Abstract
CD45, a protein tyrosine phosphatase that regulates Src family kinases, is important for regulating T cell and B cell receptor signaling; however, little is known about how CD45 regulates immunoreceptor tyrosine-based activation motif (ITAM)-dependent natural killer (NK) cell receptor signaling and the resulting effector functions. NK cells from CD45-deficient mice are relatively competent for ITAM receptor-induced cell-mediated cytotoxicity, yet completely deficient for cytokine secretion after stimulation with ligands to or antibodies against NK1.1, CD16, Ly49H, Ly49D, and NKG2D. This deficiency in cytokine/chemokine production occurs at the level of mRNA expression. After receptor engagement, extracellular signal-regulated kinase and c-Jun N-terminal kinase activation was markedly perturbed, whereas p38 activation was not substantially affected. The pattern and amounts of basal tyrosine phosphorylation were altered in freshly isolated NK cells and were surprisingly and markedly increased in IL-2-expanded NK cells from CD45−/− mice. These findings indicate that CD45-dependent regulation of ITAM-dependent signaling pathways is essential for NK cell-mediated cytokine production but not cytolytic activity.
Keywords: tyrosine phosphatase
Natural killer (NK) cells confer immune protection against certain intracellular pathogens and tumors. They recognize target cells by means of activating cell-surface receptors, which upon ligation induce target cell lysis and cytokine production. Although some ligands of the activating receptors are known, many are still undiscovered. The known ligands include Ig constant regions, viral antigens, adhesion molecules, and stress-induced glycoproteins, which are induced after viral infection or cellular transformation (1). Inhibitory receptors, which recognize self-MHC class I molecules, modulate the signals of the activating receptors to prevent damage of normal tissues.
Many of the known NK cell-activating receptors, such as CD16, NK1.1, Ly49H, Ly49D, and an isoform of NKG2D, associate with the CD3ζ, FcεRIγ, and/or DAP12 adapter proteins, which contain immunoreceptor tyrosine-based activation motifs (ITAMs) (2). Ligation of an ITAM-associated NK cell receptor results in cytokine production and killing of the target cell. Src family kinases phosphorylate the tyrosine residues in ITAMs and initiate the activating signal cascade (3). The tyrosine kinases Syk and Zap-70 then bind to the doubly phosphorylated ITAMs via their tandem SH2 domains, resulting in the activation of downstream molecules, such as phospholipase C-γ1 and -γ2, Vav-1, Vav-2, Vav3, Ras-family members, and the mitogen-activated protein kinases (MAPK) c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38. MAPKs are important regulators of NK cell-mediated cytotoxicity and cytokine production. ERK appears to play a major role in these effector functions (4–8), whereas JNK and p38 have more limited roles (4, 5, 9, 10).
The requirement for Src kinase activity for ITAM-based signaling implies that the regulation of Src kinases likely affects NK cell effector functions (11–13). Src kinases are modulated, in part, by the protein tyrosine phosphatase CD45. By dephosphorylating the negative regulatory tyrosine of Src kinases, CD45 generates a pool of “primed” Src family kinases capable of rapid activation upon receptor stimulation (14). Depending on a cell's activation status and developmental stage, CD45 can also dephosphorylate the catalytic tyrosine, leading to kinase inactivation (14). CD45−/− mice suffer from severe defects in B and T cell development and function because of deficient antigen receptor signaling, demonstrating the importance of this phosphatase (15, 16). When we began our studies, little was known about the role of CD45 in NK cell function. mAbs against CD45 had been reported to activate or inhibit human and rat NK cell cytokine production or target cell lysis (17–22). NK cells from CD45−/− mice had normal cytotoxic activity against the prototypic tumor cell line Yac-1 (23, 24); NK cells primarily use the NKG2D receptor to kill Yac-1 (25). A study showed that CD45−/− NK cells have a reduced capacity for IFNγ production when stimulated by anti-NK1.1 mAb (23). Here we examine the function of CD45 in cytokine and chemokine production and target cell killing initiated by well characterized ITAM-based activating receptors.
Results
ITAM-Based Receptor-Induced Cytotoxicity Is Intact in CD45−/− NK Cells.
We examined the role of CD45 in ITAM receptor-dependent NK cell-mediated cytotoxicity. Splenic and liver cells from WT and CD45−/− mice were stained with mAbs against NK cell receptors. CD3−, NK1.1+ cells were increased in both percentage and number in CD45−/− mice (2- to 3-fold increase in percentage compared with WT mice; data not shown), in accordance with prior reports (23, 24). Expression of NKG2D, Ly49H, CD16, Ly49G2, VLA-2 (DX5), and NK1.1 was similar between genotypes whether the NK cells were freshly isolated or IL-2 expanded (data not shown). However, the percentage of NK cells expressing Ly49A and Ly49D was reduced to 40% and 55% of WT levels, respectively.
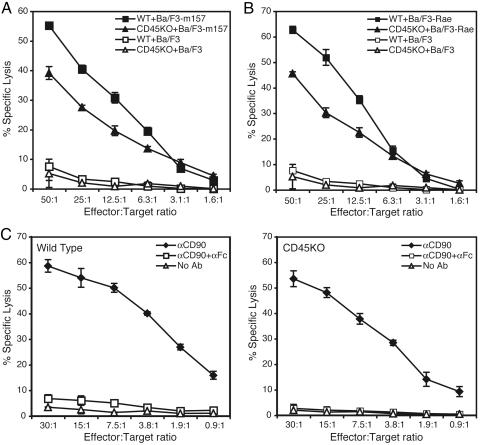
IL-2-expanded WT and CD45−/− NK cells were compared in their ability to kill target cells through specific NK cell receptors. m157 is a murine CMV-encoded ligand for the activating Ly49H receptor (26, 27). CD45−/− NK cells killed Ba/F3 mouse pro-B cells transduced with the m157 similarly to WT NK cells (71% of WT) (Fig. 1A). CD45−/− cells also efficiently killed target cells bearing RAE-1ε, a ligand for NKG2D (73% of WT) (Fig. 1B). Ly49H signals by using the ITAM-bearing DAP12 adapter (28), and NKG2D signals via the phosphatidylinositol 3-kinase activating DAP10 adapter (29), as well as through DAP12 (30).
Fig. 1.
CD45 is not required for ITAM receptor-induced NK cell-mediated cytotoxicity. (A and B) Comparison of the ability of WT and CD45−/− NK cells to kill mouse Ba/F3 cells transduced with murine CMV m157 (A) or RAE-1ε (B). Untransduced Ba/F3 cells served as a measure of background killing. (C) ADCC against RMA or anti-CD90 mAb-coated RMA targets in the presence or absence of a soluble neutralizing anti-CD16/32 mAb. Results are representative of two to four independent experiments.
To extend our findings to other ITAM adapter/receptor complexes, we examined CD16-induced cytotoxicity because this receptor signals through FcεRIγ (31, 32). We tested the function of the FcεRIγ/CD16 receptor complex in CD45−/− NK cells by testing antibody-dependent cell-mediated cytotoxicity (ADCC) against mAb-coated RMA tumor cells. 51Cr-labeled RMA cells were coated with an anti-CD90 mAb [previously shown to mediate efficient ADCC (33)] and then incubated with NK effector cells. Both WT and CD45−/− NK cells efficiently killed the anti-CD90 mAb-coated RMA cells at equivalent levels (Fig. 1C). This activity depended on signaling via CD16 because ADCC was completely blocked by the presence of a soluble neutralizing anti-CD16/32 mAb. Although ITAM-based receptor-induced killing by CD45−/− NK cells was reduced compared with WT NK cells, there was nonetheless significant and easily detectable lytic activity induced by either DAP12- or FcεRIγ-associated receptors. Therefore, NK cells lacking CD45 can still mediate cytolytic activity via ITAM-dependent receptors.
ITAM Receptor-Induced Cytokine and Chemokine Secretion Is Severely Impaired in CD45−/− NK Cells.
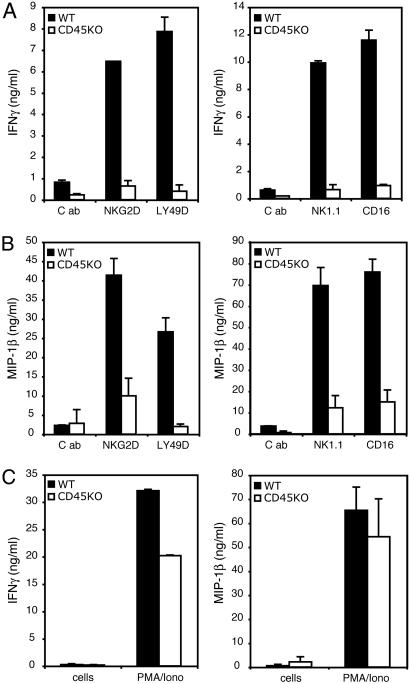
Besides directly killing target cells, NK cells also function as the main innate immune cell type producing IFNγ, as well as other cytokines and chemokines, including granulocyte–macrophage colony-stimulating factor, TNFα, macrophage inflammatory protein (MIP) 1β, MIP-1α, and RANTES. We tested the ability of CD45−/− NK cells to produce cytokines and chemokines in response to stimulation of their ITAM-based receptors. CD45−/− NK cells were defective in secreting IFNγ when stimulated with plate-bound mAbs against NKG2D, Ly49D, or NK1.1 (Fig. 2A). In contrast, WT cells made abundant amounts of cytokines. Similarly, stimulation of CD45−/− NK cells with ligands for CD16 (plate-bound IgG) or NKG2D (soluble H60) resulted in no detectable IFNγ production (Fig. 2A and data not shown). CD45−/− NK cells were also defective in MIP-1β and granulocyte–macrophage colony-stimulating factor production (Fig. 2B and data not shown). However, CD45−/− NK cells were capable of cytokine and chemokine production because abundant IFNγ and MIP-1β were produced when receptor-mediated signals were bypassed with PMA and ionomycin stimulation (Fig. 2C). Thus, CD45−/− NK cells are defective in cytokine and chemokine production when stimulated by ITAM-based receptors.
Fig. 2.
Deficient cytokine and chemokine production in response to ITAM signaling by CD45−/− NK cells. WT and CD45−/− NK cells were incubated on plate-bound mAbs for NKG2D and Ly49D (Left) or NK1.1 and mouse IgG2a (a natural ligand of CD16) (Right). C, control mAb. Supernatants were harvested at 16 h and assayed for IFNγ (A) or MIP-1β (Β) by ELISA. All mAb stimulation was performed in the presence of soluble anti-CD16/32 mAb (2.4G2) to block Fc receptor activation, except in experiments in which NK cells were stimulated via CD16 by plate-bound mouse IgG2a. (C) IFNγ (Left) and MIP-1β (Right) secretion after PMA (25 ng/ml) and ionomycin (1 μg/ml) stimulation. Results are representative of two to five independent experiments.
CD45E613R mice harbor a single amino acid mutation in the CD45 juxtamembrane wedge domain thought to play a regulatory role by inhibiting phosphatase activity upon receptor homodimerization. This point mutation results in aberrant T and B cell activation, leading to a lymphoproliferative disorder and autoimmunity (34). We hypothesized that if CD45−/− NK cells are unable to produce IFNγ, then CD45E613R NK cells might exhibit enhanced IFNγ production. However, comparison of CD45E613R and WT NK cells stimulated via CD16, NK1.1, NKG2D, or Ly49D revealed no difference in cytokine production (data not shown).
Cytokine and Chemokine mRNA Expression Is Deficient in CD45−/− NK Cells.
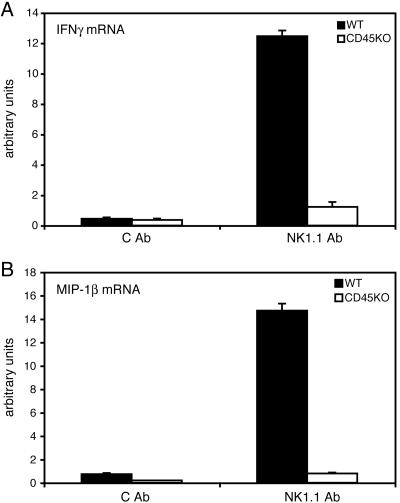
To determine the cause for the defect in cytokine and chemokine production, we isolated RNA from WT and CD45−/− NK cells stimulated with anti-NK1.1 or control mAb and performed quantitative RT-PCR. IFNγ and MIP-1β transcripts were greatly increased upon stimulation in WT NK cells but unchanged from unstimulated controls in CD45−/− NK cells (Fig. 3). Thus, the cytokine and chemokine production defect in CD45−/− NK cells is explained by the failure to accumulate mRNA from these genes.
Fig. 3.
Cytokine and chemokine mRNA levels are reduced in CD45−/− NK cells. Quantitative RT-PCR was performed on RNA harvested from WT and CD45−/− NK cells stimulated with plate-bound anti-NK1.1 mAb. IFNγ (A) and MIP-1β (B) mRNA levels were analyzed in two independent experiments with comparable results.
MAPK Activation Is Deficient upon Receptor Engagement in CD45−/− NK Cells.
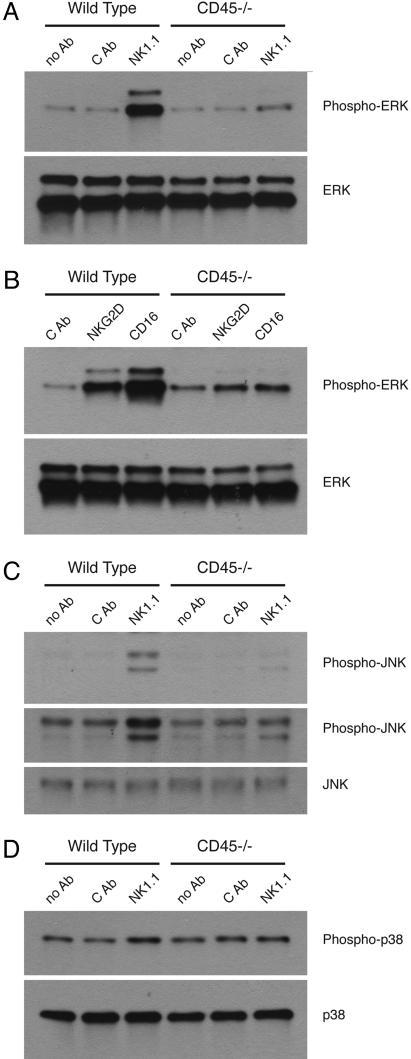
The MAPKs ERK, JNK, and p38 function at important downstream signaling branch points, where they serve as penultimate regulators of cytokine and chemokine gene expression by controlling the activation of transcription factors such as Elk-1, ATF2, and c-Jun. These MAPKs have been shown to be involved in the regulation of IFNγ, granulocyte–macrophage colony-stimulating factor, MIP-1β, MIP-1α, and TNFα gene expression (22, 35–37). Thus, we examined the activation/phosphorylation state of these MAPKs after receptor engagement in CD45−/− NK cells. ERK was markedly activated upon NK1.1, NKG2D, or CD16 stimulation in WT NK cells but was minimally activated in CD45−/− cells (Fig. 4). After NK1.1 stimulation, JNK phosphorylation was decreased in CD45−/− NK cells relative to WT controls (Fig. 4C). p38 phosphorylation was minimally (if at all) increased in WT or CD45−/− NK cells in response to anti-NK1.1 (Fig. 4D). CD45−/− NK cells are capable of activating ERK and JNK because stimulation with PMA yielded phosphorylation levels equivalent to WT NK cells (data not shown).
Fig. 4.
Defective MAPK signaling downstream of ITAM-based NK cell receptors. (A) ERK activation after stimulation of WT and CD45−/− NK cells with anti-NK1.1 mAb. (B) ERK activation after stimulation with anti-NKG2D mAb or with plate-bound mouse IgG2a to activate via CD16. (C) JNK activation after stimulation with anti-NK1.1 mAb. Two exposures (short and long) are shown. (D) p38 activation after stimulation with anti-NK1.1 mAb. Data are representative of two to seven experiments.
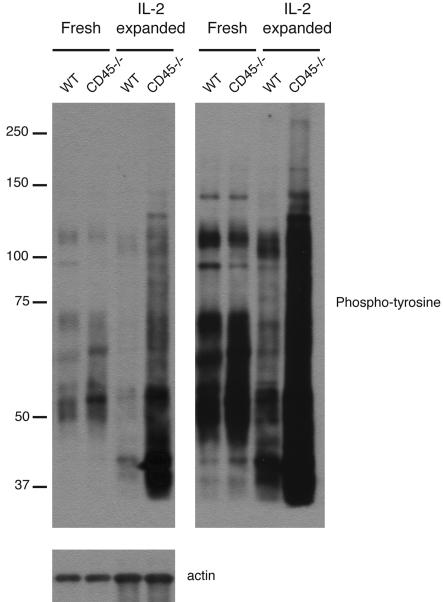
Basal Tyrosine Phosphorylation Is Increased in CD45−/− NK Cells.
Because CD45 is a transmembrane protein tyrosine phosphatase and is important for the initiation and regulation of signaling pathways, we examined CD45−/− NK cells for any alteration in tyrosine phosphorylation. Freshly isolated CD45−/− NK cells contained a number of proteins that were hyperphosphorylated relative to WT cells (Fig. 5). Examination of IL-2-expanded NK cells revealed an even more surprising and striking increase in the number of tyrosine phosphoproteins and level of tyrosine phosphorylation in CD45−/− NK cells compared with WT NK cells (Fig. 5). These changes were noted in the unstimulated state, suggesting a profound effect of CD45 in regulating the basal state of tyrosine phosphoproteins in freshly isolated and IL-2-expanded cells even before ITAM receptor-dependent pathways are activated.
Fig. 5.
Increased tyrosine phosphorylation in CD45−/− NK cells. Lysates were prepared from WT and CD45−/− NK cells that were freshly isolated or expanded in culture with IL-2. Equal cell equivalents were loaded into each lane. Left and Right are short and long exposures, respectively, of the same blot. Lower shows equal loading between WT and CD45−/− NK cells by probing for actin. Data are representative of two to nine independent experiments.
Discussion
We have shown that CD45 is not required for ITAM-dependent target cell killing induced by the CD16 or Ly49H receptors, which pair and signal through FcεRIγ or DAP12 ITAM-bearing adapters, respectively, and is not required for NKG2D-mediated cytotoxicity. Because CD45 deficiency had only a modest effect on target cell killing via NKG2D, DAP10 and DAP12 apparently do not require CD45 for activating cytotoxicity. Although ITAM-dependent killing was somewhat reduced in CD45−/− NK cells, it was still readily detectable.
In contrast, cytokine and chemokine production and mRNA accumulation are severely affected by CD45 deficiency. IFNγ production was virtually undetectable when CD45−/− NK cells were stimulated via NK1.1, CD16, NKG2D, or Ly49D. Because these receptors signal via ITAM adapters, we conclude that CD45 is essential for ITAM-based activation of cytokine and chemokine production. Although we observed a defect in mRNA accumulation of cytokine and chemokine mRNA, CD45-dependent signaling may also regulate cytokine translation, posttranslational modifications, or secretion. If there were additional stages of protein expression that CD45 controls, these would be masked by the severe defect in mRNA levels.
These findings reveal an unexpected separation in the requirement for CD45 in the activation of distinct ITAM-dependent NK cell functions. It appears that this tyrosine phosphatase is essential for ITAM receptor-induced cytokine and chemokine production but is not absolutely required for cytotoxicity. CD45 is the only molecule to date that has this activity. In contrast, Vav-1, Vav-2, and Vav-3 are required for NK cell-mediated killing but not for cytokine production (38, 39). As we were concluding our experiments, a study was published describing the same dichotomy between target cell killing and cytokine and chemokine production in CD45−/− NK cells (4).
We explored the molecular basis for this apparent dichotomy in function, focusing first on MAPKs because of their known importance in both functional outcomes. ERK1 and ERK2 have been well studied in NK cells and are activated after receptor stimulation (6–8, 40). ERK regulates both NK cell-mediated cytotoxicity and cytokine production, as demonstrated through the use of dominant-negative ERK constructs and MEK inhibitors (5–8). MEK is the upstream kinase that activates ERK. We found deficient MAPK activation in CD45−/− NK cells. ERK and JNK phosphorylation was significantly reduced, whereas p38 phosphorylation was mildly affected. Interestingly, we found upon closer comparison between various published studies that the IC50 for the MEK inhibitor, PD098059, was much higher for inhibition of NK cell target killing than was required to block IFNγ mRNA production or protein secretion (5–8). This finding suggests a relative difference in the requirement for ERK in cytotoxicity versus cytokine production. Thus, in CD45−/− NK cells, the defect in ERK activation may explain, in part, the defect seen in cytokine/chemokine production.
The role for p38 and JNK in NK cell effector function is less clear. p38 activation has been noted upon stimulation of NK cells (4, 9, 10). However, exposure of NK cells to p38 inhibitors resulted in either no or partial inhibition of target killing (9, 10), with inhibition occurring only at extremely high levels of inhibitor (10). The p38 inhibitors did not affect IFNγ secretion, and mRNA levels were only partially reduced at high inhibitor concentrations (5, 10). Thus, p38 appears to not be required for target killing or cytokine production. Although little is known about its role in cytokine secretion, JNK activation has been observed after NK cell receptor engagement (4, 10) (Fig. 4C). However, expression of a dominant-negative JNK had no effect on human NK cell-mediated cytotoxicity (10). In T cells, JNK regulates IFNγ expression (36, 37), raising the possibility that JNK performs the same function in NK cells.
To explore the basis for the dichotomous defects in CD45−/− NK cells, we examined the impact of the loss of CD45 on tyrosine phosphoprotein expression. We expected to see an effect mostly limited to Src family kinases, based on previous studies of T and B cells. Instead, we were surprised to observe an increased basal tyrosine phosphorylation of many proteins when freshly isolated CD45−/− NK cells were examined and an even more impressive effect when these cells were expanded in IL-2. Freshly isolated T cells from CD45−/− mice also demonstrate elevated basal phosphorylation of several proteins, similar to NK cells (41) (data not shown). How can one explain this effect on multiple proteins? There are several possible explanations that remain to be explored. First, some of these phosphorylated proteins are most likely substrates of CD45, such as the Src family kinases, known regulators of ITAM signaling. Second, because these Src kinases are important in inhibitory receptor signaling, it is possible that impairments or alterations in Src kinase function lead to loss of tonic inhibitory signals, resulting in higher amounts of basal activation. Finally, it is possible that the defects in MAPK activation and in cytokine production are the result of adaptation of the cells to elevated constitutive signaling and compensating inhibitory feedback loops.
Stimulation of the same receptor/ITAM adapter complex results in differential requirements for CD45 for target cytotoxicity and cytokine production. It is likely that strength and/or duration of signal may determine the requirement for CD45. Release of cytotoxic granules occurs near the cell surface, in close proximity to many cell signaling components that are activated upon receptor/ITAM activation. This process requires little time (minutes) between receptor crosslinking and granule release. Conversely, the induction of cytokines (42) by receptor engagement is a lengthier process (hours), involving signal transduction, gene transcription, RNA processing, translation, and secretion. Sustained signaling may be needed for induction of cytokines, whereas brief stimulation may be all that is needed for a robust cytotoxicity response by NK cells (43).
Materials and Methods
Mice.
Inbred C57BL/6 mice were purchased from Charles River Laboratories or the National Cancer Institute. CD45−/− mice (exon 6 disruption) were obtained from E. Brown (University of California, San Francisco) (15). All mice, including CD45E613R (34) (mice having a point mutation substituting an R for E at position 613 in CD45), were backcrossed eight generations onto the C57BL/6 genetic background. Mice were housed in a specific pathogen-free facility, and experiments were performed according to University of California, San Francisco, Institutional Animal Care and Use Committee guidelines.
Flow Cytometry.
Freshly isolated NK cells and IL-2-cultured NK cells were stained with mAbs against CD3, NK1.1, Ly49D, Ly49G2, Ly49A, and DX5 (VLA-2) (BD Biosciences); NKG2D (CX5) (eBioscience); and CD16/32 (2.4G2) (Harlan). Anti-Ly49H mAb (3D10) was a generous gift from W. Yokoyama (Washington University, St. Louis). Phycoerythrin-conjugated goat anti-rabbit and donkey anti-mouse IgG F(ab′)2 fragments were used as second-step reagents (Jackson ImmunoResearch). Flow cytometry was performed by using a FACScan (BD Biosciences).
Preparation of NK Cells.
Cells from spleen or liver were isolated, and red blood cells were lysed with ACK buffer (BioWhittaker). For the isolation of fresh cells, spleen cells were first stained with anti-CD16/32 mAb to block Fc receptors and then stained with fluorescent-labeled anti-CD3 and anti-NK1.1. NK1.1+CD3− cells were isolated by using a FACSAria (BD Biosciences). For the generation of IL-2-expanded NK cells, splenocytes were stained with anti-CD4 and anti-CD8 mAbs and then incubated with goat anti-rat IgG-coated and goat anti-mouse IgG-coated magnetic beads (Qiagen) to deplete T and B cells. NK cells were stained with phycoerythrin-conjugated DX5 mAb and positively selected by using an anti-phycoerythrin mAb-conjugated magnetic bead system (Miltenyi Biotec or StemCell Technologies). The resulting NK cells were cultured with 4,000 units/ml recombinant human IL-2 (National Institutes of Health Biological Resources Branch Preclinical Repository) and used after 6–8 days.
51Cr Release Assay.
Mouse Ba/F3 pro-B cells, Ba/F3 cells transduced with Rae-1ε (44) or murine CMV m157 (27), and RMA T cell lymphoma cells were cultured in RPMI medium 1640 containing 10% FCS, 2 mM glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. IL-2-activated NK cells were used as effector cells in a 4- to 6-h 51Cr cytotoxicity assay (45). For ADCC, RMA cells were incubated with or without 10 μg/ml anti-CD90 (Thy1) (provided by Salim Dhanji and Hung-sai Teh, University of Vancouver, Vancouver) (33), spun down, and mixed with the NK cells. NK cells were incubated with or without 10 μg/ml anti-CD16/32 mAb (2.4G2) (to block ADCC) for 30 min and then mixed with RMA or anti-CD90 mAb-coated RMA target cells at various effector:target ratios.
Stimulation of NK Cells.
Ninety-six-well plates (BD Biosciences) were coated with mAb or ligand as described (45). H60 extracellular domain–human IgG1 Fc fusion protein was generously provided by J. P. Houchins (R & D Systems) (46). CD16 stimulation was performed by incubating NK cells on tissue culture plates coated with a nonspecific mouse IgG mAb in the presence or absence of a soluble neutralizing anti-CD16/32 mAb (2.4G2) (47). In the absence of anti-CD16/32 mAb, WT NK cells gave robust cytokine production when cultured on IgG-coated plates. Addition of soluble anti-CD16/32 mAb binds CD16 and prevents binding to the IgG-coated plastic plates, thereby blocking CD16-dependent NK cell activation. A total of 2 × 105 cells per well were plated, centrifuged, and incubated for 4–18 h for cytokine and chemokine secretion assays, 4–8 h for RNA analysis, and 15, 30, 45, or 60 min for preparation of protein lysates for Western blot analysis (all time points yielded robust signals). ELISAs for IFNγ (R & D Systems), MIP-1β (BD Biosciences), and granulocyte–macrophage colony-stimulating factor (eBioscience) were performed according to the manufacturers' instructions.
mRNA Quantitation.
NK cells were stimulated with plate-bound mAb as described above. NK cell RNA was DNase-treated and converted to cDNA by using standard methods. Quantitative PCR was performed as described (48) and normalized to the amount of hypoxanthine phosphoribosyltransferase transcripts. Probe sequences were FAM-CACAGGTCCAGCGCCAAGCATTC-TAMRA (IFNγ) and FAM-CTCTGACCCTCCCACTTCCTGCTGTTT-TAMRA (MIP-1β). PCR primers were ATGCATTCATGAGTATTGCCAAGT (IFNγ sense), GCTGGATTCCGGCAACAG (IFNγ antisense), CCAGGGTTCTCAGCACCAAT (MIP-1β sense), and GCTGCCGGGAGGTGTAAGA (MIP-1β antisense).
MAPK Activation and Tyrosine Phosphorylation.
IL-2-expanded NK cells were removed from IL-2 ≈18 h before stimulation. NK cells were serum-starved for 1–2 h before lysis and analysis for tyrosine phosphorylation. NK cells stimulated with plate-bound mAb were lysed directly in the 96-well plate by the addition of 2×-reducing SDS/PAGE loading dye. Lysates were loaded onto 12% polyacrylamide gels and transferred to polyvinylidene fluoride membranes by using standard Western blotting techniques. The blots were incubated with antibodies to actin (Santa Cruz Biotechnology), phosphotyrosine (4G10), ERK, phospho-ERK, JNK, phospho-JNK, p38, and phospho-p38 (Cell Signaling Technology), probed with horseradish peroxidase-conjugated anti-rabbit IgG (Cell Signaling Technology), and developed by using ECL+ reagents (Amersham Pharmacia Bioscience).
Acknowledgments
We thank Jay Ryan, Tony DeFranco, Cliff Lowell, Clare Abram, Mark Klinger, Jessica Hamerman, Melissa Lodoen, and Susan Watson for advice; the L.L.L. and A.W. laboratories for support; and Allison Tan and Jessica Jarjoura for expert technical assistance. This work was supported by National Institutes of Health Grants AI068129 (to L.L.L.), K08 CA098418-01 (to M.L.H.), and AI35297 (to A.W.) and by the Howard Hughes Medical Research Institute (A.W.). D.G.T.H. is supported by a Cancer Research Institute Fellowship, and L.L.L. is an American Cancer Society Research Professor.
Abbreviations
- ITAM
immunoreceptor tyrosine-based activation motif
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
- JNK
c-Jun N-terminal kinase
- NK
natural killer
- MIP
macrophage inflammatory protein
- ADCC
antibody-dependent cell-mediated cytotoxicity.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lanier L. L. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Lanier L. L. Curr. Opin. Immunol. 2003;15:308–314. doi: 10.1016/s0952-7915(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 3.Samelson L. E. Annu. Rev. Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 4.Huntington N. D., Xu Y., Nutt S. L., Tarlinton D. M. J. Exp. Med. 2005;201:1421–1433. doi: 10.1084/jem.20042294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortaldo J. R., Bere E. W., Hodge D., Young H. A. J. Immunol. 2001;166:4994–4999. doi: 10.4049/jimmunol.166.8.4994. [DOI] [PubMed] [Google Scholar]
- 6.Trotta R., Puorro K. A., Paroli M., Azzoni L., Abebe B., Eisenlohr L. C., Perussia B. J. Immunol. 1998;161:6648–6656. [PubMed] [Google Scholar]
- 7.Wei S., Gamero A. M., Liu J. H., Daulton A. A., Valkov N. I., Trapani J. A., Larner A. C., Weber M. J., Djeu J. Y. J. Exp. Med. 1998;187:1753–1765. doi: 10.1084/jem.187.11.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei S., Gilvary D. L., Corliss B. C., Sebti S., Sun J., Straus D. B., Leibson P. J., Trapani J. A., Hamilton A. D., Weber M. J., Djeu J. Y. J. Immunol. 2000;165:3811–3819. doi: 10.4049/jimmunol.165.7.3811. [DOI] [PubMed] [Google Scholar]
- 9.Chini C. C., Boos M. D., Dick C. J., Schoon R. A., Leibson P. J. Eur. J. Immunol. 2000;30:2791–2798. doi: 10.1002/1521-4141(200010)30:10<2791::AID-IMMU2791>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Trotta R., Fettucciari K., Azzoni L., Abebe B., Puorro K. A., Eisenlohr L. C., Perussia B. J. Immunol. 2000;165:1782–1789. doi: 10.4049/jimmunol.165.4.1782. [DOI] [PubMed] [Google Scholar]
- 11.Einspahr K. J., Abraham R. T., Binstadt B. A., Uehara Y., Leibson P. J. Proc. Natl. Acad. Sci. USA. 1991;88:6279–6283. doi: 10.1073/pnas.88.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colucci F., Schweighoffer E., Tomasello E., Turner M., Ortaldo J. R., Vivier E., Tybulewicz V. L., Di Santo J. P. Nat. Immunol. 2002;3:288–294. doi: 10.1038/ni764. [DOI] [PubMed] [Google Scholar]
- 13.Lowin-Kropf B., Kunz B., Schneider P., Held W. Eur. J. Immunol. 2002;32:773–782. doi: 10.1002/1521-4141(200203)32:3<773::AID-IMMU773>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Hermiston M. L., Xu Z., Weiss A. Annu. Rev. Immunol. 2001;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 15.Kishihara K., Penninger J., Wallace V. A., Kundig T. M., Kawai K., Wakeham A., Timms E., Pfeffer K., Ohashi P. S., Thomas M. L., et al. Cell. 1993;74:143–156. doi: 10.1016/0092-8674(93)90302-7. [DOI] [PubMed] [Google Scholar]
- 16.Byth K. F., Conroy L. A., Howlett S., Smith A. J., May J., Alexander D. R., Holmes N. J. Exp. Med. 1996;183:1707–1718. doi: 10.1084/jem.183.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poggi A., Pardi R., Pella N., Morelli L., Sivori S., Vitale M., Revello V., Moretta A., Moretta L. Eur. J. Immunol. 1993;23:2454–2463. doi: 10.1002/eji.1830231012. [DOI] [PubMed] [Google Scholar]
- 18.Giezeman-Smits K. M., Gorter A., van Vlierberghe R. L., v Eendenburg J. D., Eggermont A. M., Fleuren G. J., Kuppen P. J. J. Immunol. 1999;163:71–76. [PubMed] [Google Scholar]
- 19.Starling G. C., Davidson S. E., McKenzie J. L., Hart D. N. Immunology. 1987;61:351–356. [PMC free article] [PubMed] [Google Scholar]
- 20.Starling G. C., Hart D. N. Immunology. 1990;71:190–195. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X., Chong A. S. J. Immunol. 1995;155:5241–5248. [PubMed] [Google Scholar]
- 22.Shen F., Xu X. L., Graf L. H., Chong A. S. J. Immunol. 1995;154:644–652. [PubMed] [Google Scholar]
- 23.Martin S. M., Mehta I. K., Yokoyama W. M., Thomas M. L., Lorenz R. G. J. Immunol. 2001;166:6066–6073. doi: 10.4049/jimmunol.166.10.6066. [DOI] [PubMed] [Google Scholar]
- 24.Yamada H., Kishihara K., Kong Y.-Y., Nomoto K. J. Immunol. 1996;157:1523–1528. [PubMed] [Google Scholar]
- 25.Zompi S., Hamerman J. A., Ogasawara K., Schweighoffer E., Tybulewicz V. L., Santo J. P., Lanier L. L., Colucci F. Nat. Immunol. 2003;4:565–572. doi: 10.1038/ni930. [DOI] [PubMed] [Google Scholar]
- 26.Smith H. R., Heusel J. W., Mehta I. K., Kim S., Dorner B. G., Naidenko O. V., Iizuka K., Furukawa H., Beckman D. L., Pingel J. T., et al. Proc. Natl. Acad. Sci. USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arase H., Mocarski E. S., Campbell A. E., Hill A. B., Lanier L. L. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 28.Smith K. M., Wu J., Bakker A. B. H., Phillips J. H., Lanier L. L. J. Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- 29.Wu J., Song Y., Bakker A. B. H., Bauer S., Groh V., Spies T., Lanier L. L., Phillips J. H. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 30.Diefenbach A., Tomasello E., Lucas M., Jamieson A. M., Hsia J. K., Vivier E., Raulet D. H. Nat. Immunol. 2002;3:1142–1149. doi: 10.1038/ni858. [DOI] [PubMed] [Google Scholar]
- 31.Arase H., Arase N., Saito T. J. Exp. Med. 1996;183:2391–2396. doi: 10.1084/jem.183.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanier L. L., Yu G., Phillips J. H. J. Immunol. 1991;146:1571–1576. [PubMed] [Google Scholar]
- 33.Dhanji S., Tse K., Teh H. S. J. Immunol. 2005;174:1253–1258. doi: 10.4049/jimmunol.174.3.1253. [DOI] [PubMed] [Google Scholar]
- 34.Majeti R., Zheng X., Parslow T. G., Olson J. L., Daikh D. I., Killeen N., Weiss A. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 35.Melchjorsen J., Sorensen L. N., Paludan S. R. J. Leukocyte Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong C., Davis R. J., Flavell R. A. Annu. Rev. Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 37.Szabo S. J., Sullivan B. M., Peng S. L., Glimcher L. H. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 38.Cella M., Fujikawa K., Tassi I., Kim S., Latinis K., Nishi S., Yokoyama W. M., Colonna M., Swat W. J. Exp. Med. 2004;200:817–823. doi: 10.1084/jem.20031847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colucci F., Rosmaraki E., Bregenholt S., Samson S. I., Di Bartolo V., Turner M., Vanes L., Tybulewicz V., Di Santo J. P. J. Exp. Med. 2001;193:1413–1424. doi: 10.1084/jem.193.12.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McVicar D. W., Taylor L. S., Gosselin P., Willette-Brown J., Mikhael A. I., Geahlen R. L., Nakamura M. C., Linnemeyer P., Seaman W. E., Anderson S. K., et al. J. Biol. Chem. 1998;273:32934–32942. doi: 10.1074/jbc.273.49.32934. [DOI] [PubMed] [Google Scholar]
- 41.Stone J. D., Conroy L. A., Byth K. F., Hederer R. A., Howlett S., Takemoto Y., Holmes N., Alexander D. R. J. Immunol. 1997;158:5773–5782. [PubMed] [Google Scholar]
- 42.Valitutti S., Dessing M., Aktories K., Gallati H., Lanzavecchia A. J. Exp. Med. 1995;181:577–584. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valitutti S., Muller S., Dessing M., Lanzavecchia A. J. Exp. Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerwenka A., Baron J. L., Lanier L. L. Proc. Natl. Acad. Sci. USA. 2001;98:11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogasawara K., Yoshinaga S. K., Lanier L. L. J. Immunol. 2002;169:3676–3685. doi: 10.4049/jimmunol.169.7.3676. [DOI] [PubMed] [Google Scholar]
- 46.O'Callaghan C. A., Cerwenka A., Willcox B. E., Lanier L. L., Bjorkman P. J. Immunity. 2001;15:201–211. doi: 10.1016/s1074-7613(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 47.Unkeless J. C. J. Exp. Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogasawara K., Hamerman J. A., Hsin H., Chikuma S., Bour-Jordan H., Chen T., Pertel T., Carnaud C., Bluestone J. A., Lanier L. L. Immunity. 2003;18:41–51. doi: 10.1016/s1074-7613(02)00505-8. [DOI] [PubMed] [Google Scholar]