Abstract
Epstein–Barr virus (EBV), an orally transmitted herpesvirus, efficiently targets B lymphocytes through binding of the viral envelope glycoprotein gp350 to the complement receptor CD21. How the virus accesses epithelial cells is less well understood, because such cells are largely resistant to infection with cell-free virus in vitro. Here, we show that, after binding to primary B cells, most Epstein–Barr virions are not internalized but remain on the B cell surface and from there can transfer efficiently to CD21-negative epithelial cells, increasing epithelial infection by 103- to 104-fold compared with cell-free virus. Transfer infection is associated with the formation of B cell–epithelial conjugates with gp350/CD21 complexes focused at the intercellular synapse; transfer involves the gp85 and gp110 viral glycoproteins but is independent of gp42, the HLA class II ligand that is essential for B cell entry. Therefore, through efficient binding to the B cell surface, EBV has developed a means of simultaneously accessing both lymphoid and epithelial compartments; in particular, infection of pharyngeal epithelium by orally transmitted virus becomes independent of initial virus replication in the B cell system.
Epstein–Barr virus (EBV), an orally transmitted herpesvirus that is widespread in human populations, shows a marked tropism for B lymphocytes. Preferential binding to B cells is mediated through interaction of the major viral envelope glycoprotein gp350 with the complement receptor CD21 on the B cell surface (1, 2). Thereafter, a second envelope glycoprotein gp42, part of a trimolecular complex with the gp85/gp25 fusion proteins (3, 4), binds to HLA class II molecules (5). This interaction initiates virus–cell fusion, a process requiring gp85/gp25 and also the gp110 glycoprotein (6), and allows viral entry. B cell infection in vitro leads to expression of EBV’s latent growth-transforming genes and the outgrowth of EBV-positive B lymphoblastoid cell lines; likewise, during primary infection in vivo as seen in infectious mononucleosis (IM) patients, the virus drives the expansion of latently infected B cells to establish persistence in the lymphoid system (7). However, EBV is not exclusively B lymphotropic. Full virus replication has been observed in squamous epithelial cells of the tongue in AIDS patients (8) and occasionally in postmortem biopsies from immunocompetent virus carriers (9). Such findings and more recent work on ex vivo explant cultures of tonsillar epithelium (10) suggest that infection of oropharyngeal epithelium is an integral part of the natural EBV–host interaction (reviewed in ref. 11). Indeed, replication at such sites could explain the high levels of infectious virus seen in the throat washings of IM patients (7).
However, exposing squamous epithelial cells (12, 13) or the apical surface of columnar epithelial cells (14) to cell-free virus preparations in vitro gives very low rates of infection. It is only when epithelial monolayers are cocultured with EBV-positive B cell lines, in which some cells have been induced into lytic cycle, that infection rates reach quantifiable levels (13). Such findings have prompted the view that, in the naive host, orally transmitted EBV must undergo an initial round of replication in primary B cells before the virus can access its permissive epithelial targets (7). Yet, new B cell infections, as visualized in the oropharyngeal lymphoid tissues of infectious mononucleosis patients, are almost entirely latent; only rare cells have been observed expressing immediate early antigens, and these cells were never seen progressing to full virus replication (15). In the present report, we explore the possibility that EBV might access epithelium not through an intermediate round of replication in B cells but through binding to the resting B cell surface and using this surface as a transfer vehicle.
Results
Transfer Infection.
In initial experiments, human epithelial cell lines were exposed either to cell-free recombinant EBV for 3 h (direct infection) or, for varying periods of time, to freshly isolated primary B cells immediately after their exposure to the same virus preparations (transfer infection). Epithelial cell infection was monitored 72 h later by GFP expression from the recombinant virus genome, and epithelial identity was confirmed by cytokeratin staining. Coculture with virus-loaded B cells for 12–24 h, followed by B cell removal by washing, dramatically increased the efficiency of epithelial cell infection. Fig. 1A shows photomicrographs from a typical experiment using the Ad-AH epithelial cell line; direct infection produced rare GFP-positive cells (<1 in 104), whereas up to 25% of cells became infected by transfer. Overall, virus-loaded B cells increased the efficiency of infection by 103- to 104-fold for multiplicities of infection between 10 and 100, and a substantial enhancement was apparent even at a multiplicity of infection (moi) of 1. Furthermore, a replication-defective EBV recombinant lacking the BZLF1 immediate early gene (16) transferred as efficiently as WT virus, indicating that the process was independent of lytic replication in donor cells. Fig. 1B records the transfer infections that were seen when different acceptor cells, all lacking CD21, were exposed for 24 h to B cells preloaded with EBV at a moi of 100. All four epithelial target cells, including cultures of normal nasopharyngeal epithelium, were highly susceptible, with 5–25% of cells infected, whereas there was never any detectable infection of normal fibroblast or endothelial cell cultures or of cocultured primary T cells.
Fig. 1.
Characteristics of transfer infection. (A) Photomicrographs show phase contrast and GFP fluorescence images of Ad-AH cultures 72 h after exposure to cell-free recombinant B95.8 EBV at a moi of 100 (direct infection) or 48 h after a 24-h cocultivation with primary B cells preexposed to the same virus (transfer infection). Histograms show efficiencies of direct versus transfer infection of Ad-AH cells in a representative experiment conducted as above but using multiplicities of infection between 1 and 100. Efficiency is expressed on a logarithmic scale as the percentage of GFP-positive cells. Gray bars show corresponding data using a BZLF1-knockout (replication-deficient) EBV. (B) Mean results from repeat assays comparing epithelial cell lines and normal nasopharyngeal epithelial cells with normal fibroblasts, endothelial cells, and primary T cells as acceptors; cells were cocultured for 24 h with primary B cells preexposed to virus at a moi of 100, and transfer infection was expressed as above (linear scale).
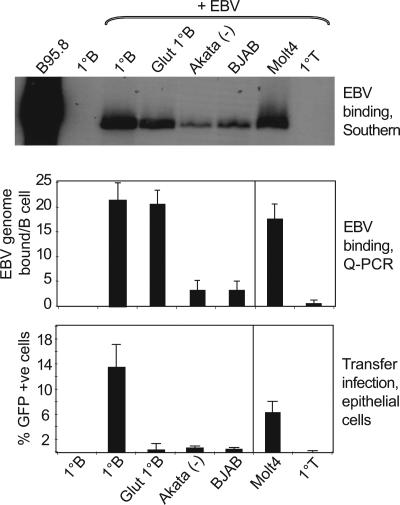
We then asked which cell types could serve as donors in transfer infection. Primary B cells, two CD21-expressing B cell lines (EBV-negative Akata-BL and BJAB), a CD21-expressing T cell line (Molt4), and primary T cells were tested as donors in combination with Ad-AH acceptor cells under the same conditions as in Fig. 1B. Virus binding to donor cells was quantitated by Southern blotting and by quantitative PCR (Q-PCR) with concordant results (Fig. 2Top and Middle); binding was proportional to the cells’ level of CD21 expression (data not shown). Transfer infection (Fig. 2 Bottom) was optimal using primary B cells; prior glutaraldehyde fixation of the B cells did not affect EBV binding but severely reduced transfer, indicating a need for active participation of the donor cell. Both B cell lines also served as donors but much less efficiently, commensurate with their lower levels of CD21 expression and EBV binding. Interestingly, the CD21-positive Molt4 T cell line was itself an efficient donor, whereas primary T cells bound virus at minimal levels and gave no transfer.
Fig. 2.
Virus binding versus transfer infection by different donor cells. Primary B cells (with and without prior glutaraldehyde fixation), EBV-negative Akata and BJAB B cell lines, primary T cells, and the Molt4 T cell line were exposed to virus at a moi of 100, and virus binding was assayed independently by Southern blotting (Top) and Q-PCR (Middle); the EBV-producing B95.8 cell line and uninfected primary B cells serve as positive and negative controls. (Bottom) These same cells were then used as donors in 24-h cocultures with Ad-AH cells, and transfer infection was expressed as in Fig. 1B. Mean results from repeat assays are shown.
Surface-Bound Virus and Transfer Infection.
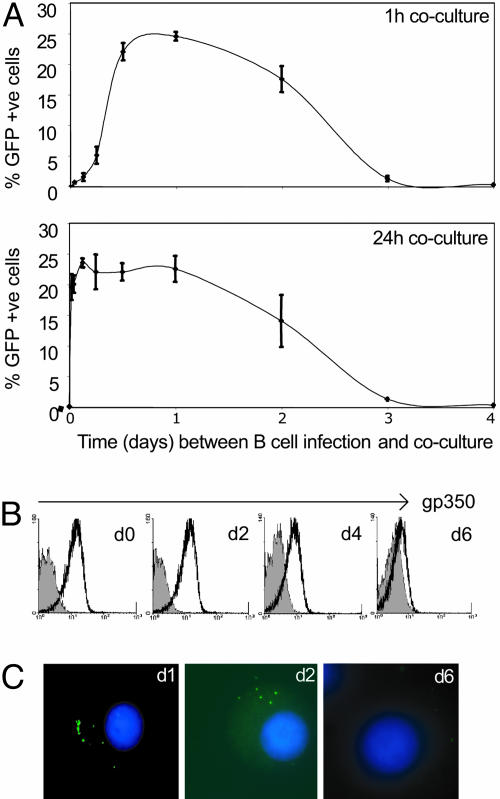
We then asked the question: How long do freshly infected B cells remain capable of mediating virus transfer? Primary B cells were harvested at different times postinfection (p.i.) and added to epithelial cells for either 1 or 24 h. They were then washed, and the epithelial cells were assayed for GFP expression 3 days after their first B cell contact. As shown in Fig. 3A, B cells were efficient virus donors when tested on days 1 or 2 p.i. and still showed some activity on day 3. Moreover, transfer infection was just as efficient with 1 h of B cell–epithelial contact as with 24 h of contact. Equally rapid transfer was also seen when EBV-negative B cell lines and CD40 ligand-activated B lymphoblasts were tested as donor cells immediately after virus loading (data not shown). A requirement for longer contact was observed only with primary B cells within the first 6 h after virus loading. These findings suggested that resting B cells need time after virus exposure to become competent for transfer infection but that the transfer process itself is rapid. Indeed, further experiments using donor B cells 1 day p.i. revealed optimal transfer with only 10 min of B cell–epithelial contact (data not shown).
Fig. 3.
Kinetics of transfer infection. (A) Duration of B cell infectivity p.i. Primary B cells were exposed to EBV (moi of 100) for 3 h and then added to Ad-AH acceptor cells either immediately after virus loading or after varying times in culture up to 4 days p.i. In each case, donor cells were left in contact with acceptors for 1 (Upper) or 24 (Lower) h, and transfer infection was assayed 72 h after the first donor–acceptor cell contact. Results are expressed as in Fig. 1B. (B) Duration of gp350 staining on the B cell membrane. Primary B cells were exposed to EBV as above and then stained with anti-gp350 mAb either immediately or up to 6 days p.i.; gp350 staining profiles are shown alongside that of an isotype-matched control mAb (shaded profile). (C) Detection of viral genomes by FISH. Primary B cells were exposed to EBV as above and then fixed either immediately or up to 6 days p.i; extranuclear viral genomes are detected by green fluorescence, and nuclei are identified by DAPI staining.
Because B cells remained infectious for epithelium up to 2 days after binding virus, we sought to discover how long EBV remained detectable on the B cell surface. mAb staining showed that surface levels of the envelope glycoprotein gp350 remained largely unchanged over the first 2 days and only after this time gradually declined (Fig. 3B). Such staining appeared to reflect persistence of some complete virions at the cell surface rather than being entirely due to discarded viral envelope or empty particles contaminating virus preparations. Thus, FISH with a sensitive EBV DNA probe (17) detected multiple extranuclear copies of viral DNA on cells within the first 2 days after virus binding, with signals apparently localized to one pole of the cell (Fig. 3C). In further experiments, we used a chymotrypsin treatment to strip the B cell surface of bound virions and found that, whereas stripping at times beyond 1 h after virus loading had no detectable effect on subsequent B cell transformation, stripping at any time up to day 3 p.i. completely abrogated transfer infection (see the supporting information, which is published on the PNAS web site). These findings strongly suggested that EBV retained on the B cell surface after virus loading was indeed the source of virus transmitted to epithelium.
Viral Glycoproteins and Transfer Infection.
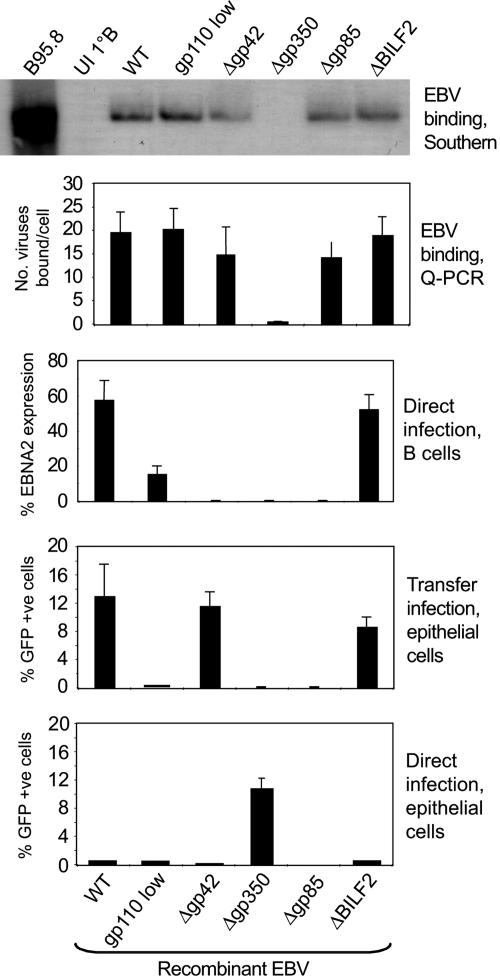
Because different sets of viral glycoproteins appear to be required for EBV entry into B cells and epithelium (18), we set up transfer infection assays using recombinant EBVs that lacked gp350, gp85, gp42, or the BILF2 glycoprotein or had low levels of gp110. The two uppermost panels of Fig. 4 show the binding of these viruses to donor B cells assayed by Southern blotting and Q-PCR as in Fig. 2. All of the viruses bound well with the exception, as anticipated, of the gp350 knockout. However, the middle panel of Fig. 4 shows that only the BILF2-knockout virus initiated B cell infection at WT levels, here assessed by staining for EBV-encoded nuclear antigen (EBNA) 2 (17). There was no detectable B cell infection by viruses lacking gp350 (the CD21 ligand), gp42 (the HLA class II ligand), or gp85 (a component of the fusion complex), and there was only poor infection by the gp110-low virus. These same viruses were then tested for transfer infection of Ad-AH acceptor cells by using as donors both resting B cells and (to control for any inability of the viruses to render B cells competent for transfer) CD40 ligand-activated B lymphoblasts. The two donor cell types gave similar results; the data with resting B cells are shown in the second panel from the bottom in Fig. 4. Transfer infection was efficient with WT virus, with the BILF2 knockout, and, importantly, also with the gp42 knockout, but it was poor with the gp110-low virus and completely undetectable with the gp350 and gp85 knockouts. As a further control, the same viruses were tested in parallel for direct epithelial infection. As expected, the BILF2 and gp42 knockouts gave the same very low rates of infection as WT virus, the gp110-low virus was even poorer, and the gp85 knockout was completely negative. Remarkably, however, the gp350 knockout consistently gave rates of direct infection equivalent to those seen by transfer infection using WT virus.
Fig. 4.
Virus binding to B cells, direct B cell infection, transfer infection, and direct epithelial cell infection by different EBV mutants. Primary B cells were exposed to recombinant EBVs (WT, gp110-low, and the glycoprotein knockouts Δgp42, Δgp350, Δgp85, and ΔBILF2) at a moi of 100, and virus binding was assayed by Southern blotting and by Q-PCR (two uppermost panels) as in Fig. 2. B cell infection was assayed by staining for EBNA2 expression after 3 days in culture (17), and the percentage of EBNA2-positive cells are shown (middle panel). Additional aliquots of these virus-loaded B cells were cocultured with Ad-AH cells for 24 h immediately p.i. to assay for transfer infection (second panel from bottom); in parallel, the same recombinant EBV preparations were assayed for direct Ad-AH cell infection at a moi of 100 (bottom panel). Results of transfer and direct epithelial cell infection are expressed as in Fig. 1B.
Conjugate Formation Between B Cells and Epithelial Cells.
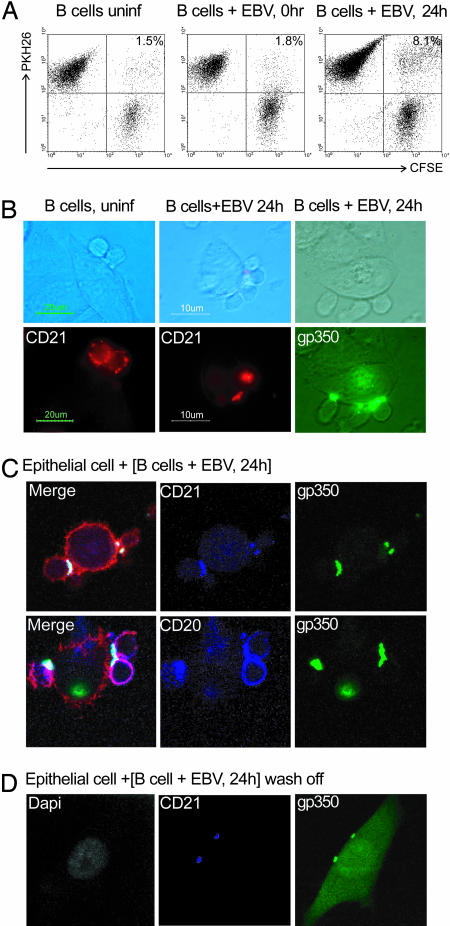
Finally, we set up an assay to monitor contacts formed between donor and acceptor cells. Primary B cells (uninfected, immediately post-EBV infection, and 24 h p.i.) were labeled with PKH26 dye, mixed with carboxyfluorescein diacetate succinimidyl ester-labeled epithelial cells in suspension, and centrifuged. After 30 min of incubation they were gently resuspended, and then B cell–epithelial conjugates were visualized by two-color FACS analysis. As shown in Fig. 5A, although uninfected B cells or cells tested immediately p.i. formed very few conjugates with epithelial cells, conjugate formation was significantly increased in B cells at 24 h p.i.
Fig. 5.
Analysis of conjugate formation. (A) FACS profiles of conjugate formation between carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled Ad-AH cells (x axis) and PKH26-labeled primary B cells (y axis) tested without EBV infection, immediately after virus binding at a moi of 100 (0 h), or at 24 h p.i. Conjugates appear in the upper right quadrant, and the percentages of cells in conjugates are shown. (B) Phase contrast and fluorescence images of B cell–epithelial conjugates formed between Ad-AH cells and primary B cells without EBV infection or at 24 h p.i. Before coculture, B cells were stained with red-labeled anti-CD21 mAb or with green-labeled anti-gp350 mAb. (C) Confocal images of 1-μm Z slices through conjugates between epithelial cells and EBV-infected B cells (labeled 24 h p.i.). All gp350 staining (green) colocalizes at the cell–cell junction with CD21 staining (blue, Upper), whereas CD20 staining (blue, Lower) remained diffusely distributed; phalloidin staining (red) shows the cell boundaries. (D) Confocal images of a 1-μm Z slice through an epithelial cell immediately after 24-h coculture with EBV-infected B cells and B cell wash-off. gp350 (green) colocalizes with CD21 (blue) at two points on the epithelial cell membrane. Light-green staining of the epithelial cell indicates GFP expression from EBV infection. DAPI staining for DNA (white) confirms that the infected cell is mononuclear and that B cells have been removed.
Pilot experiments showed that both conjugate formation and transfer infection were unaffected if B cells 24 h p.i. were labeled immediately before epithelial cell contact with mAbs to gp350, to CD21 or its associated proteins CD19 and CD81, or to other B cell surface proteins (CD20, HLA-I, HLA-II, and the Ig/CD79 complex). As a result, it was possible to visualize the cell surface distribution of these molecules in B cell–epithelial conjugates. As shown in Fig. 5B, both gp350 and CD21 localized to the point of contact between virus-loaded B cells and epithelial cells; CD19 and CD81 were similarly localized (data not shown). This capping of the receptor complex was clearly virus-induced, because CD21 remained diffusely distributed around the B cell surface in rare conjugates seen between uninfected B cells and epithelium (Fig. 5B). In further experiments, EBV-infected B cells, double-labeled with gp350-specific and cell marker-specific mAbs, were allowed to form conjugates with epithelial cells, and the conjugates were then fixed, permeabilized, and stained with phalloidin before analysis using confocal microscopy. As seen in Fig. 5C, this analysis confirmed that all gp350 and CD21 staining colocalized at the cell–cell junction, whereas staining for other markers, such as CD20 (and HLA-I, HLA-II, IgM, IgD, CD79a, and CD79b; data not shown), remained diffusely distributed. Intercellular contacts were clearly intimate because, when EBV-infected B cells were prelabeled as above, cocultured for 24 h on epithelial monolayers, and then washed, we often detected residual foci of gp350/CD21 staining on epithelial cell surfaces. Such foci are illustrated in Fig. 5D; note here that the epithelial cell, in which diffuse GFP expression reflects active infection, remains mononuclear. Although cell fusion has been suggested as a possible route of EBV entry into epithelium (19), we never observed any morphologic evidence of fusion or detected any multinucleated cells by DAPI staining.
Discussion
Although B cells remain the essential reservoir for EBV persistence in vivo, current evidence (8–10) favors the view that replication in oropharyngeal epithelium is an integral part of the normal virus–host interaction (reviewed in ref. 11). Yet, EBV entry into epithelium has remained difficult to analyze because, for almost all epithelial cell cultures, efficient rates of infection cannot be achieved by using cell-free virus; reasonably efficient rates of infection are only possible by coculture with virus-producing B cell lines (13). This observation, and the finding that B cell-deficient patients never acquire EBV even locally in the oropharynx (20), has prompted the view that epithelial infections in vivo must be secondary to EBV replication within the B cell system. Here, we present evidence to the contrary and show that virus captured on the surface of recently exposed primary B cells is highly infectious for epithelium.
Using four different epithelial target cells, virus-loaded B cells increased the efficiency of infection by 103- to 104-fold above the very low rates that we and others (12–14) see when using cell-free virus. Although these experiments used GFP as the readout, active epithelial infection was indeed confirmed by staining for EBNA1 (data not shown). Interestingly, neither fibroblasts nor endothelial cells were susceptible to viral transfer, indicating that the process is to some extent lineage-restricted. We found that, after virus loading, resting B cells become competent to serve as donor cells within 12 h p.i. and, once competent, can transfer the virus within 10 min of B cell–epithelial contact. Such kinetics argued against any requirement for virus replication in resting B cells before transfer, a point formally proven by demonstrating transfer of a replication-defective (BZLF1 knockout) virus. They also argued against any dependence upon S phase entry and blast transformation of infected B cells, because these events occur later (17). In fact, active donor cell infection proved to be entirely dispensable, because a viral mutant lacking the gp42 glycoprotein, known to be essential for B cell infection (21), transferred efficiently. We noted that virus-loaded B cells become transfer-competent at the same time that they acquire the ability to form conjugates with epithelial cells, suggesting that the two activities are related; indeed, both may be acquired as a consequence of EBV-induced cross-linking of the CD21/CD19/CD81 receptor complex. Thus, CD21 cross-linking is known to have several downstream signaling effects in resting B cells (22–24) and, in a model system, leads to cytoskeletal reorganization (25). Certainly, EBV binding induced cocapping of the gp350/receptor complex to one pole of the cell, and, after coculture with epithelium, that pole formed the point of intercellular contact. Such localization is reminiscent of the “virological synapses” seen in human retroviral systems where contacts between a productively infected or virus-loaded donor cell and an uninfected acceptor cell become the conduit for virus transfer (26, 27). Certainly, the “synapses” between EBV-loaded B cells and epithelial cells are sufficiently intimate to leave fragments of B cell membrane on the epithelial cell surface after B cell wash-off.
An initially surprising finding was that B cells remained capable of efficiently transferring EBV to epithelial cells for at least 2 days after virus loading. EBV is known to enter B cells by CD21 receptor-mediated endocytosis followed by a pH-independent fusion event within the endocytic pathway (28). However, we found by mAb detection and FISH, respectively, that most gp350 and most input EBV genomes remained at the surface for several days, consistent with our recent finding that only ≈10% of surface-loaded EBV genomes reach the B cell nucleus (17). Further evidence strongly supported a role for surface-retained virions in transfer infection. First, chymotrypsin stripping of virions from the B cell surface up to day 3 p.i. abrogated transfer infection without affecting the ongoing transformation of these cells by successfully endocytosed virus. Second, EBV loaded onto the CD21-positive T cell line Molt4 was able to infect epithelial cells, even though Molt4 cells cannot internalize virus because they lack the HLA class II coreceptor for gp42 (29). Third, the gp42-knockout virus is incapable of entering resting B cells (21), yet it transferred well to epithelium.
Thereafter, we used viral mutants in an attempt to identify which viral glycoproteins are required for transfer infection. The requirements for conventional B cell infection are already well defined [namely, gp350 (the CD21 ligand), gp42 (the HLA class II ligand), and gp85/25 and gp110 (components of the fusion machinery) (2, 3, 6)]. In contrast, infection of epithelial cells by cell-free virus is much less well understood but is hypothesized to involve gp85/gp25 acting both as a ligand for binding (to an as yet unidentified receptor) and, along with gp110, as a fusion complex (3, 11, 18). Indeed, we found both gp85 and gp110 to be important for transfer infection, whereas gp42 was clearly not required. In that regard, others have shown that the presence of gp42 in the gp85/gp25 complex actually inhibits the binding to epithelial cells (18). However, not all gp85/gp25 dimers on the viral envelope are complexed with gp42. Virions produced in HLA class II-positive B cells tend to be gp42-low, because nascent HLA class II molecules sequester gp42 into endosomes and limit its representation in virus particles. As a result, gp42-low viruses (made in B cells) preferentially target epithelium, whereas gp42-high viruses (made in epithelial cells) preferentially target B cells (18). In the present work, however, we found no obvious inhibitory effect of gp42 on transfer infection. Thus, the gp42 knockout was no more infectious than the WT virus, even though the latter (produced in the HLA class II-negative 293 cell line) is predicted to be gp42-high. Likewise, we have not seen clear differences in transfer infection comparing WT virus produced in 293 cells with that produced in B cells (data not shown). It may be that the inhibitory effects of gp42 on the binding of free virus particles to epithelium are overridden when, as in transfer infection, surface-bound virions are concentrated in a synapse between donor and acceptor cells.
As for the major envelope glycoprotein gp350, it is essential for transfer infection but apparently only as a ligand for binding to donor B cells. Thus, once bound to the B cell surface, virions can be saturated with a gp350-specific mAb that normally is neutralizing for B cell infection, and this mAb has no effect on virus transfer. Very interestingly, we found that the gp350 knockout virus, although inactive both in B cell infection and in transfer infection, was unique in its capacity to infect epithelial cells directly. This finding suggests that gp350, present in all natural EBV isolates and essential for efficient entry into the B cell system, is actually inhibitory to direct epithelial infection. We infer that the gp350–CD21 interaction relieves this inhibition and unmasks the virus envelope components that are necessary for epithelial infection. Note that this unmasking cannot be achieved by simply occluding gp350, because preincubating cell-free virus with the above gp350-specific mAb did not render it directly infectious for epithelium (data not shown).
The present findings have implications for our understanding of EBV biology. Implicit in most models of primary infection has been the requirement that, before accessing epithelium, orally transmitted EBV must first replicate in oropharyngeal B cells, a step for which there is no direct evidence. The present work removes this conceptual difficulty and shows how EBV, through efficient binding to the B cell surface, can simultaneously initiate latent infections in the B cell lineage and, through transfer infection, access the permissive epithelial compartment.
Materials and Methods
Cells and Cell Lines.
Primary B and T cells were isolated from peripheral blood as described in ref. 17. B lymphoblasts were produced by coculturing primary B cells with CD40 ligand-expressing mouse L cells in the presence of IL-4. Normal nasopharyngeal epithelial cells were grown in serum-free keratinocyte growth medium (SFM; GIBCO). Cultures of primary fibroblasts and endothelial cells and established lines of epithelial (HeLa, Ad-AH, and SVK), T cell (Molt4), and B cell (BJAB, EBV-negative Akata, and B95.8) origin were grown in RPMI medium 1640 supplemented with 10% FCS.
Virus Preparations and Binding Assays.
Virus preparations were made from 293 cells carrying recombinant B95.8 EBV genomes, either WT or deleted for gp350, gp85, gp42, or BILF2, as described in refs. 30 and 31; gp110 levels were optimized in all cases by BALF4 (gp110) transfection, except when the WT B95.8 construct was used to prepare gp110-low virus (32). Preparations were assayed for EBV genome content by Q-PCR amplifying within the BALF5 gene (17). Virus binding to cell surfaces after 3-h exposure at 4°C was quantitated by three independent methods: Southern blotting with a BALF5 gene probe, Q-PCR as above, and staining with a gp-350-specific mAb (72A-1) as described in ref. 17.
Transfer Infection.
In standard assays, donor cells were exposed to EBV at known multiplicities of infection for 3 h at 4°C and washed well, and 106 cells were added to 2-ml wells that had been seeded 24 h earlier with 3 × 105 acceptor cells. After coculture for up to 24 h, donor cells were removed from acceptors by washing; transfer infection was assayed 72 h after the initiation of coculture by counting the percentage of GFP-positive cells in trypsinized acceptor cell suspensions. Where primary T cells were tested as acceptors, donor B cells were removed by anti-CD19 beads (17). Where glutaraldehyde-fixed primary B cells were tested as donors, before EBV binding, B cells were exposed for 60 s to 0.05% glutaraldehyde. Then, the glutaraldehyde was quenched in excess 0.2 M glycine for 20 min and cells were washed extensively in 10% BSA; all solutions were in PBS and were used at room temperature. In some assays, the donor B cells were cultured after wash-off and assayed for EBV infection by mAb staining for EBNA2 expression (17).
FISH.
Viral genomes were detected by FISH with an EBV cosmid probe as described in ref. 16 but using whole-cell rather than nuclear preparations.
Conjugate Formation.
Freshly isolated primary B cells, uninfected or exposed to EBV and used either immediately after virus binding or after a further 24 h in culture, were labeled with PKH26 (Sigma-Aldrich) according to the manufacturer’s instructions. Epithelial cells were labeled with 10 nM CFSE (carboxyfluorescein diacetate succinimidyl ester) (Molecular Probes) for 10 min at 37°C. The above cells were mixed at a ratio of 1:1 and centrifuged at 100 × g. The pellet was incubated at 37°C for 30 min and then gently resuspended in 200 μl of PBS and analyzed immediately for double-labeled cell conjugates by flow cytometry. Mixtures made immediately before analysis, without centrifugation, served as controls.
Microscopy.
After virus binding and 24-h culture, B cells were labeled with individual mAbs or mAb combinations by using Cy5-conjugated mAbs to CD19, CD20, CD21 (Pharmingen), IgD, and IgM (Dako Cytomation); FITC-conjugated mAbs to CD79a and 79b (Pharmingen); and mAbs to HLA class I (W6/32), HLA class II-DP (B7/21.2), HLA class II-DQ (SPV-L3), HLA class II-DR (L243), CD81 (M83), and gp350 (ID4C-1) immediately after Alexa Fluor fluorochrome 350, 488, 546, or 647 labeling (Zenon kit; Invitrogen). Labeled B cells were then cocultured for 1 h with epithelial cells growing on coverslips, after which, nonbound B cells were washed. The remaining cells were fixed in 2% paraformaldehyde for 10 min and either viewed directly or permeabilized in 0.2% Triton X-100 for 5 min and then stained with Texas red-labeled phalloidin to visualize the actin cytoskeleton and viewed by confocal microscopy.
Supplementary Material
Acknowledgments
This work was supported by the Association for International Cancer Research and Cancer Research UK.
Abbreviations
- EBV
Epstein–Barr virus
- EBNA
EBV-encoded nuclear antigen
- moi
multiplicity of infection
- p.i.
postinfection
- Q-PCR
quantitative PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fingeroth J. D., Weis J. J., Tedder T. F., Strominger J. L., Biro P. A., Fearon D. T. Proc. Natl. Acad. Sci. USA. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemerow G. R., Wolfert R., McNaughton M. E., Cooper N. R. J. Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molesworth S. J., Lake C. M., Borza C. M., Turk S. M., Hutt-Fletcher L. M. J. Virol. 2000;74:6324–6332. doi: 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oda T., Imai S., Chiba S., Takada K. Virology. 2000;276:52–58. doi: 10.1006/viro.2000.0531. [DOI] [PubMed] [Google Scholar]
- 5.Li Q., Spriggs M. K., Kovats S., Turk S. M., Comeau M. R., Nepom B., Hutt-Fletcher L. M. J. Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McShane M. P., Longnecker R. Proc. Natl. Acad. Sci. USA. 2004;101:17474–17479. doi: 10.1073/pnas.0404535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rickinson A., Kieff E. In: Fields Virology. Knipe D. M., Howley P. M., editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2575–2627. [Google Scholar]
- 8.Greenspan J. S., Greenspan D., Lennette E. T., Abrams D. I., Conant M. A., Petersen V., Freese U. K. N. Engl. J. Med. 1985;313:1564–1571. doi: 10.1056/NEJM198512193132502. [DOI] [PubMed] [Google Scholar]
- 9.Frangou P., Buettner M., Niedobitek G. J. Infect. Dis. 2005;191:238–242. doi: 10.1086/426823. [DOI] [PubMed] [Google Scholar]
- 10.Pegtel D. M., Middeldorp J., Thorley-Lawson D. A. J. Virol. 2004;78:12613–12624. doi: 10.1128/JVI.78.22.12613-12624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutt-Fletcher L. In: Epstein Barr Virus. Robertson E., editor. Norfolk, U.K.: Caister Academic; 2005. pp. 359–378. [Google Scholar]
- 12.Sixbey J. W., Nedrud J. G., Raab-Traub N., Hanes R. A., Pagano J. S. N. Engl. J. Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 13.Imai S., Nishikawa J., Takada K. J. Virol. 1998;72:4371–4378. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tugizov S. M., Berline J. W., Palefsky J. M. Nat. Med. 2003;9:307–314. doi: 10.1038/nm830. [DOI] [PubMed] [Google Scholar]
- 15.Niedobitek G., Agathanggelou A., Herbst H., Whitehead L., Wright D. H., Young L. S. J. Pathol. 1997;182:151–159. doi: 10.1002/(SICI)1096-9896(199706)182:2<151::AID-PATH824>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Feederle R., Kost M., Baumann M., Janz A., Drouet E., Hammerschmidt W., Delecluse H.-J. EMBO J. 2000;19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon-Lowe C., Baldwin G., Feederle R., Bell A., Rickinson A., Delecluse H.-J. J. Gen. Virol. 2005;86:3009–3019. doi: 10.1099/vir.0.81153-0. [DOI] [PubMed] [Google Scholar]
- 18.Borza C. M., Hutt-Fletcher L. M. Nat. Med. 2002;8:594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 19.Bayliss G. J., Wolf H. Nature. 1980;287:164–165. doi: 10.1038/287164a0. [DOI] [PubMed] [Google Scholar]
- 20.Faulkner G. C., Burrows S. R., Khanna R., Moss D. J., Bird A. G., Crawford D. H. J. Virol. 1999;73:1555–1564. doi: 10.1128/jvi.73.2.1555-1564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Hutt-Fletcher L. M. J. Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner J., Weis J., Fearon D., Whang Y., Kieff E. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair A. J., Farrell P. J. J. Virol. 1995;69:5461–5468. doi: 10.1128/jvi.69.9.5461-5468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugano N., Chen W., Roberts M. L., Cooper N. R. J. Exp. Med. 1997;186:731–737. doi: 10.1084/jem.186.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill M. B., Roecklein-Canfield J., Sage D. R., Zambela-Soediono M., Longtine N., Uknis M., Fingeroth J. D. J. Cell Sci. 2004;117:2709–2720. doi: 10.1242/jcs.01113. [DOI] [PubMed] [Google Scholar]
- 26.Igakura T., Stinchcombe J. C., Goon P. K., Taylor G. P., Weber J. N., Griffiths G. M., Tanaka Y., Osame M., Bangham C. R. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 27.Jolly C., Sattentau Q. J. J. Virol. 2005;79:12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemerow G. R., Cooper N. R. Virology. 1984;132:186–198. doi: 10.1016/0042-6822(84)90102-8. [DOI] [PubMed] [Google Scholar]
- 29.Menezes J., Seigneurin J. M., Patel P., Bourkas A., Lenoir G. J. Virol. 1977;22:816–821. doi: 10.1128/jvi.22.3.816-821.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delecluse H. J., Hilsendegen T., Pich D., Zeidler R., Hammerschmidt W. Proc. Natl. Acad. Sci. USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janz A., Oezel M., Kurzeder C., Mautner J., Pich D., Kost M., Hammerschmidt W., Delecluse H.-J. J. Virol. 2000;74:10142–10152. doi: 10.1128/jvi.74.21.10142-10152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuhierl B., Feederle R., Hammerschmidt W., Delecluse H.-J. Proc. Natl. Acad. Sci. USA. 2002;99:15036–15041. doi: 10.1073/pnas.232381299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.