Abstract
Invasive potentials of carcinomas greatly contribute to their metastasis, which is a major threat in most cancers. We have recently shown that Arf6 plays a pivotal role in breast cancer invasive activities and identified AMAP1 as an effector of GTP-Arf6 in invasion. Expression of AMAP1 correlates well with invasive phenotypes of primary tumors of the human breast. We also have shown that AMAP1 functions by forming a trimeric protein complex with cortactin and paxillin. In this complex, AMAP1 binds to the src homology 3 (SH3) domain of cortactin via its proline-rich peptide, SKKRPPPPPPGHKRT. SH3 domains are known to bind generally to the proline-rich ligands with a one-to-one stoichiometry. We found that AMAP1/cortactin binding is very atypical in its stoichiometry and interface structure, in which one AMAP1 proline-rich peptide binds to two cortactin SH3 domains simultaneously. We made a cell-permeable peptide derived from the AMAP1 peptide, and we show that this peptide specifically blocks AMAP1/cortactin binding, but not other canonical SH3/proline bindings, and effectively inhibits breast cancer invasion and metastasis. Moreover, this peptide was found to block invasion of other types of cancers, such as glioblastomas and lung carcinomas. We also found that a small-molecule compound, UCS15A, which was previously judged as a weak inhibitor against canonical SH3/proline bindings, effectively inhibits AMAP1/cortactin binding and breast cancer invasion and metastasis. Together with fine structural analysis, we propose that the AMAP1/cortactin complex, which is not detected in normal mammary epithelial cells, is an excellent drug target for cancer therapeutics.
Keywords: cell-permeable peptide, molecular target, NMR, small-molecule inhibitor, x-ray structure
Invasive phenotypes are normally strictly restricted in adult epithelial tissues, although they are frequently used during embryogenesis and tissue remodeling and in cancer. We have previously shown that Arf6 activity is necessary for the invasive activities of different breast cancer cells (1, 2), and we have identified that AMAP1 acts as an effector of Arf6 in invasion by binding to GTP-Arf6 through its zinc-finger domain (also known as the ArfGAP domain) (3, 4). AMAP1 functions in invasion by forming a trimeric protein complex with cortactin and paxillin in which AMAP1 binds to the src homology 3 (SH3) domain of cortactin via its proline-rich sequence and to paxillin through its own SH3 domain (3). AMAP1 bears 16 repeats of proline-rich sequences, and we identified the fourth sequence, SKKRPPPPPPGHKRT, to be responsible for AMAP1 binding to the cortactin SH3 domain (see ref. 3; we call this peptide “P4” in the present paper). This peptide has six consecutive proline residues and, therefore, may be unusual compared with canonical SH3 ligands, although it contains the consensus PxxP motif (where x is any amino acid) of SH3 ligands (5). In this report, we examined the properties of AMAP1/cortactin binding regarding its impact on cancer invasion and metastasis and its fine structure and show that this interface is an excellent drug target for cancer therapeutics. Because AMAP1 expression is minimal and the AMAP1/cortactin complex is not detected in normal mammary epithelial cells, treatment by such drugs may exhibit minimal side effects on normal mammary epithelia in adults.
Results and Discussion
Blockage of Breast Cancer Cell Invasion and Metastasis by Cell-Permeable Peptide P4-TAT.
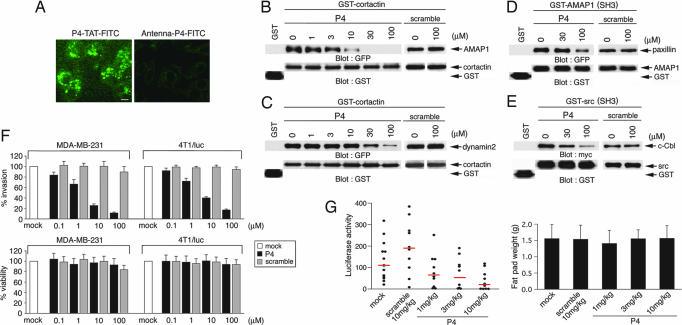
We generated cell-permeable peptides containing the P4 sequence and several of its surrounding amino acids in the AMAP1 proline-rich domain (PRD) by using the HIV Tat sequence (6) or the third α-helix of Drosophila Antennapedia (7) as cell-permeable epitopes. The AMAP1 sequence used in these peptides is common to human and mouse AMAP1 proteins. We found that P4-TAT, SSTLSKKRPPPPPPGHKRTLSD-GRKKRRQRRR, is efficiently delivered into human MDA-MB-231 and mouse 4T1/luc cells, whereas Antenna-P4, KKWKMRRNPFWIKIQRCC-SSTLSKKRPPPPPPGHKRTLSD, is not (Fig. 1A and data not shown). We then validated the efficacy of P4-TAT at blocking the binding of cortactin fused to GST with AMAP1 in vitro, as we have performed previously with the P4 peptide without a tag (3). P4-TAT efficiently inhibited AMAP1/cortactin binding (Fig. 1B). The cortactin SH3 domain is also known to bind to a canonical PxxP peptide of dynamin2, FPAPPQIPSRPVRIP (8), however we found that P4-TAT did not efficiently block cortactin/dynamin2 binding (Fig. 1C). P4-TAT also did not efficiently inhibit binding of AMAP1 and paxillin (Fig. 1D). As another control, we further examined P4-TAT and found it inefficient at inhibiting Src and c-Cbl binding (Fig. 1E). P4-TAT effectively inhibited in vitro Matrigel invasion of MDA-MB-231 and 4T1/luc cells, with an IC50 of <10 μM in each case, and a concentration of 100 μM did not affect the viability of these cells (Fig. 1F). We then examined the effects of P4-TAT on metastasis. 4T1/luc cells stably expressing firefly luciferase were injected into the mammary pad of adult female BALB/c mice, which are syngeneic to 4T1/luc cells, and their metastases into the lung were assessed by measuring luciferase activity (3). The metastatic activities were significantly suppressed by i.p. injection of P4-TAT every day for 8 days (1–10 mg/kg from 12 days after the inoculation of cancer cells) (Fig. 1G), although these conditions did not notably affect tumor growth at the originally injected sites (Fig. 1G) nor cause reduction of body weight or apparent tissue and visceral injuries (data not shown). These results suggest that AMAP1/cortactin binding is an excellent target for the prevention of breast cancer invasion and metastasis, whereas inhibition of this binding does not affect cell viability and the immediate bodily condition of mice.
Fig. 1.
Effects of P4-TAT peptide. (A) MDA-MB-231 cells were incubated with 10 μM P4-TAT or Antenna-P4, both labeled with FITC, for 20 min and analyzed by fluorescence microscopy. (Scale bar, 10 μm.) (B–E) Inhibition of AMAP1/cortactin and other SH3/proline bindings in vitro. (B and C) GST-cortactin, or GST alone, purified on glutathione beads was incubated with COS-7 cell lysates expressing GFP-AMAP1 or GFP-dynamin2 in the absence or presence of P4-TAT or its scramble peptides, and GFP fusion proteins coprecipitated with GST-cortactin were analyzed by immunoblotting as indicated. (D and E) The effects of P4-TAT on the binding of the AMAP1 SH3 domain [GST-AMAP1(SH3)] and GFP-paxillin, and the c-Src SH3 domain [GST-Src(SH3)] and myc-c-Cbl were analyzed similarly. (F) Inhibition of Matrigel transinvasion. MDA-MB-231 and 4T1/luc cells were cultured on Matrigel for 8 h in the absence or presence of P4-TAT or its scramble peptides. Percentages of cells that transmigrated through a barrier of Matrigel (Upper) and cell viability (Lower) were measured. Results shown are the means ± SEM of three experiments. (G) Inhibition of metastasis. 4T1/luc cells were injected into the mammary pads of mice and assessed for their metastases into the lung 19 days later, during which mice were i.p. injected with P4-TAT or control scramble peptides every day for the last 8 days at the amounts indicated. Mock, saline. (Left) Luciferase activity in the left lungs were plotted as activity per milligram of total proteins of the lung extracts. Red lines represent median values. (Right) Tumor weights at the originally injected fat pad on day 19. At least 10 mice were used for each experiment. Error bars indicate SEM.
Effective Blockage of Breast Cancer Cell Invasion and Metastasis by a Small-Molecule Compound, UCS15A.
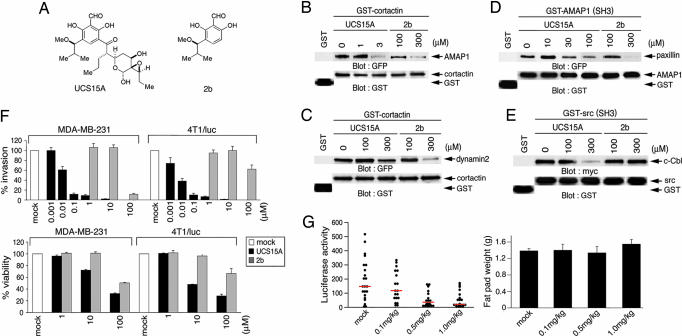
We previously isolated a small-molecule blocker of proline-rich ligand-mediated protein interactions (PLPIs), namely UCS15A, which is a product of Streptomyces sp. (Fig. 2A) (9). Here, we found that 3 μM UCS15A is sufficient to almost completely block binding of GST-cortactin and AMAP1 in vitro, although this compound is very ineffective at inhibiting the binding of GST-cortactin with dynamin2, the GST-AMAP1 SH3 domain with paxillin, and the GST-Src SH3 domain and c-Cbl (Fig. 2 B–E). UCS15A similarly blocked the binding of the GST-cortactin SH3 domain with the AMAP1 PRD (data not shown). We previously synthesized compound 2b, an analog of UCS15A (Fig. 2A) (10). A concentration of compound 2b at >100 μM was necessary to block any of these bindings (Fig. 2 B–E). These results suggest that UCS15A very selectively and efficiently inhibits PLPI between cortactin and AMAP1. We also validated the inhibitory effects of UCS15A on breast cancer invasion and metastasis, as we did above with P4-TAT. In vitro Matrigel invasion of MDA-MB-231 and 4T1/luc cells was effectively blocked by UCS15A with an IC50 of ≈10 nM in each case, whereas 10 μM compound 2b was totally ineffective (Fig. 2F). Lung metastases of 4T1/luc cells in mice were also effectively inhibited by injecting UCS15A every day for 8 days i.p. (0.1–1 mg/kg from 12 days after the inoculation of cancer cells) (Fig. 2G), whereas tumor growth at the originally injected sites (Fig. 2G) and mouse body weights were not notably affected (data not shown).
Fig. 2.
Effects of UCS15A. (A) Chemical structures of UCS15A and its derivative, 2b. (B–E) Inhibition of AMAP1/cortactin and other SH3/proline bindings in vitro. The effects of UCS15A and 2b on the in vitro binding of GST-cortactin with GFP-AMAP1 (B) or GFP-dynamin2 (C) or the binding of GST-AMAP1 (SH3) with GFP-paxillin (D) and GST-Src (SH3) with myc-c-Cbl (E) were analyzed as in Fig. 1, in which GST beads were preincubated with the chemical compounds as described in refs. 9 and 10. (F) The effects of UCS15A and 2b on Matrigel transinvasion (Upper) and cell viability (Lower) of MDA-MB-231 and 4T1/luc cells were analyzed as in Fig. 1F. The mock included dimethyl sulfoxide (DMSO) used as a solvent. Results shown are the means ± SEM of three experiments. (G) (Upper) Effects of UCS15A on lung metastasis of 4T1/luc cells were measured as in Fig. 1G (red lines represent median values). (Lower) Tumor weights at the originally injected fat pad on day 19 also are shown. More than 20 mice were used for each experiment. Error bars show SEM.
Atypical Properties of the AMAP1 and Cortactin-Binding Interface.
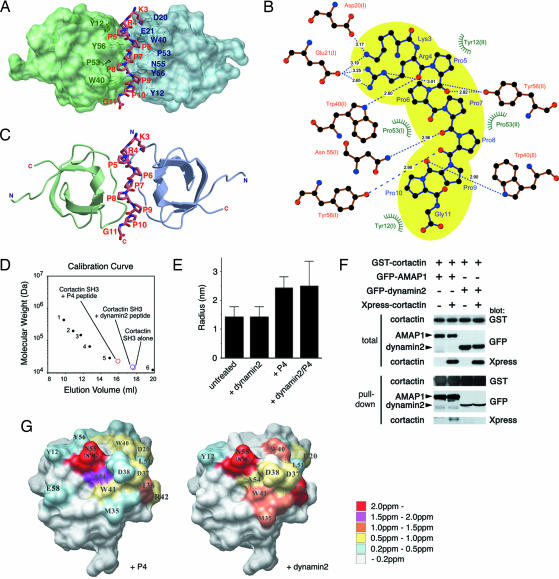
The very high efficacy of UCS15A on inhibition of cortactin/AMAP1 binding was surprising because we previously judged UCS15A as a weak PLPI blocker: 300 μM UCS15A is necessary to block other PLPIs in vitro, such as between the Fyn SH3 domain and the Sam68 PRD (10), similar to what we observed here with cortactin/dynamin2 and AMAP1/paxillin binding. These results prompted us to investigate the fine structure of the interface between the cortactin SH3 domain and P4. For this purpose, we made their cocrystal and determined its x-ray structure at 1.9-Å resolution (Fig. 3 A–C). SH3 domains generally bind to peptides of an all-trans, left-handed, polyproline type II helix with a one-to-one stoichiometry (5). Among the 15 amino acids of P4, the core 9 amino acids, KRPPPPPPG, were determined in the x-ray structure and found to take this configuration in binding to the cortactin SH3 domain. However, we found that a single molecule of P4 binds to two molecules of the cortactin SH3 domain in the crystal, in which the two SH3 domains were related to each other by noncrystallographic twofold symmetry. This interaction appeared to involve 10 hydrogen bonds and a hydrophobic pocket formed by Tyr-12, Trp-40, and Pro-53 of both SH3 domains (Fig. 3B). Interestingly, two amino acids of P4 interact with both SH3 domains simultaneously: Pro-7 of P4 makes hydrophobic interactions with Pro-53 of both SH3 domains, and Arg-4 forms hydrogen bonds with Glu-21 of one SH3 domain (Fig. 3B, left side) and with Tyr-56 of the other (Fig. 3B, right side). Two classes of SH3 ligands have been defined, class I and class II, that possess consensus RxxPxxP and PxxPxR motifs, respectively, and class I ligands bind to SH3 domains in a direction opposite to that of class II ligands (5). From the binding geometries, P4 was judged to act as a class I ligand in its binding to one of the cortactin SH3 domains (Fig. 3 A and C, left side), whereas the same peptide was considered a class II ligand in its binding to the other cortactin SH3 domain (Fig. 3 A and C, right side). However, this class II binding is atypical because the consensus basic amino acid, which is C-terminal to the PxxP motif, was not present. Size exclusion chromatography and dynamic light scattering measurements supported the one-to-two binding stoichiometry of P4 and the cortactin SH3 domain in solution (Fig. 3 D and E). We also confirmed that two full-length cortactin molecules can bind to one full-length AMAP1 molecule within cells by coexpressing Xpress-cortactin and GST-cortactin with GFP-AMAP1. Pull-down of GST-cortactin by glutathione-beads coprecipitated Xpress-cortactin when GFP-AMAP1, but not GFP-dynamin2, was present (Fig. 3F).
Fig. 3.
Structure and properties of binding of the cortactin SH3 domain and P4. (A) Crystal structure of an SH3 dimer intercalated with a single P4. SH3 molecules in the class I and pseudoclass II orientations binding to P4 are colored in green and light blue, respectively. (B) Schematic representation of binding site geometries. Residues of P4 peptide are colored blue, and the SH3 domains are colored orange. Hydrogen bonds are shown as dashed lines, with bond distances in angstroms. Hydrophobic contacts are shown as green rays. The space occupied by the core nine amino acids of P4 is colored yellow. (C) Ribbon representation. The structures of SH3 (class I orientation) (green) and SH3 (pseudoclass II orientation) (light blue) are shown at the top and bottom. The peptide sequence KRPPPPPPG is shown in stick representation. (D) Size-exclusion chromatography to assess apparent molecular masses of the cortactin SH3 protein in the presence and absence of the P4 or dynamin2 peptides. Positions of standard proteins, apoferritin, β-amylase, alcohol dehydrogenase, BSA, carbonic anhydrase, and cytochrome C, are shown as dots 1–6, respectively. (E) Dynamic light scattering measurements of the cortactin SH3 domain in the presence and absence of the P4 or dynamin2 peptides. (F) Dimerization of cortactin in the presence of AMAP1. One microgram of pEBG-cortactin (encoding GST-cortactin) was cotransfected with 2 μg each of pEGFP-dynamin2 (GFP-dynamin2) or pEGFP-AMAP1 (GFP-AMAP1) or with 1 μg of pcDNA His C-cortactin (Xpress-cortactin) in COS-7 cells, as indicated. GST- cortactin was pulled down from 500 μg of cell lysate, and proteins coprecipitated were then analyzed by immunoblotting, as indicated. Total includes 20 μg of total cell lysate. (G) Residues of the cortactin SH3 domain in which amide resonances were perturbed upon interaction with PxxP ligands of P4 (Left) and dynamin2 (Right).
We also analyzed this interaction by use of NMR, in which the amide chemical shifts in 15N–1H-heteronuclear single quantum correlation spectra of the 15N-labeled cortactin SH3 domain was measured with or without nonlabeled P4. The locations of resonance that undergo chemical shift perturbations are then mapped on the crystal structures of the cortactin SH3 domain (Fig. 3G). These locations represent the putative binding sites for P4, although mixed signals from the class I and II bindings might have been detected. Consistent with the above results, interfacial residues of the cortactin SH3 domain, including Tyr-12, Trp-40, Asn-55 (δ2), and Tyr-56, exhibited resonances that were significantly perturbed upon the addition of P4. We then compared the interaction of the cortactin SH3 domain toward P4 with the interaction toward the canonical PxxP peptide. Unlike P4, dimerization of the cortactin SH3 domain was not induced by the dynamin2 PxxP peptide (Fig. 3 D–F). Mapping the locations of chemical shift perturbations induced by the dynamin peptide revealed that surface areas of the cortactin SH3 domain used for binding to P4 and to the dynamin2 peptide largely overlap but are not identical (Fig. 3G). Therefore, compared with canonical SH3/PxxP bindings, interaction of the cortactin SH3 domain with P4 is unique in its atypical binding stoichiometry and manner of ligand binding. In addition, the use of several sites, such as Tyr-56, are potentially unique to this binding. Such unique features of the cortactin/AMAP1 interface may have made UCS15A, an otherwise weak PLPI inhibitor, very effective at blocking this binding.
Potential Binding of Other SH3 Domains with P4 and the Effects of UCS15A on Their Binding.
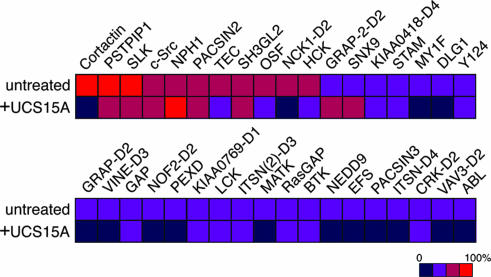
UCS15A exhibits toxicity to mice when injected i.p. at 2 mg per kilogram of body weight (ref. 11 and our unpublished observation). Moreover, P4 contains the consensus PxxP sequence for SH3 binding and may interact with SH3 domains other than the cortactin SH3 domain. Identification of other SH3 domains with potential P4 binding and investigation of the possible inhibition of such bindings by UCS15A might help in modifying UCS15A to be safer and more effective for cancer therapeutics. We assessed this issue by using commercially available SH3 domain array filters. We found that not a small number of different human SH3 domains also have the potential to interact with P4 peptide, although to a much lesser extent in most cases compared to the binding of the cortactin SH3 domain to P4 (Fig. 4). Among the SH3 domains we examined, SH3 domains, such as those of PSTPIP1 (proline–serine–threonine phosphatase-interacting protein) and SLK, exhibited affinities to P4 similar to that of the cortactin SH3 domain. PSTPIP1 is a cytoskeletal protein previously identified as a binding partner of the PEST-type tyrosine phosphatase (12). SLK is also known as Fyn tyrosine kinase, and it plays pivotal roles in different intracellular signalings, including cell adhesion and migration in both normal and transformed cells (13). In contrast, our results also show that the binding of PSTPIP1 and SLK to P4 is inhibited only marginally by UCS15A (Fig. 4). However, we noticed that the second SH3 domain of Nck1 (NCK-D2) exhibited a lower but still appreciable affinity to P4, and this binding was significantly affected by UCS15A (Fig. 4). Nck is a scaffold adaptor molecule also involved in receptor signaling and cell adhesion and migration in both normal and transformed cells (14). These results nevertheless confirmed the appreciable high specificity of UCS15A in blocking cortactin/AMAP1 binding but simultaneously revealed that UCS15A might potentially interfere with some SH3/proline interactions other than that of AMAP1 and cortactin.
Fig. 4.
Human SH3 domains with the potential to bind to P4 peptide and their blockage by UCS15A. Membrane filters spotted with various SH3 domains were incubated with biotinylated P4 peptide in the absence or presence of UCS15A. Binding strengths of the SH3 domains were calculated by normalizing the binding of P4 and cortactin SH3 as 100% and are indicated as colored squares. The SH3 domains we examined that exhibited <25% of binding compared with that of P4 and cortactin SH3 are as follows: ABL2, ABL2B, Amphiphysin, ARGBP2-D1, ARGBP2-D2, ARGBP2-D3, ARH6, ARHGEF9, ARHGEF16, BAIAP2, BCA1, BIN1, BLK, BMX, BPAG1, BZRAP1-D1, CACNβ2, CCBA, CRK-D1, CRKL-D1, CRKL-D2, CSK, CSKP, CXorf9-D1, DDEF2, DKFZp434D0215-D3, DKFZp434D0215-D4, DKFZP434D146, DLG2, EMP55, EPS8, FGR, FYB-D1, FYB-D2, GRAF, GRAP-D1, GRB2L-D1, GRB2-D1, GRB2-D2, HIP-55, HS1, ITK, ITSN-D1, ITSN-D2, ITSN-D3, ITSN (2)-D1, ITSN (2)-D2, JIP1, JIP2, KIAA0418-D3, KIAA0418-D5, KIAA0456, KIAA0554, KIAA0790, KIAA1139, KIAA1249, LASP1, M3KA, MIA, MLPK3, MY1E, MY7A, NCF1-D1, NCF1-D2, NCF4, NCK1-D1, NCK1-D3, NCK2-D1, NCK2-D2, NCK2-D3, NEB1, Nebulin, NE-DLG, NOF2-D1, OTOR, PI3a, PI3b, PIG2, PLCγ, PPP1R13B, PRMT2, PSD95, RHG4, RIZ, SH3GL3, SH31, SJHUA, SLA, SORBS-D3, SP93, SPCN, SPIN90, SRGAP2, STAC, STAM2, TIM, TRIO, TRIO (2), TRIP10, TXK, UAS3, VAV-D1, VAV-D2, VAV2-D1, VAV2-D2, VAV3-D1, VINE-D1, VINE-D2, YES1, and ZO2.
Conclusion
Most breast tumors arise from luminal epithelial cells, which are normally surrounded by basement layers of the extracellular matrices (15). Thus, invasion into the basement layers might be a prerequisite for the metastasis of most breast carcinomas, although this step is sometimes helped by stromal cells (15–19). Here, we provide several lines of evidence suggesting that the AMAP1/cortactin binding interface is an excellent molecular target for prevention of breast cancer invasion and metastasis. In contrast, our results also indicate that UCS15A might potentially interfere with SH3/proline interactions other than that of AMAP1 and cortactin. Our structural results will help in modifying UCS15A to be more specific to the binding interface of the cortactin SH3 domain and AMAP1 P4 peptide and to screen and logically design chemical compounds for breast cancer therapeutics. Moreover, we also found that the cell-permeable P4 peptide can inhibit the invasive activities of different cancer cells in addition to breast cancer cells (Table 1). Thus, the AMAP1/cortactin binding interface may hold promise as an excellent molecular target for prevention of the invasive activities of not only breast cancer cells but also other types of cancers.
Table 1.
Inhibition of Matrigel invasion of different cancer cells by P4-TAT and UCS15A
| Cell line | Origin | P4, IC50 μM ± SEM |
UCS15A, IC50 μM ± SEM |
||
|---|---|---|---|---|---|
| Invasion | Viability | Invasion | Viability | ||
| MDA-MB-231 | Human breast cancer | 7.9 ± 3.12 | >100 | 0.0128 ± 0.0047 | 11.0 ± 1.46 |
| Hs578T | Human breast cancer | 12.2 ± 4.78 | >100 | 0.0117 ± 0.0053 | 18.1 ± 2.53 |
| MDA-MB-435s | Human breast cancer | 17.6 ± 5.83 | >100 | 0.0154 ± 0.0045 | 13.3 ± 2.12 |
| 4T1/luc | Mouse breast cancer | 4.2 ± 1.97 | >100 | 0.0082 ± 0.0021 | 10.4 ± 0.61 |
| CCF-STTG1 | Human glioblastoma | 12.5 ± 3.12 | >100 | 0.0100 ± 0.0031 | 9.8 ± 2.14 |
| IN351 | Human glioblastoma | 7.9 ± 2.72 | >100 | 0.0106 ± 0.0025 | 15.3 ± 1.68 |
| T98G | Human glioblastoma | 32.6 ± 9.42 | >100 | 0.0115 ± 0.0013 | 8.6 ± 1.91 |
| U373 | Human glioblastoma | 18.7 ± 4.17 | >100 | 0.0150 ± 0.0019 | 10.9 ± 1.36 |
| H1299 | Human lung cancer | 11.7 ± 3.91 | >100 | 0.0524 ± 0.0037 | 6.5 ± 1.78 |
| Lu99 | Human lung cancer | 6.8 ± 2.34 | >100 | 0.0183 ± 0.0026 | 12.6 ± 2.64 |
Materials and Methods
Peptides.
Peptides were synthesized by Sigma Genosys (The Woodlands, TX). P4 peptides tagged with cell-permeable epitopes, and their control scrambled peptides were each conjugated with FITC at their C termini to monitor their delivery into cells.
cDNAs and Cells.
cDNAs for AMAP1, cortactin, dynamin2, and paxillin and their expression in cultured cells were performed as described in refs. 3 and 20. A cDNA for c-Src was a gift from M. Sudol (Geisinger Clinic, Danville, PA), and a cDNA for c-Cbl was a gift from T. Kurosaki (RIKEN Research Center for Allergy and Immunology, Yokohama, Japan). MDA-MB-231 cells were obtained from the American Type Culture Collection and cultured as described in refs. 2 and 3. 4T1/luc cells (from T. Yoneda, Osaka University), human glioblastoma cell lines, and COS-7 cells were cultured in DMEM with 10% FCS (HyClone). Human lung cancer cell lines were cultured in RPMI medium 1640, supplemented with 10% FCS. COS-7 cells (5 × 105) were transfected with 3 μg of cDNAs encoding AMAP1, cortactin, dynamin2, paxillin, or c-Cbl, using PolyFect (Qiagen, Valencia, CA).
Cellular Extracts, Protein Binding, and Immunoblotting.
For the in vitro protein binding assay, similar amounts of GST fusion proteins and cell lysates were used as described in refs. 9 and 10. Preparation of cell lysates in 1% Nonidet P-40 buffer, protein binding assays using glutathione-Sepharose beads (Amersham Pharmacia Biosciences), and immunoblotting analyses were performed as described in refs. 4 and 20. Antibodies against GFP and GST were from Upstate Biotechnology (Lake Placid, NY), Xpress-tag was from Invitrogen, and myc-tag was from Babco (Richmond, CA). Horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG were from Jackson ImmunoResearch. Immunoreactive bands were visualized by an enzyme-linked chemiluminescence method (Amersham Pharmacia).
Invasion and Metastasis.
Matrigel chemoinvasion and cell viability were measured by using Biocoat Matrigel chambers (Becton Dickinson) as described in refs. 2 and 3. Data were collected from three independent experiments, each done in duplicate (>100 cells were scored in each experiment). IC50 values were calculated by linear regression analysis of the percentage inhibition in duplicate.
Metastasis studies in mice were performed as described in ref. 3. Briefly, female BALB/c mice (6–8 weeks old; SLC, Shizuoka Ken, Japan) were anesthetized with pentobarbital (0.05 mg per gram of body weight), a 2-mm skin incision was made in the right inguinal mammary fat pad, and 1 × 106 4T1/luc cell clones in a volume of 0.1 ml in PBS were injected into the tissue through a 27-gauge needle. From the 12th day after the injection of cells, 0.1, 0.5, or 1.0 mg/kg UCS15A; 1, 3, or 10 mg/kg P4-TAT; or 10 mg/kg of control scrambled peptide was injected i.p. every day for 8 days. Twelve hours after the last injection on the 19th day, left lungs were excised under deep anesthesia with ether, homogenized, and determined for luciferase activity with a luciferase assay kit (Promega). The protocols used for all animal experiments in this study were approved by the Animal Research Committee of Osaka Bioscience Institute.
X-Ray and NMR Analyses.
A cDNA fragment encoding the human cortactin SH3 domain (amino acid residues 490–550) was cloned into pGEX-6P-1 (Amersham Pharmacia Biosciences), expressed as a GST fusion protein in Escherichia coli BL21, and isolated with glutathione-Sepharose beads. After cleavage with PreScission Protease (Amersham Pharmacia) for separation from the GST and further purification on a Superdex 200 HR10/30 gel filtration column (Amersham Pharmacia), the SH3 protein was concentrated to ≈10 mg/ml.
For the x-ray analysis, the purified SH3 protein was mixed with synthetic P4 peptide at a molar ratio of 1:2 and subjected to crystallization using the hanging drop vapor diffusion method under an equilibrium condition of 100 mM Hepes (pH 7.25), 2.2 M ammonium sulfate, and 100 mM NaCl at 20°C for 24 h. The crystals generated were cryoprotected by soaking in 20% glycerol, harvested in nylon loops, and immediately flash-cooled in liquid nitrogen. Data were measured in nitrogen at the SPring-8 beamline BL44XU (RIKEN, Hyogo, Japan) at 100 K using a DIP6040 imaging plate detector (MAC Science/Bruker AXS, Yokohama, Japan). Structure of the SH3–peptide complex was determined by molecular replacement using the molrep program (21), with a search model of the Abp1 SH3 domain (Protein Data Bank ID code 1JO8). Crystallographic data and refinement statistics are shown in Table 2, which is published as supporting information on the PNAS web site.
NMR analysis on the interaction of the cortactin SH3 domain with its ligands was performed according to a method described in ref. 22.
Size Exclusion Chromatography.
Analytical size exclusion chromatography was carried out at 4°C by using a Superdex 200 HR 10/30 gel filtration column attached to an AKTA system (Amersham Pharmacia Biosciences). Purified cortactin SH3 protein (50 μg) was analyzed in the presence and absence of 1 mM P4 or the dynamin2 PxxP peptides.
Dynamic Light-Scattering Analysis.
Dynamic light-scattering analysis was carried out by using a DynaPro MSK/PCL molecular-sizing instrument equipped with a microsampler (Protein Solutions, which is now Wyatt Technology, Santa Barbara, CA) as described in ref. 23. The concentrations used were 0.1 mM for the cortactin SH3 protein and 0.5 mM for the P4 and dynamin2 PxxP peptides. The data were analyzed with the dynamics 6.0 software.
SH3 Domain Binding.
TranSignal SH3 domain arrays spotted with peptides representing 146 different human SH3 domains were purchased from Panomics, Redwood City, CA, and tested for their binding to P4 peptide according to the manufacturer's instruction. Briefly, membrane filters were incubated with 3 μM biotinylated P4 peptide in the absence or presence of 3 μM UCS15A. After washing, peptide binding was visualized by incubation with streptavidin conjugated to horseradish peroxidase, coupled with enzyme-linked chemiluminescence detection (Amersham Pharmacia). After exposure to x-ray films, the density of each SH3 domain spot was measured by a densitometer (GT-8700 Scanner; Seiko Epson, Nagano, Japan) using imagej 1.34 software (National Institutes of Health, Bethesda). This assay was carried out in duplicate, and the mean values are shown as colored squares in Fig. 4.
Supplementary Material
Acknowledgments
We are grateful to Manami Hiraishi, Yumiko Shibata, and Eriko Chikaishi for technical assistance; Mayumi Iwahara for secretarial work; Toshiyuki Yoneda for 4T1/luc cells; Kuniaki Tatsuta (Kyowa Hakko BioFrontier Laboratories) for compound 2b; Marius Sudol and Tomohiro Kurosaki for cDNAs; and Helena Akiko Popiel for critical reading of the manuscript. This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan; a grant from the Astellas Foundation for Research on Metabolic Disorders; and a grant from the Northern Osaka Saito Biomedical Cluster.
Abbreviations
- SH3
src homology 3
- PLPI
proline-rich ligand-mediated protein interaction
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2D1X).
References
- 1.Sabe H. J. Biochem. 2003;134:485–489. doi: 10.1093/jb/mvg181. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto S., Onodera Y., Hashimoto A., Tanaka M., Hamaguchi M., Yamada A., Sabe H. Proc. Natl. Acad. Sci. USA. 2004;101:6647–6652. doi: 10.1073/pnas.0401753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onodera Y., Hashimoto S., Hashimoto A., Morishige M., Mazaki Y., Yamada A., Ogawa E., Adachi M., Sakurai T., Manabe T., et al. EMBO J. 2005;24:963–973. doi: 10.1038/sj.emboj.7600588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto S., Hashimoto A., Yamada A., Onodera Y., Sabe H. Methods Enzymol. 2005;404:216–231. doi: 10.1016/S0076-6879(05)04021-8. [DOI] [PubMed] [Google Scholar]
- 5.Mayer B. J. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 6.Vives E., Brodin P., Lebleu B. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 7.Derossi D., Joliot A. H., Chassaing G., Prochiantz A. J. Biol. Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 8.McNiven M. A., Cao H., Pitts K. R., Yoon Y. Trends Biochem. Sci. 2000;25:115–120. doi: 10.1016/s0968-0004(99)01538-8. [DOI] [PubMed] [Google Scholar]
- 9.Oneyama C., Nakano H., Sharma S. V. Oncogene. 2002;21:2037–2050. doi: 10.1038/sj.onc.1205271. [DOI] [PubMed] [Google Scholar]
- 10.Oneyama C., Agatsuma T., Kanda Y., Nakano H., Sharma S. V., Nakano S., Narazaki F., Tatsuta K. Chem. Biol. 2003;10:443–451. doi: 10.1016/s1074-5521(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S. V., Oneyama C., Yamashita Y., Nakano H., Sugawara K., Hamada M., Kosaka N., Tamaoki T. Oncogene. 2001;20:2068–2079. doi: 10.1038/sj.onc.1204296. [DOI] [PubMed] [Google Scholar]
- 12.Spencerl S., Dowbenko D., Cheng J., Li W., Brush J., Utzig S., Simanis V., Lasky L. A. J. Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas S. M., Brugge J. S. Annu. Rev. Cell Dev. Biol. 1997;136:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 14.Li W., Fan J., Woodley D. T. Oncogene. 2001;20:6403–6417. doi: 10.1038/sj.onc.1204782. [DOI] [PubMed] [Google Scholar]
- 15.Liotta L. A., Kohn E. C. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 16.Bissell M. J., Radisky D. Nat. Rev. Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiseman B. S., Werb Z. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard J. W. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 19.Bissell M. J., Labrge M. A. Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto S., Hashimoto A., Yamada A., Kojima C., Yamamoto H., Tsutsumi T., Higashi M., Mizoguchi A., Yagi R., Sabe H. J. Biol. Chem. 2004;279:37677–37684. doi: 10.1074/jbc.M404196200. [DOI] [PubMed] [Google Scholar]
- 21.Vagin A., Teplyakov A. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 22.Kojima C., Hashimoto A., Yabuta I., Hirose M., Hashimoto S., Kanaho Y., Sumimoto H., Ikegami T., Sabe H. EMBO J. 2004;23:4413–4422. doi: 10.1038/sj.emboj.7600442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shogren R., Jamieson A. M., Blackwell J., Carrino D. A., Caplan A. I. J. Biol. Chem. 1982;257:8627–8629. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.