Abstract
Deformylase inhibitors belong to a novel antibiotic class that targets peptide deformylase, a bacterial enzyme that removes the formyl group from N-terminal methionine in nascent polypeptides. Using the bacterium Salmonella enterica, we isolated mutants with resistance toward the peptide deformylase inhibitor actinonin. Resistance mutations were identified in two genes that are required for the formylation of methionyl (Met) initiator tRNA (tRNAi) fMet: the fmt gene encoding the enzyme methionyl-tRNA formyltransferase and the folD gene encoding the bifunctional enzyme methylenetetrahydrofolate-dehydrogenase and -cyclohydrolase. In the absence of antibiotic, these resistance mutations conferred a fitness cost that was manifested as a reduced growth rate in laboratory medium and in mice. By serially passaging the low-fitness mutants in growth medium without antibiotic, the fitness costs could be partly ameliorated either by intragenic mutations in the fmt/folD genes or by extragenic compensatory mutations. Of the extragenically compensated fmt mutants, approximately one-third carried amplifications of the identical, tandemly repeated metZ and metW genes, encoding tRNAi. The increase in metZW gene copy number varied from 5- to 40-fold and was accompanied by a similar increase in tRNAi levels. The rise in tRNAi level compensated for the lack of methionyl-tRNA formyltransferase activity and allowed translation initiation to proceed with nonformylated methionyl tRNAi. Amplified units varied in size from 1.9 to 94 kbp. Suppression of deleterious mutations by gene amplification may be involved in the evolution of new gene functions.
Keywords: compensatory evolution, gene amplification, Salmonella typhimurium, actinonin, peptide deformylase
Peptide deformylase (PDF) inhibitors such as actinonin are a new class of antibiotic that in vitro and in animal models have shown good activity against several bacterial species, including Staphylococcus aureus and Streptococcus pyogenes (1–3). Actinonin is synthesized by actinomycetes (4), and it inhibits bacterial growth in a bacteriostatic manner (5, 6). Unlike protein synthesis in eukaryotes, where translation is initiated with a nonformylated methionyl (Met) initiator tRNA (tRNAi), most eubacteria initiate translation with a formylated Met-tRNAi (5, 7). When translation is complete, the formyl group is removed by PDF, encoded by the def gene (1, 8), and, for many proteins, an amino peptidase subsequently removes the N-terminal methionine. These steps are often required for the nascent polypeptide to mature into a functional protein (1, 8). Actinonin inhibits PDF, leading to accumulation of toxic formylated polypeptides in the cell and subsequent growth inhibition (5, 6).
The most common, previously identified mutations causing actinonin resistance are loss-of-function mutations in the fmt gene (2, 5). This gene encodes methionyl-tRNA formyltransferase (FMT), an enzyme that adds the formyl group to the Met-tRNAi. Resistance is conferred in fmt mutants because Met-tRNAi is unformylated, and, as a result, the need for the PDF function is bypassed. When FMT activity is absent, protein synthesis has to be initiated with unformylated Met-tRNAi. As a consequence of this abnormal initiation, both translation and growth rates are reduced (9). Such reductions in fitness (reduced growth rate and/or virulence) are typical for most types of resistance mechanisms and bacterial species (10–14). Furthermore, it has been demonstrated that these costs can be compensated by second-site mutations, often without loss of resistance (10, 15–21).
In both eubacteria and eukaryotes, gene amplification is a common mechanism of adaptation in response to different types of selective pressures. Thus, when over-expression of a gene product confers a phenotypic advantage needed for fitness and survival, cells with specific gene amplifications might be selected. For example, in Escherichia coli over-expression (via increased gene dosage) of ampC or blaA, encoded β-lactamases may confer resistance to penicillins, and, in Salmonella typhimurium and Acinetobacter, amplification of the araBADC operon and cat genes can enhance the growth rates on arabinose and benzoate, respectively (22–25). Similarly, adaptive mutability in the lac system has been proposed to be mediated via selected stepwise increases in the copy number of the partly functional lac operon (26, 27). Because of the high intrinsic instability of tandem amplifications, haploid segregants will rapidly appear and take over the population when the selective condition disappears (28, 29). Gene amplification is also thought to be important for the creation of new genes. One classical and widely accepted hypothesis suggests that novel gene functions can evolve from duplication of a preexisting gene, and, over time, one gene copy diverges genetically to perform a different function (30). For this process to occur, the duplicated genes need to rise to an appreciable frequency and be maintained long enough in the population to allow for functional divergence and selection for the new function before one of the copies is lost or mutationally inactivated. It is still unclear how often and by which mechanism(s) this process occurs (27, 31–36).
We isolated a set of actinonin-resistant Salmonella enterica serovariant (var.) typhimurium LT2 mutants, identified the resistance mutations, and determined their effect on bacterial fitness. Actinonin-resistant mutants were subjected to compensatory evolution, and it was shown that selective gene amplification and subsequent overproduction of initiator tRNA could compensate for the fitness cost of resistance to actinonin.
Results
Resistance Mutations.
We isolated 31 independent actinonin-resistant mutants. To identify the resistance mutations, we sequenced the fmt and def genes, which previously had been identified as targets for actinonin resistance (2, 3). No mutations were found in the def gene (encoding PDF), whereas, in 11 of the 31 resistant strains, mutations were identified in the fmt gene (Table 1). Transfer of the mutated fmt genes by P22 phage transduction into a wild-type strain confirmed that these mutations were sufficient to confer the resistance (data not shown).
Table 1.
Resistance mutations identified in actinonin-resistant strains
| Gene | Identified mutations | No. of independent mutants isolated |
|---|---|---|
| fmt | aca→aga, T12R | 1 |
| tgt→tat, C107Y | 1 | |
| tgg→tga, W129Stop | 1 | |
| gcg→gag, A213E | 1 | |
| tgg→cgg, W217R | 1 | |
| tgc→tga, C227Stop | 1 | |
| tgg→tga, W238Stop | 1 | |
| gcg→ccg, A251P | 1 | |
| deletion of base pairs 363–367 | 3 | |
| folD | tat→tct, C50S | 1 |
| ctg→ccg, L86P | 1 | |
| ggt→cgt, G129R | 1 | |
| acg→acc, T185R | 1 | |
| ggc→agc, G264S | 1 | |
| deletion of base pairs 403–413 | 1 | |
| Unidentified | Mutation linked to fimC::Tn10dTet | 6 |
| Unidentified | Unknown | 8 |
Nucleotide substitutions, amino acid substitutions (one-letter code), and their positions are indicated.
To identify additional resistance mutations, we transduced a pool of random mini transposon Tn10 (miniTn10) insertions into one actinonin-resistant strain (DA8325). Transductants that had lost the actinonin-resistance phenotype, indicating that the transposon was genetically linked to the actinonin-resistance mutation, were isolated. The transposon insertion point was located to the fimC gene. All actinonin-resistant mutants without mutations in fmt were screened for linkage of the resistance mutation to the transposon in fimC. In 12 resistant mutants, we found a genetic linkage between the resistance mutation and the transposon located in fimC. The folD gene is situated ≈2 kbp from the transposon. This gene encodes a bifunctional enzyme required for production of 10-formyl-H4folate (37). 10-formyl-H4folate is the donor of the formyl group to the Met-tRNAi (37). We could identify mutations in the coding sequence of the folD gene in 6 of the 12 strains displaying genetic linkage to the transposon (Table 1). The 6 strains with an intact folD coding sequence, but with a resistance mutation linked to fimC, might carry mutations that reduce expression of the folD gene. Loss of folD function is likely to decrease the level of formylated Met-tRNAi in the cell by reducing biosyntheis of the precursor molecule 10-formyl-H4folate (37). Similar to mutations in fmt, loss of folD function is expected to cause initiation of translation with an unformylated tRNAi and thereby bypass the requirement for the PDF function.
Fitness of Mutants Measured by Growth Rates in LB Broth and Mice.
Most resistant mutants showed severe reductions in growth rate in LB broth (Fig. 1). The results for the fmt mutants are in agreement with previous observations in E. coli, showing that formylation of Met-tRNAi is required for efficient translation initiation and rapid growth (9). For three strains, fitness was also determined by performing competition experiments in mice. Growth rates in mice were drastically reduced (Table 2), demonstrating that actinonin resistance also imposes a fitness burden on the resistant bacteria during growth in a host organism.
Fig. 1.
Fitness of actinonin-resistant strains measured as growth rates in LB. All values are presented as a relative fitness calculated as the generation time of the wild type divided by the generation time of the tested strain. Susceptible wild-type strain, open circle; actinonin-resistant fmt mutant, filled triangles; actinonin-resistant folD mutant, filled squares; actinonin-resistance linked to folD, filled diamonds; unknown resistance mutations, filled circles. Each data point represents one specific strain. The standard error was less than ±5% of the mean value.
Table 2.
MIC values and fitness in vitro and in mice for actinonin-resistant and compensated mutants
| Strain index | Genotype | MIC (mg/L) | Relative growth rate in LB | Competition in mice |
|---|---|---|---|---|
| DA6192 | Wild type | 64 | 1 | 1 |
| Resistant | ||||
| DA8325 | folD (G129R) | >1,024 | 0.58 | 0.0007 |
| DA8326 | fmt (deletion of base pairs 363–367) | >1,024 | 0.24 | 4 × 10−6 |
| DA8340 | fmt (T12R) | >1,024 | 0.38 | 5 × 10−5 |
| Compensated | ||||
| DA8771 | fmt (deletion of base pairs 363–367), amplification metZW (Fig. 3) | >1,024 | 0.4 | 4 × 10−6 |
| DA8773 | fmt (deletion of base pairs 363–367), amplification metZW (Fig. 3) | >1,024 | 0.85 | 2 × 10−4 |
| DA8797 | fmt (T12R), amplification of metZW (Fig. 3) | >1,024 | 0.81 | 0.02 |
| DA8809 | fmt (T12R), fmt (G176S) | 64 | 0.57 | 0.003 |
| DA8616 | folD (G129R), folD (R129S) | 64 | 0.94 | 0.06 |
Compensatory Evolution.
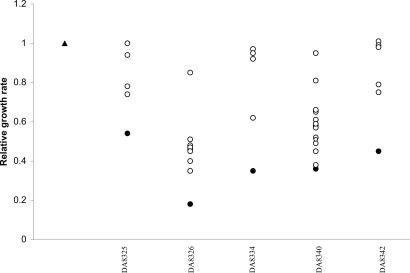
Five slow-growing resistant mutants were subjected to compensatory evolution (four fmt mutants and one folD mutant). For each resistant strain, 4–15 independent lineages were serially passaged in rich medium. When growth-compensated cells constituted the majority of the population (50–150 generations of growth), one compensated mutant from each lineage was isolated and saved. All compensated lineages exhibited increased growth rates compared with the resistant parental strain (Fig. 2). A subset of the compensated mutants was also tested with regard to fitness in mice by competition experiments against the parental susceptible strain. The compensated mutants only showed a partial restoration of fitness in mice (Table 2).
Fig. 2.
Relative fitness of growth-compensated strains measured as growth rates in LB. Values are presented as the wild-type generation time divided by the generation time of the tested strain. Susceptible wild-type strain, filled triangle; actinonin-resistant parental strains, filled circles; growth-compensated strains, open circles. The DA numbers indicate the actinonin-resistant parental strain used for compensatory evolution. Each data point represents one specific strain. The standard error was less than ±5% of the mean value.
Intragenic Compensatory Mutations.
We identified intragenic compensatory mutations by sequencing the fmt and folD genes. Several fast-growing mutants derived from the resistant strains (DA8325, DA8334, DA8340, and DA8342) contained intragenic compensatory mutations that were either intracodonic (reversion of a nonsense codon to sense) or extracodonic (Table 3).
Table 3.
Mutations identified in growth-compensated mutants
| Parental strain | Resistance mutation | Compensatory mutation | No. of independent mutants isolated |
|---|---|---|---|
| DA8326 | fmt (del bp363-367) | Amplification of metZW | 6 |
| unknown | 4 | ||
| DA8334 | fmt (W129Stop) | fmt (tga→aga, Stop129R) | 2 |
| fmt (tga→gga, Stop129G) | 1 | ||
| unknown | 1 | ||
| DA8340 | fmt (T12R) | fmt (R12T), reversion | 1 |
| fmt (ggc→agc, G176S) | 1 | ||
| Amplification of metZW | 2 | ||
| unknown | 11 | ||
| DA8342 | fmt (W238Stop) | fmt (tga→aga, Stop238R) | 3 |
| unknown | 2 | ||
| DA8325 | folD (G129R) | folD (cgt→agt, R129S) | 5 |
Extragenic Compensatory Mutations.
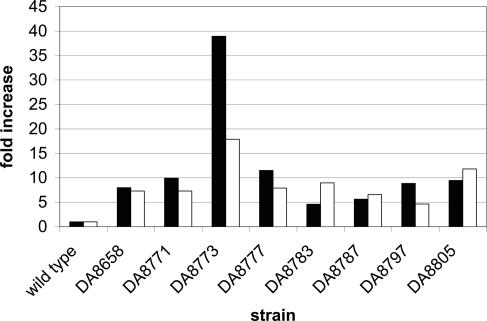
We isolated miniTn10s adjacent to the compensatory mutation in strain DA8773 (evolved from strain DA8326). By sequencing outward from the transposon, the insertion point was identified to the ygdI gene. The metZ and metW genes are situated ≈6 kbp from the insertion point. These identical, tandemly repeated genes encode tRNAi and were possible targets for compensatory mutations. For the eight independent growth-compensated mutants examined, no mutations were found in the metZW structural genes, but Southern and Northern blot hybridizations showed that the gene copy number and the levels of tRNAi were increased 5- to 40-fold (Fig. 3).
Fig. 3.
Increase in copy number of metZW genes (black bars) and tRNAi levels (white bars) in the compensated mutants as measured by Southern and Northern blot hybridization, respectively. The standard error was less than ±30% of the mean value.
Overproduction of tRNAi Alone Can Suppress the Growth Defect Caused by fmt Inactivation.
To prove that overproduction of tRNAi is sufficient for suppressing the growth defect associated with loss of FMT function, we constructed a fmt::kanR knockout mutation. As expected, this mutant showed a very slow growth rate (tgen = 110 min) as compared with the wild type (tgen = 22 min). When introducing a plasmid with a low copy number carrying the metZW genes and their native promoter into the fmt::kanR strain, the generation time was reduced to 27 min (the plasmid itself had no effect on growth rate). Similarly, when the metZW plasmid was introduced into other actinonin-resistant fmt mutants (DA8326 and DA8340), the generation times were reduced from 78 to 27 and from 67 to 24 min, respectively. This result shows that an increased level of tRNAi is sufficient to confer suppression of the fmt growth defect, similar to what was previously reported for E. coli fmt mutants (9, 38).
Minimal Inhibitory Concentrations (MIC) in Resistant and Compensated Mutants.
S. typhimurium showed a relatively high intrinsic resistance toward actinonin that could be further increased by mutations in the fmt or folD genes (Table 2). The different compensatory mutations had different effects on the MIC. Thus, fmt and folD mutants that were compensated by intragenic mutations showed a decreased MIC (identical to the susceptible wild-type strain), indicating that the respective functions had been restored. In contrast, fmt mutants that were compensated by tRNA amplification retained high MIC values, as would be expected because the FMT function was still defective.
Size of the Amplified Units.
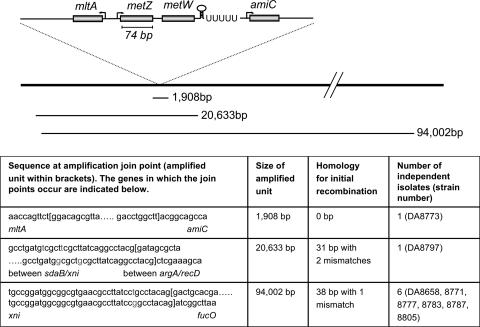
To determine the size of the amplified units and the structure of the duplication junctions, we performed PCRs with primer combinations facing outward from the metZW genes. To give rise to a PCR product, the primers need to face each other over a duplication join point. Using this procedure, we mapped the duplication join points for the eight independently isolated mutants (Fig. 4). The sizes of the amplified units varied from 1.9 to 94 kbp, and, for one of the junctions, no sequence homology was observed (39). Seven of the amplifications occurred via longer repeats; one had a 31-bp repeat sequence with two mismatches (size of amplified unit was 20,633 bp), and the remaining six had identical join points with a 38-bp repeat sequence with one mismatch (size of amplified unit was 94,002 bp).
Fig. 4.
Schematic organization of the metZW region and the amplified regions. Arrows indicate promoters and transcription directions. The stem-loop followed by a stretch of U's is a potential Rho-independent transcription terminator. Region is not drawn to scale. The size of the amplified units, the sequence of the duplication join points, the potential homology used to form the initial duplication, and the number of independent mutants isolated are indicated.
Discussion
One general mechanism to increase resistance against deformylase inhibitors is to reduce formylation of Met-tRNAi (2, 5). We identified two specific mechanisms that conferred reduced formylation. Mutations in the fmt gene were identified in ≈30% of the independently isolated resistant strains. Similar resistance mutations previously have been described in, for example, S. aureus and E. coli (2, 5). We also discovered a resistance mechanism caused by mutations in or near the folD gene. These mutations are most likely reducing the amount of the formyl donor 10-formyl-H4folate required for the formylation reaction.
Most resistance mutations substantially reduced bacterial fitness during growth in vitro and in mice. For the fmt and folD mutants, the fitness reduction likely results from the decreased rate of formylation and translation initiation (9, 38). Functional compensation of the defective formylation and slow translation rate could occur via two different mechanisms: by restoring the capacity to formylate Met-tRNAi or by bypassing the need for formylated Met-tRNAi to initiate translation. Regaining the ability to formylate the Met-tRNAi mainly depends on intragenic compensatory mutations that restore FMT activity, whereas creation of a bypass pathway involves extragenic compensatory mutations. For the extragenically compensated fmt mutants, amplification of the metZW genes was found in 8 of the 25 growth-compensated strains. The resulting increase in tRNAi level could potentially, by mass-action, compensate for the reduced translation initiation rate in at least two different ways. If the mutated FMT enzyme has some residual activity, the rate of formylation could rise as a result of an increase in the level of Met-tRNAi substrate. Alternatively, if FMT activity is completely absent, an increased level of tRNAi could increase the level of unformylated Met-tRNAi that could then abnormally bind to the ribosomal P site. Because growth of the mutant with a completely inactivated fmt gene was restored by tRNAi overproduction, our results suggest that overproduced unformylated Met-tRNAi can bind directly to the ribosomal P site (9, 38). However, this finding does not exclude the possibility that some fmt mutants have a residual formylation-activity or that there exists an FMT-independent pathway that provides low levels of formylated Met-tRNAi.
Most bacterial genomes are in a dynamic state with regard to gene copy number, and spontaneous duplications form and segregate at high rates. For example, in S. typhimurium under nonselective growth conditions, most genes are duplicated with a frequency of 10−5 to 10−2, suggesting that at any given time ≈10% of all cells in a population have a duplication somewhere in the chromosome (24, 26). The highest frequency and largest duplications are typically seen between rrn operons, where extensive homologies are available to form the duplication (26). Certain duplications may be further amplified in response to a novel selection pressure when over-expression of a gene product included in the duplication confers a fitness advantage. Formation of the first duplication is probably the rate-limiting step in high level gene amplification (28, 40).
RecA-dependent intra- and interchromosomal recombination requires at least 25 bp of homology to function efficiently (39). As expected, most (7 of 8) of the independent duplication join points occurred via longer repeats (30–38 bp) and were therefore likely to be formed in a RecA-dependent manner. One of the identified duplication join points did not show any DNA homology, indicating that the initial duplication was formed via a more infrequent RecA-independent recombination mechanism. Other studies have also shown that the initial duplication may be formed by a RecA-independent pathway, whereas the subsequent high-level gene amplification is mediated through RecA-dependent homologous recombination of the duplicated region (25, 40, 41). The size of the amplified units varied from 1.9 to 94 kbp, and it is notable that, for the latter class, the observed copy number of ten would result in the addition of almost 1 Mbp of extra DNA per chromosome.
There are several broader implications of our data. The first implication relates to genome evolution, where our findings provide a potential solution to a problem associated with the duplication-divergence model for the evolution of new genes (31–36, 42). After a gene duplication has occurred, it is more likely that one copy is silenced by degenerative mutations or that both copies are partially compromised by complementary degenerative mutations, rather than one copy acquiring a novel, beneficial function that is preserved by selection. However, if an increased gene dosage is required to compensate for the fitness effect of a deleterious mutation, positive selection could raise the frequency of and maintain an amplified array free of degenerative mutations long enough for one of the copies to acquire a novel adaptive function (see Fig. 5, which is published as supporting information on the PNAS web site). This adaptation could occur either by accumulating by chance a rare mutation that creates the new function or by selection for improvement of a weak secondary activity already preexistent in the ancestral gene product. In addition, amplification increases the number of gene copies that can accumulate weakly selected mutations that can then be reassorted by recombination to generate an increasingly beneficial gene variant. Finally, the amplified arrays can be quite large (up to 94 kbp in this system) and therefore include many additional genes apart from the one that confers the compensation. Thus, all of the genes within the amplified unit are potential targets for acquiring rare beneficial mutations that result in novel functions. To examine the feasibility of the suggested model for gene evolution, we performed a population genetic analysis where the rate of fixation of a beneficial mutation that occurs in a compensatory gene amplification was calculated (see Supporting Text, which is published as supporting information on the PNAS web site). Using realistic assumptions for the population genetic parameters, the proposed process is under certain circumstances expected to provide a substantial driving force for introducing new genes into the genome.
Finally, in a medical context, the present study shows that resistance to deformylase inhibitors is associated with substantial fitness costs. Furthermore, even though these costs can be partly compensated in vitro, the compensated mutants are still severely impaired for growth in mice. With regard to the rate of resistance development in clinical settings, these findings would appear beneficial to this class of drugs (43). Thus, it is expected that, because of their low fitness, the resistant mutants, even with compensatory mutations, will have difficulties growing inside a human host. This finding implies that the rate of appearance and spread of mutants resistant to deformylase inhibitors could be low in a clinical environment (10–14).
Materials and Methods
Isolation of Actinonin-Resistant Strains.
For all experiments, S. enterica serovariant (var.) typhimurium LT2 was used. Thirty-one independent actinonin-resistant mutants were isolated on rich agar (LA) plates supplemented with 200 mg/liter of actinonin and stored at −80°C.
Measurements of Growth Rates.
Exponential growth rates were measured in LB broth at 37°C. On a bioscreen plate, 104 cells were inoculated into 400 μl LB, and the absorbance at 600 nm was read by using a BioscreenC reader (Labsystems). Relative generation times were calculated as the generation time of the reference strain divided by the generation time of the tester strain.
Competition Experiments in Mice.
All mouse experiments were carried out according to institutional and national guidelines (Ethical permit number N318/03). For a full description of the mouse model, see refs. 11 and 18. Briefly, each resistant- and growth-compensated strain tested was tagged with a neutral kanamycin resistance marker and competed with the susceptible strain. The strain tested and the reference strain were mixed at a 1:1 ratio, and 2 × 105 cells of the mixture were injected i.p. into BALB/c mice. After 2 days, the mice were killed, and the liver and spleen were homogenized in PBS. Dilutions of the homogenates were plated on LA and LA supplemented with 50 mg/liter of kanamycin. Competition indexes were calculated as the ratio of the reference and tester strains in the inoculum divided by the ratio of the two strains recovered after 2 days of growth in mice.
Compensatory Evolution.
Several independent lineages were inoculated with 4 × 106 cells from a culture of the slow-growing resistant parental strain grown to saturation. The lineages were then serially passaged as follows: Each day, 2 μl (corresponding to ≈8 × 106 cells) from a 1.5-ml culture grown to saturation were transferred to fresh LB broth. The culture was grown to saturation at 37°C (≈4 × 109 cells per ml), and the procedure was repeated. Every second cycle, a sample of each culture was spread on LA plates and, after incubation at 37°C for 24 h, examined for fast-growing variants (large colonies). When the fast-growing variants constituted the majority of the population, the experiment was stopped. One random growth-compensated colony was saved from each lineage.
PCR and Sequencing.
Primers and probes used for PCR, sequencing, and Southern and Northern blot hybridization are available in Table 4, which is published as supporting information on the PNAS web site. For sequencing, the gene of interest was PCR-amplified and purified from solution using GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech). The purified PCR product was used as template in a sequencing reaction using BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems), and the sequences were read with an Prism 3100 genetic analyzer (Applied Biosystems).
Construction of the fmt Insertion Mutant and Cloning of the tRNAi Genes.
The fmt gene was inactivated by linear transformation as described (44). A kanamycin-resistance cassette from plasmid pKD4 was PCR-amplified by using primers with 40 nucleotides of flanking homology toward the fmt gene. This PCR fragment was electroporated into S. typhimurium, and transformants were selected on LA, kanamycin (30 mg/liter) agar plates. PCR and DNA sequencing confirmed the correct insertion site. The tRNAi genes, including their native promoter, were amplified by PCR from genomic DNA isolated from a wild-type strain. The PCR-amplified DNA fragment was 300 bp in length and extended 63 bp upstream of the metZ gene and 57 bp downstream of the metW gene. The PCR fragment and the pBAD30 plasmid (ampR; ref. 45) were then cut with SacI and XbaI, ligated, and transformed into top10 competent E. coli cells (Invitrogen). Transformants were then selected on LA plates supplemented with 100 mg/liter ampicillin.
Isolation of Tn10 Insertions.
Transposon pools with random insertions of the miniTn10 Δ16Δ17 (tetR) were constructed as described in ref. 46.
Isolation of miniTn10s linked to actinonin-resistance mutations.
A P22 phage lysate grown on a pool of randomly inserted miniTn10 insertions in a wild-type genetic background was prepared. The phage lysate was used to infect the actinonin-resistant strain of interest. Tetracyclin-resistant colonies were selected on LA plates supplemented with 30 mg/liter of tetracyclin, and loss of actinonin resistance was screened for by replica printing to LA plates with and without 200 mg/liter of actinonin. Tetracyclin-resistant and actinonin-susceptible clones were identified, and linkage of the tetracyclin marker to the actinonin-resistance mutation was confirmed by backcrosses to the actinonin-resistant strain.
Isolation of miniTn10s linked to compensatory mutations.
P22 phage was grown on a pool of random miniTn10 insertions prepared in a compensated mutant strain. The phage lysate was used to infect the slow-growing resistant strain. Tetracyclin-resistant colonies were selected as described above. The transductants were then screened for acquisition of the growth-compensated phenotype by visual examination of colony size. Linkage between the growth-compensated phenotype and the tetracyclin-resistance marker was established by backcrossing to the slow-growing mutant.
Identification of insertion points.
Arbitrary primed PCR directed outward from the miniTn10 was performed in two steps. First, a PCR was set up with one specific primer for the transposon and a mix of arbitrary primers. The arbitrary primers used consisted of a defined part (20 bp) and a randomly variable part (15 bp). Using the PCR products from the first reaction as template, a second nested PCR was performed. The product from the second PCR was then used as a template for sequencing.
Southern Blot Hybridization.
Genomic DNA was isolated using Wizard Genomic DNA purification kit (Promega). The DNA was cut with BamHI (Amersham Pharmacia Biotech), and the fragments were separated on 1% Seakem LE agarose gels (Cambrex) in 1× TBE buffer. DNA was transferred to Hybond-N+ nylon membranes (Amersham Pharmacia Biotech) by vacuum blotting. DNA probes were prepared from purified PCR products and labeled with 32P by using Rediprime II random primer labeling system (Amersham Pharmacia Biotech). Quantification of the radioactivity in the fragments was performed on a Molecular Dynamics PhosphorImager, and the intensities of the fragments containing the tRNAi genes were normalized to an intrinsic control gene (STM0329).
Northern Blot Hybridization.
Total RNA was purified from exponential phase cultures (OD600 ≈ 0.5) by using SV total RNA isolation kit (Promega). The RNA was separated on denaturing 8% polyacrylamide gels (7 M urea, 0.5× TBE) and transferred to Hybond-N+ nylon membranes (Amersham Pharmacia Biotech) by electroblotting. The membranes were hybridized with specific 32P-end-labeled primers. Quantification of the radioactivity in the bands was performed on a Molecular Dynamics PhosphorImager, and the tRNAi bands were normalized to the 5S rRNA band.
Identification of Duplication Junctions.
To identify duplication join points, a set of primers facing outward from the metZW genes were constructed. PCRs were then performed with different combinations of the primers. A PCR product will be produced only if the reverse and forward primers are brought to face each other over a duplication junction. PCR products unique for the strains carrying the amplifications were sequenced and the join points identified.
Supplementary Material
Acknowledgments
We thank Lynn Miesel (Schering-Plough Research Institute, Kenilworth, NJ) for a generous gift of actinonin and Andrea Hinas and Fredrik Söderbom (both of the Swedish University of Agricultural Sciences, Uppsala) for help with Northern blot hybridizations. This work was supported by grants from the Swedish Research Council and Uppsala University (to D.I.A.).
Abbreviations
- FMT
methionyl-tRNA formyltransferase
- LA
rich agar
- Met
methionyl
- MIC
minimal inhibitory concentrations
- miniTn10
mini transposon Tn10
peptide deformylase
- tRNAi
methionyl initiator tRNA.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Giglione C., Pierre M., Meinnel T. Mol. Microbiol. 2000;36:1197–1205. doi: 10.1046/j.1365-2958.2000.01908.x. [DOI] [PubMed] [Google Scholar]
- 2.Margolis P. S., Hackbarth C. J., Young D. C., Wang W., Chen D., Yuan Z., White R., Trias J. Antimicrob. Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margolis P., Hackbarth C., Lopez S., Maniar M., Wang W., Yuan Z., White R., Trias J. Antimicrob. Agents Chemother. 2001;45:2432–2435. doi: 10.1128/AAC.45.9.2432-2435.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umezawa H., Aoyagi T., Tanaka T., Suda H., Okuyama A., Naganawa H., Hamada M., Takeuchi T. J. Antibiot. 1985;38:1629–1630. doi: 10.7164/antibiotics.38.1629. [DOI] [PubMed] [Google Scholar]
- 5.Apfel C. M., Locher H., Evers S., Takacs B., Hubschwerlen C., Pirson W., Page M. G., Keck W. Antimicrob. Agents Chemother. 2001;45:1058–1064. doi: 10.1128/AAC.45.4.1058-1064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D. Z., Patel D. V., Hackbarth C. J., Wang W., Dreyer G., Young D. C., Margolis P. S., Wu C., Ni Z. J., Trias J., et al. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- 7.Newton D. T., Creuzenet C., Mangroo D. J. Biol. Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 8.Bandow J. E., Becher D., Buttner K., Hochgrafe F., Freiberg C., Brotz H., Hecker M. Proteomics. 2003;3:299–306. doi: 10.1002/pmic.200390043. [DOI] [PubMed] [Google Scholar]
- 9.Guillon J. M., Mechulam Y., Schmitter J. M., Blanquet S., Fayat G. J. Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson D. I., Levin B. R. Curr. Opin. Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 11.Bjorkman J., Hughes D., Andersson D. I. Proc. Natl. Acad. Sci. USA. 1998;95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacVanin M., Johanson U., Ehrenberg M., Hughes D. Mol. Microbiol. 2000;37:98–107. doi: 10.1046/j.1365-2958.2000.01967.x. [DOI] [PubMed] [Google Scholar]
- 13.Nilsson A. I., Berg O. G., Aspevall O., Kahlmeter G., Andersson D. I. Antimicrob. Agents Chemother. 2003;47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bjorkholm B., Sjolund M., Falk P. G., Berg O. G., Engstrand L., Andersson D. I. Proc. Natl. Acad. Sci. USA. 2001;98:14607–14612. doi: 10.1073/pnas.241517298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maisnier-Patin S., Berg O. G., Liljas L., Andersson D. I. Mol. Microbiol. 2002;46:355–366. doi: 10.1046/j.1365-2958.2002.03173.x. [DOI] [PubMed] [Google Scholar]
- 16.Macvanin M., Ballagi A., Hughes D. Antimicrob. Agents Chemother. 2004;48:3877–3883. doi: 10.1128/AAC.48.10.3877-3883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macvanin M., Bjorkman J., Eriksson S., Rhen M., Andersson D. I., Hughes D. Antimicrob. Agents Chemother. 2003;47:3743–3749. doi: 10.1128/AAC.47.12.3743-3749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjorkman J., Nagaev I., Berg O. G., Hughes D., Andersson D. I. Science. 2000;287:1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 19.Nagaev I., Bjorkman J., Andersson D. I., Hughes D. Mol. Microbiol. 2001;40:433–439. doi: 10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds M. G. Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maisnier-Patin S., Andersson D. I. Res. Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Edlund T., Normark S. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- 23.Seoane A., Sanchez E., Garcia-Lobo J. M. Antimicrob. Agents Chemother. 2003;47:682–688. doi: 10.1128/AAC.47.2.682-688.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonti R. V., Roth J. R. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reams A. B., Neidle E. L. Mol. Microbiol. 2003;47:1291–1304. doi: 10.1046/j.1365-2958.2003.03342.x. [DOI] [PubMed] [Google Scholar]
- 26.Anderson P., Roth J. Proc. Natl. Acad. Sci. USA. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth J. R., Andersson D. I. Res. Microbiol. 2004;155:342–351. doi: 10.1016/j.resmic.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 28.Reams A. B., Neidle E. L. Annu. Rev. Microbiol. 2004;58:119–142. doi: 10.1146/annurev.micro.58.030603.123806. [DOI] [PubMed] [Google Scholar]
- 29.Romero D., Palacios R. Annu. Rev. Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 30.Ohno S. Evolution by Gene Duplication. Heidelberg: Springer; 1970. [Google Scholar]
- 31.Prince V. E., Pickett F. B. Nat. Rev. Genet; 2002. pp. 827–837. [DOI] [PubMed] [Google Scholar]
- 32.Hughes A. L. Proc. Biol. Sci; 1994. pp. 119–124. [DOI] [PubMed] [Google Scholar]
- 33.Wagner A. Genome Biol. 2002;3:1012. doi: 10.1186/gb-2002-3-5-reviews1012. reviews. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krakauer D. C., Nowak M. A. Semin. Cell Dev. Biol. 1999;10:555–559. doi: 10.1006/scdb.1999.0337. [DOI] [PubMed] [Google Scholar]
- 35.Taylor J. S., Raes J. Annu. Rev. Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 36.Lynch M., Conery J. S. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 37.Matthews R. G. In: Cellular and Molecular Biology, Escherichia coli and Salmonella. Neidhardt F. C., Curtiss R., editors. Vol. 1. Washington, DC: Am. Soc. Microbiol; 1996. pp. 600–611. [Google Scholar]
- 38.Guillon J. M., Heiss S., Soutourina J., Mechulam Y., Laalami S., Grunberg-Manago M., Blanquet S. J. Biol. Chem. 1996;271:22321–22325. doi: 10.1074/jbc.271.37.22321. [DOI] [PubMed] [Google Scholar]
- 39.Shen P., Huang H. V. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reams A. B., Neidle E. L. J. Mol. Biol. 2004;338:643–656. doi: 10.1016/j.jmb.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Tlsty T. D., Albertini A. M., Miller J. H. Cell. 1984;37:217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- 42.Kimura M., Ohta T. Genet. Res. 1970;16:145–150. doi: 10.1017/s0016672300002378. [DOI] [PubMed] [Google Scholar]
- 43.Johnson K. W., Lofland D., Moser H. E. Curr. Drug Targets Infect. Disord. 2005;5:39–52. doi: 10.2174/1568005053174618. [DOI] [PubMed] [Google Scholar]
- 44.Datsenko K. A., Wanner B. L. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guzman L. M., Belin D., Carson M. J., Beckwith J. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altman E., Roth J. R., Hessel A., Sanderson K. E. In: Cellular and Molecular Biology, Escherichia coli and Salmonella. Neidhardt F. C., Curtiss R., editors. Vol. 2. Washington, DC: Am. Soc. Microbiol; 1996. pp. 2613–2626. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.