were separated from each other by using HLA-B-specific coding region polymorphisms. The specificity of the capture of each haplotype in heterozygous samples was validated, and the accuracy of haplotype assignments was demonstrated in a large blinded analysis of phase-known samples.
Results
Our method for haplotype separation was developed in samples heterozygous at HLA-B and heterozygous at HLA-A and/or HLA-DRB1 to prove definitive haplotype separation. The method was designed with several key features in mind: (i) use of genomic DNA as a template for maximal applicability in different research and clinical applications and sample sources, (ii) broad utility of the classical three-locus HLA-A, HLA-B, HLA-DR haplotype for population-based anthropologic and disease-mapping applications, and (iii) a platform that provides scalability for application to large unrelated study populations. The HLA haplotyping approach includes the following steps: preparation of 2-Mbp-long human genomic DNA, chemical treatment of the glass array surface, immobilization of HLA-B-specific probes, haplotype-specific hybridization of genomic DNA, harvest of captured DNA, PCR amplification and genotyping of HLA-A and HLA-DRB1 from captured DNA, and image analysis (Fig. 1). The method was refined through careful optimization of each of these steps.
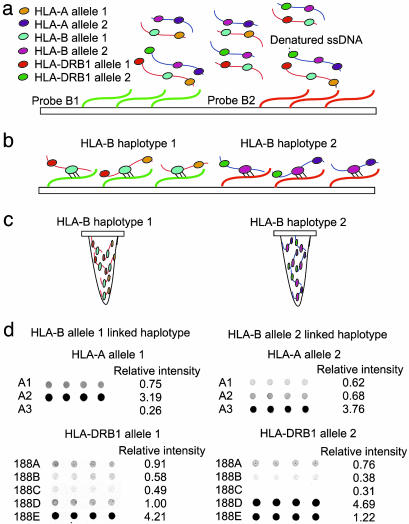
Fig. 1.
Procedure for haplotyping 2-Mbp-long human genomic DNA. (a) Genomic DNA was hybridized with two HLA-B oligonucleotide probes immobilized on glass surface. (b) Excess genomic DNA was removed from surface through stringent washes, retaining template DNA with an HLA-B sequence complementary to the probe. (c) Captured genomic DNA fragments were collected into separate tubes, each containing one haplotype. (d) HLA-A and HLA-DRB genes were PCR-amplified and typed by using oligonucleotide arrays. HLA-A and HLA-DRB1 alleles were assigned by evaluating the relative signal intensities of quadruplicate probe hybridizations derived from the quantarray software program. For illustration purposes, hybridization images and relative intensities for probes A1–A3 for HLA-A and 188A–188E for HLA-DRB1 are shown (complete probes are listed in Tables 4 and 5).
Concepts Underlying Assay Design.
We chose the classical HLA-A, HLA-B, HLA-DRB1 genes to define the three-locus haplotype. This decision was motivated by the importance of HLA-A, HLA-B, HLA-DRB1 haplotype associations in anthropologic and disease studies and in the selection of potential donors for solid organ and hematopoietic cell transplantation. These genetic loci have well known associations to HLA-C and HLA-DQB1 (29). Hence, a haplotyping approach based on HLA-A, HLA-B, HLA-DRB1 is anticipated to provide genetic information relevant to all five classical HLA genes. Our method represents a step toward a comprehensive haplotyping method for this region of the genome. The haplotyping method is based on prior knowledge of the HLA-A, HLA-B, and HLA-DRB1 genotype of the sample. It was not our intention to develop a haplotyping method for blinded genotyping, although that will be feasible in the future.
We chose HLA-B as the genetic locus for separation because of its central location 1.4 Mbp centromeric to HLA-A and 1.2 Mbp telomeric to HLA-DRB1. To verify that human genomic DNA contains fragments that span at least the length between HLA-A and HLA-B, and HLA-B to HLA-DRB1, we first demonstrated that our genomic DNA samples contain fragments measuring at least 2 Mbp in length by using pulse-field electrophoresis (Fig. 2, which is published as supporting information on the PNAS web site). Therefore, phase is determined by the hybridization signals from fragments containing HLA-A and HLA-B and fragments containing HLA-B and HLA-DRB1.
Quality of Genomic DNA.
Human genomic DNA measuring at least 2 Mbp in length was a necessity for linking HLA-A and HLA-DRB1 alleles to the HLA-B allele of each haplotype. Compared with commercially available approaches for DNA preparation, the salting-out method yielded intact 2-Mbp-long DNA suitable for capture at HLA-B as described below (30).
HLA-B Oligonucleotide Probes for Haplotype Capture.
All samples in this study were previously genotyped for HLA-A, HLA-B, and HLA-DR alleles by using published methods (31, 32). Capture of HLA-B haplotype-specific genomic DNA to the array was accomplished by using two oligonucleotide probes, each specific for the two known HLA-B alleles encoded in the sample. We previously reported a method for typing HLA-B alleles by using oligonucleotide arrays (ref. 33; for probe sequences please refer to Tables 1 and 2, which are published as supporting information on the PNAS web site). We first determined the probes informative for exon 2 and exon 3 polymorphisms unique to each HLA-B allele in a given sample (Table 3, which is published as supporting information on the PNAS web site). All positions at which the two HLA-B alleles differed by at least three nucleotide bases were identified, and the pair of probes with the largest number of base-pair differences was selected to construct the array. From the original panel of 137 20-mer HLA-B probes (33), 43 unique probes in 44 different pairwise combinations were used to separate the two haplotypes at HLA-B in the 528 study samples.
HLA-B Haplotype-Specific Capture of Genomic DNA.
Six parameters affected the efficiency and specificity of HLA-B-specific capture of haplotypes.
Treatment of glass surface with 3-aminopropyltrimethoxylsilane.
Before the attachment of HLA-B probes to the glass surface, a prepreparation step with 3-aminopropyltrimethoxylsilane was necessary to create a DNA-reactive surface for probe immobilization. The temperature and time of this chemical reaction affected the hydrophobicity, reactivity, and surface tension of the glass array (34) and therefore were critical determinants for HLA-B haplotype-specific capture of genomic DNA. We tested 80°C, 100°C, 120°C, 140°C, 160°C, 180°C, and 200°C reaction temperatures and 2-, 4-, 8-, 16-, and 24-h reaction times as described (33). Reaction temperatures >160°C and reaction times <2 h deposited an excess of 3-aminopropyltrimethoxylsilane that increased the hydrophobicity of the glass and prevented attachment of the probes. Similarly, temperatures <100°C led to incomplete evaporation of 3-aminopropyltrimethoxylsilane and erratic coating of the glass surface that prevented immobilization of the HLA-B probes. Overnight treatment of the glass surface with the chemical reagent vapor at 120°C under 30 mm Hg vacuum produced appropriate hydrophobicity and high reactivity, leading to reproducible capture of genomic DNA and successful amplification of HLA-A and HLA-B genes from seven genomic DNA samples (B*1501/0702 and B*4402/5501-heterozygous samples captured with 2L1 and 2L2 HLA-B probes, respectively; B*0702/0801, B*3501/1801, B*4402/4001, B*0801/1402, and B*0801/5501-heterozygous samples captured with 2L1/2L7, 3A2/3A11, 3D2/3D10, 3J2/3J6, and 3A1/3A3 probes, respectively).
Probe concentration and volume.
The optimal concentration of HLA-B probes applied on the glass surface was determined to be 250 pmol/μl by testing 100-, 250-, 500-, 1,000-, 1,500-, and 2,000-pmol/μl concentrations of probe solution on the glass. Concentrations >1,000 pmol/μl saturated the surface and caused the DNA to be inaccessible for hybridization, thereby producing low efficiency of capture. A 100-pmol/μl probe concentration significantly reduced the quantity of immobilized probes, resulting in poor DNA capture. The optimal volume of the immobilized probe spots was determined to be 10 μl by testing 1, 3, 10, 20, and 40 μl of probe solutions. Volumes <10 μl caused rapid evaporation and inadequate spot size for capture. Volumes >10 μl produced irregularly shaped spots that suffered from irreproducibility. Each HLA-B probe was spotted in quadruplicate, and the captured DNA from each of the four spots was combined into a single tube (see below). This process yielded sufficient material of haplotype-separated DNA for subsequent steps of the protocol.
Blocking of nonspecific binding of genomic DNA to the array.
Sonicated undenatured salmon-sperm DNA was applied to the glass slide by pipette. Slides were left at room temperature overnight (please refer to Materials and Methods). The specificity of the captured genomic DNA was evaluated in two experiments. First, genomic DNA from eight HLA-B*0702/0801-heterozygous individuals was captured by using HLA-B7- and HLA-B8-specific probes. The HLA-A genes of these haplotype-separated samples were PCR-amplified; however, no amplification of the PART1 gene located on chromosome 5 (35) could be detected, indicating that the salmon-sperm DNA prevented nonspecific binding of chromosome 5 DNA to the HLA-B probe (Fig. 3, which is published as supporting information on the PNAS web site).
The second experiment used 10 each of HLA-B*0702- and HLA-B*0801-homozygous and HLA-B*0702/0801-heterozygous samples. Genomic DNA was captured with the 2L1 and 2L7 probes specific for HLA-B*0702 and HLA-B*0801, respectively. Haplotype-specific DNA was collected from each probe and amplified by using B7- and B8-specific Taqman assays. Only HLA-B*0702 alleles could be amplified from HLA-B7-homozygous samples and HLA-B*0801 alleles could be amplified from B8-homozygous samples. Only HLA-B*0702 and not B*0801 could be amplified from heterozygous samples captured with B7-specific probes; likewise only HLA-B*0801 and not B*0702 alleles could be amplified from B8-captured heterozygous samples (Fig. 4, which is published as supporting information on the PNAS web site). These results demonstrate that one dominant HLA haplotype was captured by using the oligonucleotide probe with complementary sequence to the HLA-B allele of interest.
Denaturation of genomic DNA.
Denaturation of genomic DNA was evaluated at 98°C for 1, 2, 3, and 5 min. Denaturation for 3 and 5 min yielded single-stranded DNA suitable for HLA-A and HLA-B amplification.
Hybridization of denatured genomic DNA to immobilized probes.
The speed of hybridization and nonspecific binding of template DNA to the HLA-B probe affected the hybridization signal. A range of hybridization temperatures and times were evaluated (room temperature, 30°C, and 37°C over 2, 3, 6, and 16 h); 30°C for 3 h conditions yielded successful amplification of HLA-A and HLA-B genes.
The stringency of the wash conditions for the collection of the haplotype-specific DNA from each probe was evaluated for a range of temperatures (room temperature, 30°C, 37°C, 45°C, and 50°C). Low-stringency conditions (room temperature) failed to remove mismatched fragments from the glass surface, whereas high-stringency conditions (50°C) removed both matched templates from their probes and mismatched fragments from the surrounding glass surface. HLA-A and HLA-B locus-specific amplification from 14 haplotype-separated control DNAs was achieved by washing slides twice with 2× standard saline phosphate/EDTA (SSPE) (3 M NaCl/200 mM Na2HPO4/20 mM EDTA) at 37°C for 10 min.
Elution of HLA-B-specific captured DNA from array.
The haplotype-specific DNA from each probe was removed by applying water to each probe spot and recollecting the fluid by pipette. The temperature and volume of the heated water were critical in collecting sufficient quantity of haplotype-separated DNA from the probes. A range of water temperatures (room temperature, 40°C, 50°C, 60°C, 80°C, and 100°C) and volumes (2.5, 5, 7.5, 10, 12.5, and 15 μl) were tested. When water at room temperature was used to elute the template, the genomic DNA did not denature from its probes, whereas water temperatures in excess of 100°C caused rapid evaporation and insufficient material for collection. A minimal volume of 5 μl was required to cover the hybridized probes; 15-μl volume diluted the captured samples and precluded PCR amplification of HLA genes. HLA-A and HLA-B locus-specific amplification was achieved by collecting samples twice with 7.5 μl of 50°C water each time.
Typing of HLA-A and HLA-DRB1 from Captured DNA.
Single-stranded PCR products for hybridization to HLA-A and HLA-DRB1 oligonucleotide arrays could be prepared by using either a one-step amplification from captured genomic DNA samples or a two-step amplification approach. We favored the two-step approach in which genomic DNA was amplified into a double-stranded PCR (dsPCR) product and subsequently used as template for asymmetric PCR to produce single-stranded DNA. The dsPCR products provided DNA sufficient for several hybridizations. DNA fragments captured with HLA-B probes were amplified separately by using primers specific for exon 3 of HLA-A and exon 2 of HLA-DRB genes. The dsPCR products were purified before single-stranded PCR amplification. Asymmetric PCR with a fluorescently labeled primer was used to generate the single-stranded DNA for hybridization.
The choice of HLA-A- and HLA-DRB1-specific oligonucleotide probes was based on the known genotypes of the samples. For the 67 HLA-A and 90 HLA-DRB1 phenotypes in the study population, 57 unique HLA-A and 64 unique HLA-DRB1 probes were designed (Tables 4 and 5, which are published as supporting information on the PNAS web site). Because the HLA genotypes of our samples were known, it was not necessary for the oligonucleotide probe hybridization patterns to be informative for all theoretical heterozygous combinations, as would be the case for blinded typing.
There are currently several different technologies available for genotyping HLA alleles (36, 37). We favor an oligonucleotide approach because relative signal intensities of probe hybridization can be quantitated. The separation of the positive/negative ratios for each positive and negative signal demonstrated that one dominant allele was present on each captured fragment (Tables 6 and 7, which are published as supporting information on the PNAS web site). Nucleotide diversity of HLA genes is clustered into hypervariable regions; alleles were assigned by first evaluating the quantitated signal intensities of probes within each hypervariable region, followed by the pattern of signal intensities across all hypervariable regions. Because our samples were heterozygous for two different HLA-A and HLA-DRB1 alleles, the probe hybridization patterns of one haplotype-specific genotype within each probe region served as the control for the genotype encoded on the second haplotype.
Haplotyping of Study Samples.
The haplotyping method was validated in three groups of samples. The first set consisted of 32 individuals representing two Centre d’Etude du Polymorphisme Humain (CEPH) families. Only the HLA-B genotypes of the CEPH samples were made known to the laboratory personnel to select the appropriate HLA-B probes for haplotype separation. The personnel were blinded to the HLA-A and HLA-DRB1 genotypes and to the relationship between CEPH family members. Phase was determined for each sample, and pedigrees were reconstructed from the CEPH material (Fig. 5, which is published as supporting information on the PNAS web site). All assigned haplotypes agreed with pedigree-defined haplotypes.
The second test set consisted of 199 phase-known patients referred to the Fred Hutchinson Cancer Research Center for consideration of hematopoietic cell transplantation. A third set of samples consisted of 297 phase-unknown patients and unrelated transplant donors. As described above, laboratory personnel were blinded to the HLA haplotypes of the 199 phase-known patient samples. The HLA-A, HLA-B, HLA-DRB1 haplotype assignments for the 199 phase-known and 297 phase-unknown samples were concordant between three observers. Furthermore, the experimentally derived haplotypes for the 199 phase-known samples were concordant with the pedigree-defined haplotypes.
PCR amplification failure of HLA-A and HLA-DRB genes from haplotype-separated DNA occurred in 32 of the 528 (6%) of samples. These 32 samples were captured and assayed with other samples that amplified successfully with the identical probe solution, hybridization buffer, PCR reagents, and conditions for DNA denaturation and PCR amplification, thus ruling out these variables as a cause of the PCR failure. All 32 samples were recaptured by using the original DNA specimen, and HLA-A and HLA-DR were successfully amplified. These observations suggest that variation within a given glass slide with respect to the 3-aminopropyltrimethoxylsilane chemical reaction step may have led to their random PCR failure.
Of the 528 samples, 43 (8%) had evidence of a second allele on HLA-A and HLA-DRB1 probe hybridization. The incomplete haplotype separation was randomly distributed across different glass slides and different probes and assays. Repeat capture, HLA-A and HLA-DR amplification, and probe hybridization using the original DNA sample and reagents for capture, PCR, and probing yielded a single allele. We surmise that the admixture of the second unwanted haplotype for these 43 samples was caused by uneven coating of the slide with salmon-sperm DNA, which led to nonspecific binding of DNA to the glass surface.
Discussion
An unmet need of population-based disease association studies is a definitive long-range haplotype phasing approach in unrelated individuals. The definition of long-distance haplotypes that contain several tagSNP haplotype blocks (3) might permit disease association signals to be ascertained without the selection of individual tagSNPs and thereby increase mapping efficiency; once an association signal is obtained, finer mapping with SNP haplotypes could be applied to localize the causative gene. Experimentally derived phase might permit matching of controls to cases for the marker of interest so that cis/trans relationships of candidate markers can be defined. Broad applications for understanding human evolution (28) and for population-based haplotypes for clinical purposes can be envisioned (38).
Haplotyping the MHC offers a unique challenge because of the extreme polymorphism of class I and II genes and the great genetic distance between loci. Unlike the biallelic SNP, exonic diversity of HLA genes is characterized by multiple, unevenly distributed substitutions at hypervariable positions. As an illustration, our study population encoded 76 unique polymorphic positions in HLA-A exons 2 and 3, 92 in HLA-B exons 2 and 3, and 104 in HLA-DRB1 exon 2. Of these 272 positions, 9 displayed 4-nt substitutions, and 127 positions displayed 3-nt substitutions. To determine HLA-A, HLA-B, HLA-DRB1 phase, the method must not only be informative for the extensive coding region polymorphisms but must also link these polymorphisms across 2.6 Mbp. Hence, many of the currently available approaches for haplotyping SNPs may not be suitable for haplotyping HLA genes. Several haplotyping methods use either individual or pooled samples and are based on statistical methods and provide inferred haplotypes (39–42). To study SNPs evenly distributed over long distances, DNA fragments containing adjacent SNPs must be generated to retain the phase information. Methods such as single-molecule PCR and polymerase colony and cloning-based systematic haplotyping can be used to analyze linkage of SNPs separated by thousands of nucleotides (7, 8, 16, 17, 23). More recently, the polymerase colony method has been used to generate haplotypes by using multiple amplifications from single molecules (23). This powerful method for haplotyping pooled DNAs provides high throughput capacity; its application to the HLA genetic region may be challenging because of the need for multiplexing many hundreds of combinations of polymorphic sites of class I and II genes.
Allele-specific PCR approaches can link SNPs within 10 kb but are limited by the maximal length of the PCR product and the biallelism of SNPs and SNP density (9–12). Single-molecule PCR has high sensitivity for 1-kb-sized fragments, albeit at lower efficiency (7, 8). High throughput and sensitivity can be achieved with pyrosequencing techniques for 100-bp templates (13–15). Sperm typing provides definitive haploid data and has been successfully used for MHC microsatellite mapping (43), but its practicality limits application to large populations (44), a similar situation with bacterial artificial chromosome cloning (16). Finally, several technologies using atomic force microscopy, rolling circle DNA amplification, and mass spectrometry offer highly efficient parallel sequence analysis and may in the future have relevance to haplotyping HLA region genes (18–21).
In this study, we demonstrate that high-quality 2-Mbp-long genomic DNA fragments can be specifically captured by using HLA-B polymorphisms for separating the haplotypes. Future modification of our protocol to span the entire 7.6-Mbp extended MHC region is possible but will require “reconstructing” the MHC into several overlapping segments. The choice of genes that could be considered for use in the initial capture of genomic DNA would be dictated by the polymorphism and heterozygosity of the loci. There are many candidate genes that could serve as a separation point between HLA-A and HLA-DPB1. The current map telomeric to HLA-A, however, has a paucity of informative markers (45) and will likely be the more challenging region for future extension of the haplotyping approach.
The specificity of the capture of a 2-Mbp-long genomic DNA fragment was not only chromosome 6-specific but also specific for only one of the two HLA-B haplotypes (Figs. 3 and 4). Two key steps for increasing the specificity of HLA haplotype-specific annealing were the preparation of the glass slide with 3-aminopropyltrimethoxylsilane and the use of salmon-sperm DNA to block nonspecific hybridization. Furthermore, the use of relative signal intensities to assign linkage of HLA-A and HLA-DRB1 alleles to HLA-B provided quantification of the single haplotype relative to any potential background hybridization contributed by low levels of contamination from the second haplotype. The haplotyping method was designed to use the positive and negative probe hybridizations from each separated haplotype as the control for the second haplotype. This process provided internal “checks” to the interpretation of positive and negative hybridization signals and contributed to the high concordance of phase assignments among different observers (Tables 6 and 7). In this regard, the haplotype assignment was no more complicated than interpreting oligonucleotide probe hybridization signals common to many genotyping systems for individual class I and II genes (36) and suggests that translation of this method to large-scale clinical applications is feasible.
Because the goal of our method was to definitively ascertain the phase of HLA-A, HLA-B, and HLA-DRB1 alleles in samples for which high-resolution HLA typing was known, a complete panel of oligonucleotide probes for haplotype capture and HLA genotyping was not required. When the HLA-A, HLA-B, HLA-DRB1 genotype is known, 40 samples can be haplotyped in 7 days (4 days for DNA preparation, chemical treatment of the glass surface, immobilization of HLA-B specific probes, hybridization of genomic DNA, and harvest of captured DNA; 3 days for HLA-A and HLA-DR PCR amplification, oligonucleotide probe typing, and image analysis). Optimization of the DNA capture step alone is anticipated to shorten the haplotyping procedure to 4 days. Future modifications for haplotyping blinded samples will increase the utility of this method for studies that require highly accurate phase information for large unrelated populations.
The availability of a haplotype map of the human genome provides the research community with a tool for localizing genes responsible for complex human diseases (3). Long-distance haplotype phasing could be integrated into mapping strategies by using a minimal set of known heterozygous markers by linking several haplotype-tagging SNP blocks that contain different densities of SNPs or number of unique SNP haplotypes. Once a positive association is found to the 2-Mbp haplotype, then further characterization could ensure the localization of the specific gene. Common and rare variations that are contained within the 2-Mbp region could be characterized, and genetic mapping in diverse populations for whom complete haplotype information is not yet available may be feasible by using a few known anchor markers. With respect to the MHC, the genome-wide haplotype map showcases the similarities and differences in genetic properties between the MHC region and other chromosomal regions (3, 46). For MHC-associated diseases, long-range haplotyping methods may be useful for defining genes that cause or modify susceptibility to autoimmunity, defend against infections, govern graft acceptance in solid organ transplantation, and participate in alloimmune reactions after hematopoietic cell transplantation (24–26).
Materials and Methods
Study Population.
A total of 528 individuals were studied and included: CEPH families 1331 and 1362, each representing 16 individuals; 199 phase-known patients referred to the Fred Hutchinson Cancer Research Center for hematopoietic cell transplantation; and 297 phase-unknown transplant patients and unrelated donors. All individuals were previously typed for HLA-A, HLA-B, and HLA-DRB1 alleles as described (32) and were heterozygous at HLA-B and at HLA-A and/or HLA-DRB1. All research samples and data were collected according to human subjects guidelines under institutional review board-approved protocols.
Capture of DNA with HLA-B Probes for Haplotype Separation.
Genomic DNA was extracted from cultured lymphoblastoid cell lines by using the salting-out method (30). DNA concentration was measured with a UV spectrometer. The size of the DNA was determined by pulse-field electrophoresis in a 1% agarose gel with 0.5× TBE buffer (45 mM Tris-borate/0.5 mM EDTA, pH 8.3) for 20 h at 6 V/cm at 14°C with switching intervals of 60 s and visualized with ethidium bromide staining.
Two HLA-B oligonucleotide probes differing by a minimum of 3 nt in their exon 2 or 3 sequence were chosen for each sample based on the HLA-B genotype (ref. 33 and Tables 1–3). Glass slides were chemically treated as described (33), with the following exceptions: precleaned microscope slides (Becton Dickinson) were placed in a vacuum chamber with 750 μl of 3-aminopropyltrimethoxysilane (Aldrich Chem, Metuchen, NJ), and the vacuum chamber was maintained at 120°C and 30 ppm Hg pressure overnight. The slides were subsequently washed four times with acetone, 5 min per wash, and then treated for 3 h with a solution of 0.2% 1,4-phenylene diisothiocyanate (Aldrich) in 10% pyridine/dimethyl formamide and washed with acetone and then methanol five times, 5 min per wash. All washes were performed with an orbital shaker (Barnstead/Lab-Line model MaxQ4000, Dubuque, IA) at 85 rpm unless otherwise stated. The activated glass slides were stored in a desiccator. HLA-B probes [250 pmol/μl in sterile doubly distilled H2O (ddH2O)] were diluted 1:1 with DMSO, and 10 μl was deposited onto the slide in quadruplicate. The slides were incubated at 90°C for 3 h and washed once in 1% NH4OH and four times in distilled H2O at room temperature for 5 min each. Salmon-sperm DNA solution [10-μl mixture of 2 μg of undenatured sonicated salmon-sperm DNA (Stratagene), 2.5 μl of 20× SSPE (Amresco, Solon, OH)] was deposited onto each probe spot. Slides were placed flat on the top of an empty 96-tip, 10- to 200-μl pipette tipbox filled with 150 ml of ddH2O, covered with a lid (henceforth referred to as “humid hybridization chamber”), and maintained for 16 h at room temperature. Because the salmon-sperm DNA was not covalently bound to the glass surface, no part of the protected surface was touched by hand to avoid disruption of the layer of the salmon-sperm DNA. After hybridization, the glass slides were washed twice in 2× SSPE for 5 min at room temperature. Slides were quickly dried by centrifugation (Eppendorf model 5810) at 700 rpm for 1 min.
Genomic DNA (32 μl of 100 ng/μl in 10 mM Tris·HCl/0.1 mM EDTA, pH 7.5) was denatured in a thermal cycler at 98°C for 3 min, followed by 4°C for 10 min, and suspended in 8 μl of 20× SSPE. All thermal cycler reactions were performed in 96-well plates with GeneAmp PCR System 9700 (Applied Biosystems) unless otherwise stated. DNA/SSPE mixture (5 μl) was applied into the center of each probe spot. The glass slides were placed in the humid hybridization chamber and incubated at 30°C for 3 h, washed twice with 200 ml of 2× SSPE for 10 min at 37°C, and air-dried. The slides were placed in the humid hybridization chamber filled with 50°C water. To collect the DNA from each probe, 7.5 μl of 50°C sterile ddH2O was deposited by pipette onto each of the quadruplicate probe spots, and the humid hybridization chamber was placed in 50°C water bath for 3 min. After this period, the DNA from all quadruplicate spots was pipetted and pooled into a single tube. The collection step was repeated, and the DNA from the second collection was pooled with that from the first collection.
The specificity of the separation of the two HLA haplotypes was tested with the Taqman assay by using unlabeled forward primer 5′-ACCCGGTTTCATTTTCAGTTG-3′ [intron 2 position 621–641, per the ImMunoGeneTics/HLA sequence database (www.ebi.ac.uk/imgt/hla)] and unlabeled reverse primer 5′-CCCACTGCCCCTGGTACC-3′ (exon 3 position 619 followed by intron 3 position 993-1010) to amplify a 329-bp region of the HLA-B exon 3. The assay used two carboxytetramethylrhodamine-quenched Taqman probes informative for positions 554–573 in exon 3, one specific for HLA-B*0702 (HLA-B7/6-FAM-CCACTCCACGCACTCGCCCT-3′) and the other for HLA-B*0801 (HLA-B8/VIC-CCACTCCACGCACGTGCCCT-3′). The assay was tested in uncaptured genomic DNA from 10 B*0702-homozygous samples, 10 B*0801-homozygous samples, 10 B*0702/0801-heterozygous samples, and 10 B*0702/0801 samples captured with 2L1 (HLA-B*0702-specific) and 2L7 (HLA-B*0801-specific) probe. The assay was conducted in a 10-μl volume containing 5 μl of Universal master mix with AmpErase uracil N-glycosylase (Applied Biosystems), a 700 nM concentration of each unlabeled forward and reverse primer, a 100 nM concentration of each labeled probe, and either 5 μl of HLA-B probe-captured DNA template or 1 μl of uncaptured DNA (100 ng/μl). Reactions were amplified with the 7500 Real-Time PCR system (Applied Biosystems) with the following conditions: 50°C for 2 min, 95°C for 10 min followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescence release was measured with the ABI 7500 Real-Time PCR system using the allelic discrimination setting of sequence detection 1.2.3 software (Applied Biosystems).
HLA-A and HLA-DRB PCR Amplification of Captured DNA.
HLA-A and HLA-DRB genes were first amplified from genomic DNA followed by asymmetric PCR using a fluorescently labeled primer to produce ssDNA. HLA-A primers (5′-AGTTTAGGCCAAAAAT[C/T]CGCC-3′ and 5′-GTGGCCCCTGGTACCCGTG-3′) were locus-specific and amplified a 371-bp region of exon 3. HLA-DRB generic primers (5′-CCCCACAGCACGTTTCTTG-3′ and 5′-CCGGATCCTTCGTGTCCCCACAGCACG-3′) amplified a 230-bp fragment shared by all DRB exon 2 genes (DRB1, DRB3, DRB4, and DRB5) (www.ihwg.org/tmanual/TMcontents). The amplification was conducted in a 50-μl reaction containing 5 μl of 10× Fast Start PCR buffer [500 mM Tris·HCl/100 mM KCl/50 mM (NH4)2SO4/20 mM MgCl2, pH 8.3] (Roche Diagnostics), 4 μl of 10 mM dNTP (Roche Diagnostics), 50 pmol of each primer, 2 units of Fast Start TaqDNA polymerase (Roche Diagnostics), and either 20 μl (for HLA-A PCR) or 10 μl (for HLA-DRB PCR) of each of the captured genomic DNA. The PCR profile consisted of 4 min at 95°C followed by 35 cycles of 95°C for 30 s, 60°C for 30 s for HLA-A or 53°C for 30 s for DRB, 72°C for 1 min. The quality of amplified products (5 μl) was verified on a 1% agarose gel in 1× TBE (89 mM Tris-borate/2 mM EDTA) (Amresco). After amplification, dsPCR products were purified with a PCR purification kit (Qiagen, Valencia, CA) following the manufacturer’s protocol and eluted in 80 μl of sterile ddH2O.
Purified HLA-A and HLA-DRB PCR products were amplified with bodipy 630/650 dye (Bdy) 5′-labeled primer. The HLA-A and HLA-DRB fragments were separately amplified in 50-μl reactions containing 5 μl of 10× PCR buffer (100 mM Tris·HCl, pH 8.3/500 mM KCl) (Applied Biosystems), 2 μl of 25 mM MgCl2 (Applied Biosystems), 4 μl of 10 mM dNTP (Roche Diagnostics), 25 pmol of primer (HLA-A: 5′-BdyTCTCCAGGTATCTGCGGAGC-3′ or HLA-DRB: 5′-BdyCCCCACAGCACGTTTCTTG-3′), 2 units of TaqDNA polymerase (Applied Biosystems), and 20 μl of the purified dsPCR product. The PCR profile consisted of 3 min at 96°C followed by 30 cycles of 96°C for 30 s, 64°C for 30 s for HLA-A or 62°C for 30 s for HLA-DRB, 72°C for 1 min. Quality of amplified single-stranded PCR product (8 μl) was verified on a 1% agarose gel in 1× TBE (89 mM Tris-borate/2 mM EDTA) (Amresco).
Oligonucleotide HLA-A and HLA-DRB1 Array Design.
HLA-A and HLA-DRB1 alleles carried on each captured fragment were determined by hybridization with oligonucleotide probes descriptive of HLA-A exon 3 and HLA-DRB1-specific exon 2 polymorphisms (Tables 4 and 5, respectively). A theoretical hybridization pattern was generated for each allele. Each probe contained a 5′-amino group for immobilization chemistry, a 6-mer poly(dT) spacer, followed by the hybridization sequence (33). The polymorphic sequence was situated near the center of each hybridization sequence.
Construction of Oligonucleotide Array.
HLA-A and HLA-DRB1 probes (500 pmol/μl in sterile ddH2O) were diluted 1:1 with DMSO. Each probe/DMSO mixture (20 μl) was deposited into a 96-well microtiter plate and placed in a SpotArray microarray spotter (PerkinElmer) together with 20 preactivated glass slides (33). Probes measuring 100–150 μm in diameter were spotted onto glass slides with a separation distance of 400 μm at 55% humidity and incubated at 90°C for 3 h. After immobilization, the glass slides were washed once in 1% NH4OH and four times in distilled H2O at room temperature for 5 min each.
Hybridization of Captured DNA with HLA-A and HLA-DRB1 Oligonucleotide Arrays.
Oligonucleotide array slides were labeled with a barcode to link the sample number with the HLA-B capture probe and the amplified gene (HLA-A or HLA-DRB). PCR hybridization solution (30 μl of single-stranded PCR product, 8 μl of 20× SSPE, and 2 μl of 10% SDS) was added onto the array, sealed with a cover glass, placed in the humid hybridization chamber, and incubated at 30°C for 3 h. After hybridization, the array slides were washed twice at 37°C in 2× SSPE, 0.2% SDS buffer for 15 min. The washed slides were placed in a ScanArray Express HT Microarray scanner (PerkinElmer) for quantification of hybridization signals by using quantarray software (PerkinElmer).
HLA-A, HLA-B, HLA-DRB1 Haplotype Assignment.
Hybridization signal intensities of the four duplicate spots were averaged within each HLA-A or HLA-DRB1 probe group. The highest signal within each group was designated as the positive hybridization of that genomic region. Positive hybridization patterns for each group were compared with theoretical hybridization patterns of HLA-A and HLA-DRB1. The HLA-B allele type was determined by the HLA-B probe used to capture the DNA sample and linked to the assigned HLA-A and HLA-DRB1 alleles. The presence of one HLA-A, HLA-B, and HLA-DRB1 allele in each of the captured samples indicated that the HLA-A and HLA-DRB1 alleles were physically encoded on the same haplotype.
Supplementary Material
Acknowledgments
We thank Sharie Cheng, Mark Gatterman, and Michelle Baillie for excellent technical assistance; Dr. John Hansen and Eric Mickelson (Fred Hutchinson Cancer Research Center) for providing cell lines; and Dr. Leland Hartwell for helpful discussions. This work was supported by National Institutes of Health Grants AI18029 and CA100019 (haplotyping method development) and National Institutes of Health Grants AI49213 and AI33484 (13th International Histocompatibility Working Group sample repository).
Abbreviations
- SSPE
standard saline phosphate/EDTA
- dsPCR
double-stranded PCR
- CEPH
Centre d’Etude Polymorphisme Humain
- ddH2O
doubly distilled H2O.
Footnotes
Conflict of interest statement: Z.G., L.H., and E.W.P. declare that they have applied for a U.S. nonprovisional 111 (a) patent for the haplotyping method, serial number 10/843,985.
References
- 1.Nagel R. L., Fabry M. E., Pagnier J., Zohoun I., Wajman H., Baudin V., Labie D. N. Engl. J. Med. 1985;312:880–884. doi: 10.1056/NEJM198504043121403. [DOI] [PubMed] [Google Scholar]
- 2.Drysdale C., McGraw D., Stack C., Stephens J., Judson R., Nandabalan K., Arnold K., Ruano G., Liggett S. Proc. Natl. Acad. Sci. USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The International HapMap Consortium. Nature. 2005;437:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 4.Excoffier L., Slatkin M. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 5.Fallin D., Schork N. J. Am. J. Hum. Genet. 2000;67:947–959. doi: 10.1086/303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tishkoff S. A., Pakstis A. J., Ruano G., Kidd K. K. Am. J. Hum. Genet. 2000;67:518–522. doi: 10.1086/303000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul P., Apgar J. BioTechniques. 2005;38:553–559. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- 8.Kwok P. Y., Xiao M. Hum. Mutat. 2004;23:442–446. doi: 10.1002/humu.20020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding C., Cantor C. R. Proc. Natl. Acad. Sci. USA. 2003;100:7449–7453. doi: 10.1073/pnas.1232475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W. M., Tsai H. J., Pang J. H., Wang T. H., Wang H. S., Hong H. S., Lee Y. S. Hum. Mutat. 2005;26:393–394. doi: 10.1002/humu.9369. [DOI] [PubMed] [Google Scholar]
- 11.Wetmur J. G., Kumar M., Zhang L., Palomeque C., Wallenstein S., Chen J. Nucleic Acids Res. 2005;33:2615–2619. doi: 10.1093/nar/gki556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu C. E., Devlin B., Galloway N., Loomis E., Schellenberg G. D. Genomics. 2004;84:600–612. doi: 10.1016/j.ygeno.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Hurley J., Engle L., Davis J., Welsh A., Landers J. Nucleic Acids Res. 2004;32:e186. doi: 10.1093/nar/gnh187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettersson M., Bylund M., Alderborn A. Genomics. 2003;82:390–396. doi: 10.1016/s0888-7543(03)00177-0. [DOI] [PubMed] [Google Scholar]
- 15.Odeberg J., Holmberg K., Eriksson P., Uhlen M. BioTechniques. 2002;33:1104–1108. doi: 10.2144/02335dd02. [DOI] [PubMed] [Google Scholar]
- 16.Burgtorf C., Kepper P., Hoehe M., Schmitt C., Reinhardt R., Lehrach H., Sauer S. Genome Res. 2003;13:2717–2724. doi: 10.1101/gr.1442303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitra R. D., Butty V. L., Shendure J., Williams B. R., Housman D. E., Church G. M. Proc. Natl. Acad. Sci. USA. 2003;100:5926–5931. doi: 10.1073/pnas.0936399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner J. H., Cheung C. L., Woolley A. T., Lieber C. M. Prog. Biophys. Mol. Biol. 2001;77:73–110. doi: 10.1016/s0079-6107(01)00011-6. [DOI] [PubMed] [Google Scholar]
- 19.Woolley A. T., Guillemette C., Li Cheung C., Housman D. E., Lieber C. M. Nat. Biotechnol. 2000;18:760–763. doi: 10.1038/77760. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X. B., Lizardi P. M., Huang X. H., Bray-Ward P. L., Ward D. C. Proc. Natl. Acad. Sci. USA. 2001;98:3940–3945. doi: 10.1073/pnas.061026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wjst M., van den Boom D. Methods Mol. Biol. 2005;311:125–137. doi: 10.1385/1-59259-957-5:125. [DOI] [PubMed] [Google Scholar]
- 22.Douglas J. A., Boehnke M., Gillanders E., Trent J. M., Gruber S. B. Nat. Genet. 2001;28:361–364. doi: 10.1038/ng582. [DOI] [PubMed] [Google Scholar]
- 23.Zhang K., Zhu J., Shendure J., Porreca G. J., Aach J. D., Mitra R. D., Church G. M. Nat. Genet. 2006;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- 24.Shiina T., Inoko H., Kulski J. K. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 25.Petersdorf E. W., Hansen J. A., Martin P. J., Woolfrey A., Malkki M., Gooley T., Storer B., Mickelson E., Smith A., Anasetti C. N. Engl. J. Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 26.Takemoto S. K., Terasaki P. I., Gjertson D. W., Cecka J. M. N. Engl. J. Med. 2000;343:1078–1084. doi: 10.1056/NEJM200010123431504. [DOI] [PubMed] [Google Scholar]
- 27.Carrington M. Immunol. Rev. 1999;167:245–256. doi: 10.1111/j.1600-065x.1999.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 28.Cavalli-Sforza L., Bodmer W. The Genetics of Human Populations. San Francisco: Freeman; 1971. [Google Scholar]
- 29.Imanishi T., Akaza T., Kimura A., Tokunaga K., Gojobori T. In: HLA 1991, Proceedings of the Eleventh International Histocompatibility Workshop and Conference. Tsuji K., Aizawa M., Sasazuki T., editors. Oxford: Oxford Univ. Press; 1992. pp. 1065–1220. [Google Scholar]
- 30.Miller S. A., Dykes D. D., Polesky H. F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersdorf E. W., Smith A. G., Mickelson E. M., Martin P. J., Hansen J. A. Immunogenetics. 1991;33:267–275. doi: 10.1007/BF00230505. [DOI] [PubMed] [Google Scholar]
- 32.Petersdorf E. W., Gooley T. A., Anasetti C., Martin P. J., Smith A. G., Mickelson E. M., Woolfrey A. E., Hansen J. A. Blood. 1998;92:3515–3520. [PubMed] [Google Scholar]
- 33.Guo Z., Gatterman M. S., Hood L., Hansen J. A., Petersdorf E. W. Genome Res. 2002;12:447–457. doi: 10.1101/gr.206402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Z., Guilfoyle R. A., Thiel A., Wang R., Smith L. Nucleic Acids Res. 1994;22:5456–5465. doi: 10.1093/nar/22.24.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin B., White J. T., Ferguson C., Bumgarner R., Friedman C., Trask B., Ellis W., Lange P., Hood L., Nelson P. S. Cancer Res. 2000;60:858–863. [PubMed] [Google Scholar]
- 36.Cao K., Chopek M., Fernandez-Vina M. A. Rev. Immunogenet. 1999;1:53–84. [PubMed] [Google Scholar]
- 37.Petersdorf E. W., Hansen J. A. Tissue Antigens. 1995;46:73–85. doi: 10.1111/j.1399-0039.1995.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 38.Mori M., Beatty P. G., Graves M., Boucher K. M., Milford E. L. Transplantation. 1997;64:1017–1027. doi: 10.1097/00007890-199710150-00014. [DOI] [PubMed] [Google Scholar]
- 39.Salem R. M., Wessel J., Schork N. J. Hum. Genomics. 2005;2:39–66. doi: 10.1186/1479-7364-2-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fishelson M., Dovgolevsky N., Geiger D. Hum. Hered. 2005;59:41–60. doi: 10.1159/000084736. [DOI] [PubMed] [Google Scholar]
- 41.Kwok P. Y., Xiao M. Cold Spring Harbor Symp. Quant. Biol. 2003;68:65–67. doi: 10.1101/sqb.2003.68.65. [DOI] [PubMed] [Google Scholar]
- 42.Inbar E., Yakir B., Darvasi A. Nucleic Acids Res. 2002;30:e76. doi: 10.1093/nar/gnf075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cullen M., Perfetto S. P., Klitz W., Nelson G., Carrington M. Am. J. Hum. Genet. 2002;71:759–776. doi: 10.1086/342973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H. Y., Luo M., Tereshchenko I. V., Frikker D. M., Cui X., Li J. Y., Hu G., Chu Y., Azaro M. A., Lin Y., et al. Genome Res. 2005;15:276–283. doi: 10.1101/gr.2885205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton R., Wilming L., Rand V., Lovering R. C., Bruford E. A., Khodiyar V. K., Lush M. J., Povey S., Talbot C. C., Jr., Wright M. W., et al. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 46.Miretti M. M., Walsh E. C., Ke X., Delgado M., Griffiths M., Hunt S., Morrison J., Whittaker P., Lander E. S., Cardon L. R., et al. Am. J. Hum. Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.