Abstract
Nuclear receptors (NRs) are a large family of transcription factors. One hallmark of this family is the ligand-binding domain (LBD), for its primary sequence, structure, and regulatory function. To date, NRs have been found exclusively in animals and sponges, which has led to the generally accepted notion that they arose with them. We have overcome the limitations of primary sequence searches by combining sequence profile searches with structural predictions at a genomic scale, and have discovered that the heterodimeric transcription factors Oaf1/Pip2 of the budding yeast Saccharomyces cerevisiae contain putative LBDs resembling those of animal NRs. Although the Oaf1/Pip2 LBDs are embedded in an entirely different architecture, the regulation and function of these transcription factors are strikingly similar to those of the mammalian NR heterodimer peroxisome proliferator-activated receptor α/retinoid X receptor (PPARα/RXR). We demonstrate that the induction of Oaf1/Pip2 activity by the fatty acid oleate depends on oleate’s direct binding to the Oaf1 LBD. The alteration of two amino acids in the predicted ligand-binding pocket of Oaf1 abolishes both ligand binding and the transcriptional response. Hence, LBDs may have arisen as allosteric switches, for example, to respond to nutritional and metabolic ligands, before the animal and fungal lineages diverged.
Keywords: evolution, fatty acids, structure prediction, yeast, bioinformatics
The nuclear receptor (NR) superfamily is characterized by two unique domains. Almost all members contain a very highly conserved DNA-binding domain (DBD) consisting of two Cys4 zinc fingers, as well as a somewhat less conserved ligand-binding domain (LBD) comprising ≈250 aa at the C terminus. These building blocks confer the potential to act as both an intracellular receptor and a ligand-regulated transcription factor. Although only a minority of NRs have known ligands, it appears that LBDs evolved as allosteric switches to control NR activities as transcription factors (1–3).
Homologous sequences have been identified in a large number of species by performing searches with DBD and/or LBD sequences (3, 4). Because only animals and sponges have recognizable NR sequences, it has been concluded that NRs evolved in a common animal or urmetazoan ancestor (5, 6). However, because these primary sequence searches were all based on one of the available blast algorithms (7, 8), more distant homologs with poor primary sequence similarity could not have been identified. We have now combined such searches with structural predictions and have uncovered additional potential homologs in yeast. Our results challenge the assumption that NRs are an animal-specific family of transcription factors and argue that allosteric regulation by NR LBDs is more ancient than animals.
Results
Bioinformatic Discovery of LBD Candidates.
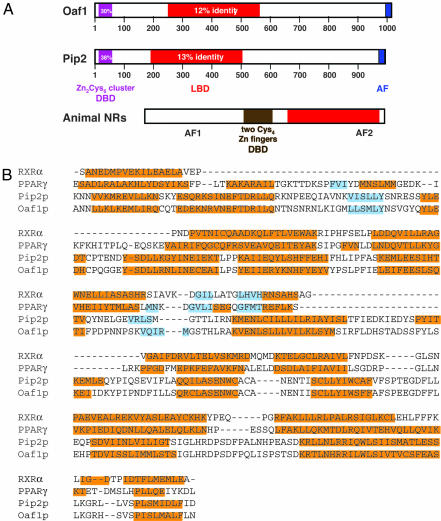
We have applied the Genome Threader algorithm (9–11), which uses a threading approach to protein structure prediction, at a genomic scale. This method allows the identification of proteins that are predicted to have a similar structural fold, although they may not share significant sequence homology. The structural similarity requirement enables the identification of very distant members of a functional family with a confidence that cannot be achieved by using sequence similarity alone. A combination of sequence similarity that is limited to a few functionally important residues with an overall conservation of the structural fold has been used successfully as a criterion to assign the proper function to distantly related sequences (12–14). In the course of attempting to identify all proteins predicted to have the NR LBD fold (1, 2), we discovered two proteins from Saccharomyces cerevisiae, Oaf1 and Pip2 (Fig. 1), in the dataset. We predict two known structural folds in each of the proteins: (i) an N-terminal Zn2Cys6 DBD and (ii) an LBD fold C-terminal to the DBD (Fig. 1A). As expected, the DBD prediction is of high confidence for both proteins because they exhibit substantial sequence identity to the Zn2Cys6 DBD family of proteins. The LBD predictions are of a lower confidence, reflecting the substantial divergence of these domains. The best LBD structural assignments for Oaf1 and Pip2 are to the LBD of human estrogen receptor β (Protein Data Bank ID code 1QKM) and the LBD of retinoic acid-related orphan receptor β (RORβ) (Protein Data Bank ID code 1K4W) at 78% and 68% confidence, respectively. The sequence identities to these mammalian proteins are 12% and 13% for Oaf1 and Pip2, respectively. Given the large number of NR LBD structures in public databases, there were many more assignments with various degrees of confidence for both proteins (data not shown). Fig. 1B shows a structural alignment of Oaf1 and Pip2 with the LBDs of retinoid X receptor α (RXRα) and peroxisome proliferator-activated receptor γ (PPARγ). On the basis of position-specific scoring matrices (15), the predicted secondary structures of the Oaf1 and Pip2 LBDs align well with the known secondary structure features of RXRα and PPARγ. Based on a partial sequence alignment with the peroxisomal membrane protein Pex11 from yeast, it had previously been speculated that animal LBDs arose from an older protein (16). However, no functional support was provided, and a substantial portion of the LBD was not aligned to Pex11, which may explain why we could not confirm this hit with our own bioinformatic tools. The relationship of LBDs to Pex11, although intriguing, remains to be further explored.
Fig. 1.
Oaf1 and Pip2 contain a domain with structural homology to the LBD of NRs. (A) Schematic representation of the domain structures of Oaf1 and Pip2, compared with those of NRs from animals. The LBDs are shown in red, the yeast-specific DBD in purple, and activation functions (AF) in blue. The indicated primary-sequence identities are to structures with Protein Data Bank (PDB) ID codes 1PYI and 1HWT for the DBDs and to the human estrogen receptor β (PDB ID code 1QKM) and RORβ (PDB ID code 1K4W) for the LBDs of Oaf1 and Pip2, respectively. (B) Structural alignment of the Oaf1 (amino acids 254–563) and Pip2 (amino acids 190–497) sequences with the LBD sequences of RXRα (PDB ID code 1DKF) and PPARγ (PDB ID code 1PRG), showing α-helices in orange and β-sheets in blue (predicted for Oaf1/Pip2, experimental for RXR/PPAR). RXR and PPAR were chosen because of the numerous functional analogies that are discussed in the text.
Spectrum of in Vivo Responses of Oaf1/Pip2 to Fatty Acids.
Oaf1 and Pip2 are essential heterodimeric transcription factors for the utilization of the fatty acid oleate as an external carbon source (17–20). In the absence of glucose, the preferred carbon source of S. cerevisiae, oleate induces the expression of the entire panel of genes required for uptake and breakdown of fatty acids in an OAF1/PIP2-dependent fashion (18–21). Moreover, the transcriptional activity of Oaf1 fused to a heterologous DBD is stimulated by oleate (22). In conjunction with our structural predictions, these findings suggested that oleate might be a ligand and direct activator of Oaf1/Pip2.
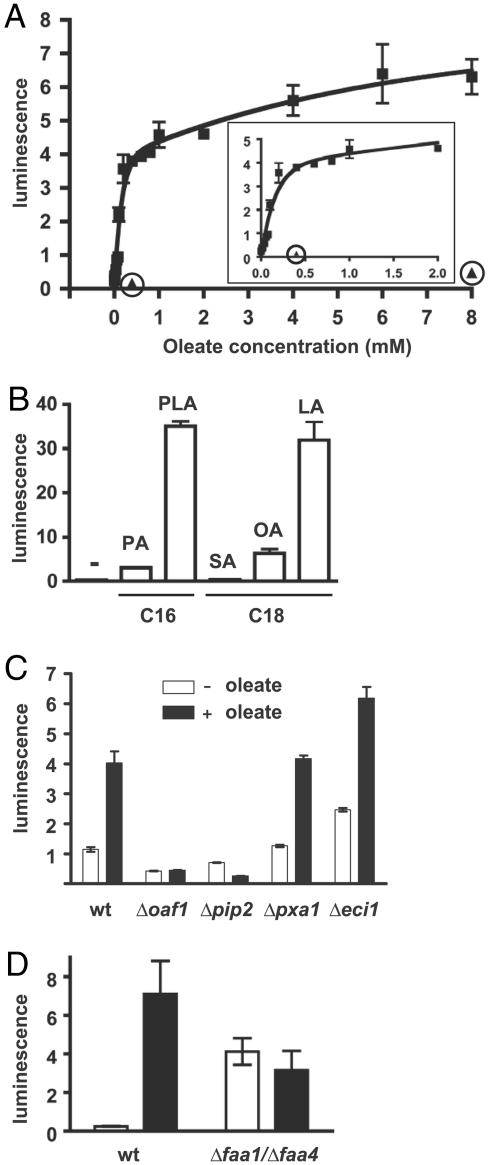
To test this hypothesis, we began by further characterizing the in vivo response to fatty acids. To facilitate our analysis, we constructed a luciferase reporter gene whose expression is under the control of the oleate response element from the FOX3 gene (23) upstream of a minimal promoter. Moreover, we performed these experiments under conditions in which the Oaf1/Pip2-dependent activating effects of fatty acids could be separated from their nutritional contribution as a carbon source. Hence, cells were cultured in the presence of a mixture of carbon sources (raffinose, glycerol, and ethanol) that ensure cell growth irrespective of the addition of fatty acids and that, unlike glucose, do not elicit catabolite repression. As expected, the Δ9-unsaturated C18 fatty acid oleate induces the response robustly (Fig. 2A). Half-maximal induction (EC50) is achieved with ≈0.24 mM oleate. A survey of a panel of various saturated and unsaturated fatty acids revealed that Δ9-unsaturated fatty acids are stronger activators than saturated ones (Fig. 2B). Linoleate, a double-unsaturated C18 fatty acid, produces an even stronger transcriptional response than oleate, whereas the saturated C18 fatty acid stearate produces no response at all. The shorter saturated fatty acid palmitate is able to elicit a weak response. Taken together with the previously reported observation that the saturated C12 fatty acid laurate activates Oaf1/Pip2 target genes (24), these data suggest that short and/or compact fatty acids may be preferred as activators. Despite the strong activating potential of several different fatty acids, oleate was used in all subsequent experiments because it is one of the most abundant physiological fatty acids in yeast membranes (25), and because it has traditionally been used to induce the responses to unsaturated fatty acids, including utilization as a carbon source and peroxisome proliferation (26).
Fig. 2.
Fatty acid response profile of Oaf1/Pip2 reporter gene. (A) A dose–response experiment with oleate. Activities of the luciferase reporter gene in the wild-type strain are indicated in normalized arbitrary luciferase units. (Inset) Enlarged portion of the graph for concentrations up to 2 mM. The circled triangles represent the activities obtained with the isogenic Δoaf1 strain. (B) Responses to a panel of fatty acids (added to 6 mM). PA, palmitic acid; PLA, palmitoleic acid; SA, stearic acid; OA, oleic acid (oleate); LA, linoleic acid. (C and D) Oleate responses of different mutant strains; strain backgrounds, and hence the two corresponding wild-type strains, are different for the mutants used in the two panels. wt, wild type.
Oleate Itself Induces Oaf1/Pip2 Activity.
Next, we determined whether oleate first needs to be metabolized to activate this pathway. An earlier study (24) had suggested that intact peroxisomes may not be needed for the response, but the requirement for the metabolism of unsaturated fatty acids had not been examined directly. Therefore, the response of the reporter gene was tested in a panel of mutant strains that are all defective in oleate utilization but are comparably competent for the induction of a galactose-inducible luciferase reporter gene (data not shown). As expected, the oleate response depends on both OAF1 and PIP2. In contrast, induction still works in Δpxa1 and Δeci1 strains that are defective in two key steps for β-oxidative metabolism of fatty acids: peroxisomal import and enoyl isomerization (Fig. 2C). The Δfaa1/Δfaa4 double-mutant strain is defective in fatty acid uptake and activation to CoA derivatives, the first enzymatic step toward metabolism, because it lacks the two main acyl-CoA synthetases for C12–C18 fatty acids (27, 28). It is conceivable that the levels of endogenously synthesized oleate in these mutant cells are elevated because they are trying to cope with their inability to take up fatty acids. Indeed, we found that there is no further induction by added oleate beyond a dramatically elevated basal activity. This finding also suggests that acyl-CoA synthetases may contribute to keeping the basal activity of Oaf1/Pip2 low by controlling the concentration of unconjugated oleate. At this point, the oleate response profile was compatible with the idea that oleate acts directly as a ligand and activator of the Oaf1/Pip2 heterodimer.
Oleate Is a Ligand of the Oaf1 LBD.
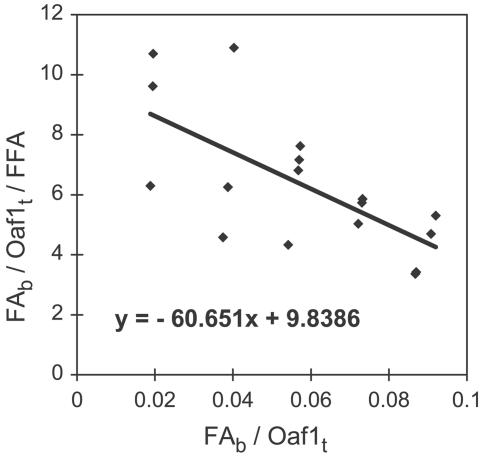
Direct binding of oleate to the hypothesized LBDs was then assessed in vitro by a ligand-binding assay with purified recombinant Oaf1 and Pip2 LBDs (Fig. 3 and Table 1). The Oaf1 LBD binds oleate with a Kd of ≈17 nM, whereas it does not bind the noninducing saturated fatty acid stearate. Moreover, the corresponding Pip2 domain fails to bind oleate. The much higher affinity of the Oaf1 LBD compared with the EC50 of the in vivo response may indicate that the oleate concentration that is freely available to Oaf1 inside cells is much lower than what is added to the medium.
Fig. 3.
Oaf1 binds oleate. Presented are results of a ligand-binding assay with recombinant Oaf1 LBD protein. For the Scatchard analysis, the ratio of bound fatty acid (FAb) to total Oaf1 protein (Oaf1t) to free fatty acid (FFA) (y axis) is plotted against the ratio of FAb to Oaf1t. The parameters of the fitted curve are shown inside the graph area.
Table 1.
Summary of the ligand-binding assays
| LBD | Fatty acid | Slope of fitted curve | Affinity,* nM |
|---|---|---|---|
| Oaf1 | Oleate | −60.651 | 16.5 ± 4.5 |
| Stearate | 10.046 | ND | |
| Pip2 | Oleate | 10.777 | ND |
| Oaf1 E543A/A544S | Oleate | −10.102 | ND (99 ± 150) |
*Affinities (Kds) with SEs were calculated from the slope of the fitted curves of Scatchard analyses, such as the analysis shown in Fig. 3. ND, not defined, indicates no binding (positive slopes or large SE).
Further Genetic Validation of the Structural Prediction.
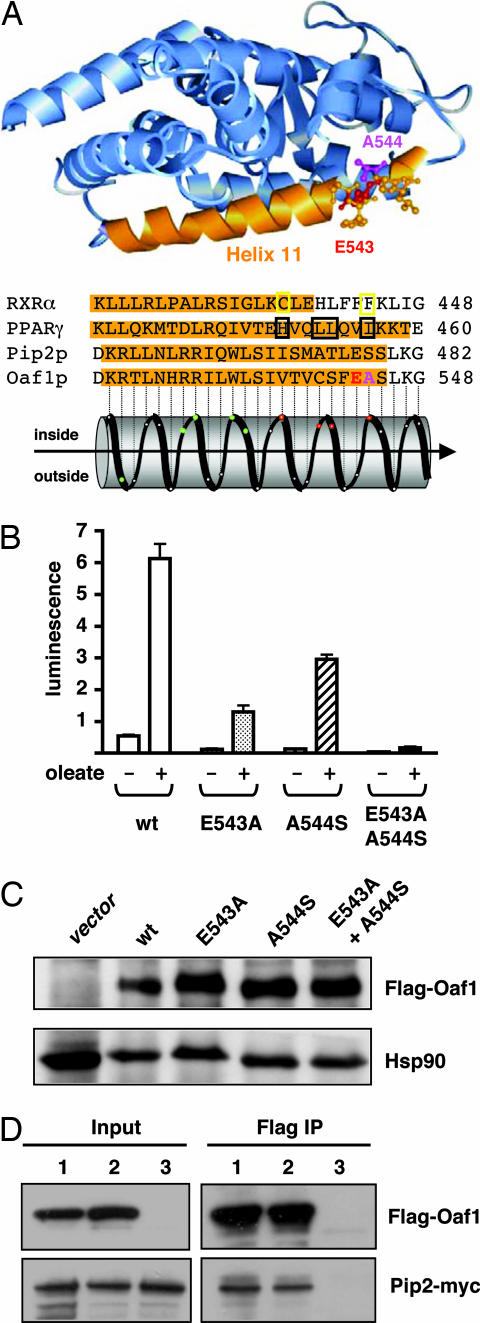
Our results strongly suggested that oleate might stimulate Oaf1 activity by acting as a ligand of the hypothesized LBD. We therefore sought further genetic evidence for the validity of the structural prediction. In animal NRs, helix 11 of the LBD forms the bottom of the ligand-binding pocket (1, 2). Because the topology of the residues making up the putative helix 11 of Oaf1 could not be definitively modeled due to the distant nature of the structural relationship, we mutated all residues along the relevant portion of the predicted helix 11 from V537 to A544 (Fig. 4A). The single-point mutants E543A and A544S are the only ones that display a diminished response to oleate (Fig. 4B and data not shown). Their induced activities are reduced to 25–50% of wild type. Their basal activities are reduced as well, consistent with the observation that the presence of wild-type Oaf1 (and Pip2) results in appreciable basal activity of the reporter gene, which may be due to endogenously produced, albeit low levels of, oleate. In the absence of any exogenously added oleate, the point mutants E543A and A544S might be too insensitive to respond at all. Remarkably, the response of the double mutant E543A/A544S, which combines both of the partially defective amino acid replacements, is completely defective (Fig. 4B). As shown in Fig. 3B, the ability of the double mutant E543A/A544S to bind oleate in vitro is correspondingly reduced. Thus, changing two amino acids in the predicted helix 11 at the bottom of the LBD abolishes both ligand binding and transcriptional response. These mutants still accumulate to normal levels, and even the defective double mutant is still able to heterodimerize with Pip2 (Fig. 4C and D), indicating structural integrity. This finding is consistent with the fact that Oaf1/Pip2 heterodimers can form and bind DNA in the absence of oleate (17–19).
Fig. 4.
Alteration of two amino acids within the predicted ligand-binding pocket abolishes the oleate response of Oaf1. (A) Schematic representation of the Oaf1 LBD, with magnification of a region of helix 11 that was probed by mutagenesis. The Oaf1 sequence is threaded onto the PPARγ structure (Protein Data Bank ID code 1PRG). The ribbon representation of helix 11 is broken up around E543 (red) and A544 (purple) to reveal more detail. The C and F boxed in yellow are oleate-binding residues in the RXRα. The residues boxed in black in the PPARγ sequence (and marked with a red dot in the helical representation) represent ligand-binding residues of both PPARγ itself and other NRs. (B) Oleate responses of Oaf1 point mutants. Both wild type and mutants were expressed with a FLAG epitope in a Δoaf1 strain. (C) Expression of wild-type and mutant Oaf1 variants. Total extracts of the same strains analyzed in B were immunoblotted with antibodies against the FLAG epitope (Upper) and Hsp90 as an unrelated control protein (Lower). (D) The Oaf1 double mutant still heterodimerizes with Pip2. Association of FLAG-Oaf1 with Pip2-Myc was assessed by a coimmunoprecipitation experiment with an antibody directed against the FLAG tag. Immunoprecipitated proteins were revealed by immunblotting with antibodies against the FLAG (Upper) and Myc (Lower) epitopes. Pip2-Myc was present in all three extracts, whereas wild-type and double-mutant Oaf1 and only the FLAG epitope were present for the samples of lanes 1, 2, and 3, respectively.
Discussion
We have incorporated structural predictions in a bioinformatic screen of all available genome sequences for potential homologs of a particular protein domain. Having found candidate homologs for the LBD of NRs, we then identified a ligand and demonstrated that this ligand binds the predicted LBD in vitro and that predicted key residues are important for ligand binding in vitro and for function in vivo.
The correlation between in vitro ligand binding and the in vivo response strongly supports the conclusion that the S. cerevisiae protein Oaf1 contains a functional domain hitherto thought to be restricted to animals (3, 5). By analogy, it is very likely that the putative LBD of Pip2 is a genuine LBD as well. Its LBD may either bind oleate with much lower affinity or bind an as yet unidentified ligand supporting oleate induction of the Oaf1-Pip2 heterodimer, or it may play a structural role within Pip2 itself or within the heterodimer. Note that only a minority of animal NRs have known ligands and that some NRs cannot bind any ligand at all because their “ligand-binding” pocket is filled with bulky amino acid side chains (29). The structural, molecular, and physiological analogies between the yeast Oaf1/Pip2 and the vertebrate PPAR/RXR heterodimers (30) are striking. In both groups of organisms, these heterodimeric transcription factors bind unsaturated fatty acids such as oleate and activate the metabolic pathway for fatty acids in response to a nutritional input of fatty acids. Surprisingly, their corresponding acyl-CoA esters behave as antagonists of PPAR/RXR, and acyl-CoA synthetase inhibitors result in increased PPAR/RXR activity in mammalian cells (31, 32), which is highly reminiscent of the effects of deleting the FAA1/FAA4 genes on Oaf1/Pip2 activity in yeast. Although we have not determined whether the acyl-CoA esters are able to bind Oaf1/Pip2 as well, it is possible that they contribute directly to lowering the basal activity as antagonists, in the absence of exogenously added fatty acids such as oleate. Thus, although the LBDs are embedded within transcription factors of a different architecture (Fig. 1A), they likely provide a common ancestral function as an allosteric switch.
The mechanistic details of how LBDs regulate the activities of the proteins within which they reside may have diverged considerably. Indeed, although many animal NRs (3) carry within their LBD a ligand-induced transcriptional activation function, referred to as AF-2, the activation domains of Oaf1 and Pip2 appear to reside exclusively at the very C terminus of the proteins (ref. 22 and data not shown) (Fig. 1A). It is conceivable that allosteric regulation by oleate relieves an inhibitory mechanism that blocks some step after DNA binding of the heterodimer by intra- and/or intermolecular interactions. The presence of auxiliary and inhibitory domains has previously been proposed on the basis of mutagenesis (22) and sequence (33) analyses. These domains partially overlap with the LBDs that we have defined here as a distinct structural and functional unit. Taken together, these findings point to an intricate intramolecular interplay between multiple domains.
The same technology that enabled us to discover the Oaf1 and Pip2 LBDs by predicting a similar structural fold, despite poor primary sequence conservation, produced no significant hits beyond animals and fungi. Highly conserved orthologs are present in several other fungi of the phylum ascomycetes, notably of the order Saccharomycetales (budding yeasts), including in the genera Candida, Eremothecium, and Saccharomyces, and possibly also in Aspergillus, an ascomycete that is in an entirely different class (data not shown). It is noteworthy that, unlike plants, fungi and animals both belong to the opisthokonts, one of the eight groups of eukaryotes (34). In light of the structural and functional similarities, common ancestry between animal and yeast LBDs is the most parsimonious hypothesis. However, more genome sequences, notably of organisms filling the gap between animals and fungi, as well as enhanced versions of our bioinformatic tools, will be necessary to exclude convergent evolution and to determine whether LBDs are restricted to opisthokonts, when they appeared, and perhaps, in some species, when they disappeared again.
Materials and Methods
Plasmids.
Full-length OAF1 and PIP2 coding sequences were cloned from yeast strain W303. Our Oaf1 sequence differs in three amino acids (R70, Q447, and K588) from that deposited in the Saccharomyces Genome Database (www.yeastgenome.org). The latter sequence, derived from yeast strain S288C, may be deviant because our “variations” are conserved in several other Saccharomyces species. Both wild-type and point-mutant Oaf1 and Pip2, tagged with FLAG and 13 copies of the c-Myc epitope, respectively, were expressed from the TDH3 promoter in CEN/ARS plasmids. For expression in bacteria, sequences encoding the Oaf1 (amino acids 244–573) and Pip2 (amino acids 180–507) LBDs were cloned into vector pET-32Ek/LIC (Novagen). The luciferase reporter plasmid p2UG-2XORE-Luc was built from plasmid p2UG (35); it is a 2μ-based episome containing two tandem copies of the oleate response elements from the FOX3 gene inserted upstream of a minimal CYC1 promoter driving the expression of firefly luciferase.
Yeast Experiments.
Strain BY4741 was used as the wild-type strain and as background for all yeast deletion strains except for the Δfaa1 Δfaa4 strain YB525 (27). For luciferase assays and the preparation of protein extracts, transformants were grown in rich medium with 1.5% raffinose, 1.5% glycerol, and 1% ethanol as carbon sources. Induction was done with 6 mM oleate overnight, unless indicated otherwise. Luciferase activities were determined in triplicate, essentially as described (36), except that cells were first washed once with water. Luciferase activities were normalized to the optical densities of the cultures.
Ligand-Binding Assays.
Recombinant LBDs were purified from Escherichia coli as thioredoxin fusion proteins with His6 tags by affinity chromatography. Ligand-binding assays were done with oleate and stearate by using the ADIFAB free fatty acid indicator (Molecular Probes) as a fluorescent probe, as described (37). Briefly, the fluorescence of the modified fatty acid binding protein ADIFAB changes upon binding fatty acids. From this change, the free fatty acid concentration in the test solution at binding equilibrium can be calculated, as can, indirectly, the concentration of the fatty acid bound for a known total amount of the test LBD (for example, Oaf1t). Data from three to four replicates were analyzed by the Scatchard plot method.
Acknowledgments
We thank Natasha Kralli for having brought together the partners of this study. We thank Gisou van der Goot for help with the fluorometer, Jeffrey I. Gordon for yeast strains, Katharina Strub for critical comments on the manuscript, Matt Couch for help with the bioinformatics analysis, and Richard Kidd for homology modeling. We are indebted to Pierre-André Briand for technical assistance. Work in the laboratory of D.P. was supported by the Canton de Genève and the Swiss National Science Foundation.
Abbreviations
- NR
nuclear receptor
- LBD
ligand-binding domain
- PPAR
peroxisome proliferator-activated receptor
- RXR
retinoid X receptor
- DBD
DNA-binding domain
- ROR
retinoic acid-related orphan receptor.
Footnotes
Conflict of interest statement: C.P. and R.J.F. contributed to this work as employees of Inpharmatica Ltd.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nagy L., Schwabe J. W. Trends Biochem. Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Renaud J. P., Moras D. Cell. Mol. Life Sci. 2000;57:1748–1769. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson-Rechavi M., Escriva Garcia H., Laudet V. J. Cell Sci. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- 4.Thornton J. W., Need E., Crews D. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 5.Escriva H., Delaunay F., Laudet V. BioEssays. 2000;22:717–727. doi: 10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 6.Wiens M., Batel R., Korzhev M., Muller W. E. J. Exp. Biol. 2003;206:3261–3271. doi: 10.1242/jeb.00541. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S. F., Gish W. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 8.McGinnis S., Madden T. L. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones D. T. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 10.Swindells M., Rae M., Pearce M., Moodie S., Miller R., Leach P. Philos. Trans. R. Soc. London A. 2002;360:1179–1189. doi: 10.1098/rsta.2002.0987. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu L., White J. V., Smith T. F. Protein Sci. 1998;7:2499–2510. doi: 10.1002/pro.5560071203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chmiel A. A., Radlinska M., Pawlak S. D., Krowarsch D., Bujnicki J. M., Skowronek K. J. Protein Eng. Des. Sel. 2005;18:181–189. doi: 10.1093/protein/gzi019. [DOI] [PubMed] [Google Scholar]
- 14.Fagan R., Swindells M., Overington J., Weir M. Trends Biochem. Sci. 2001;26:213–214. doi: 10.1016/s0968-0004(01)01789-3. [DOI] [PubMed] [Google Scholar]
- 15.Jones D. T. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 16.Barnett P., Tabak H. F., Hettema E. H. Trends Biochem. Sci. 2000;25:227–228. doi: 10.1016/s0968-0004(00)01579-6. [DOI] [PubMed] [Google Scholar]
- 17.Rottensteiner H., Kal A. J., Filipits M., Binder M., Hamilton B., Tabak H. F., Ruis H. EMBO J. 1996;15:2924–2934. [PMC free article] [PubMed] [Google Scholar]
- 18.Rottensteiner H., Kal A. J., Hamilton B., Ruis H., Tabak H. F. Eur. J. Biochem. 1997;247:776–783. doi: 10.1111/j.1432-1033.1997.00776.x. [DOI] [PubMed] [Google Scholar]
- 19.Karpichev I. V., Luo Y., Marians R. C., Small G. M. Mol. Cell. Biol. 1997;17:69–80. doi: 10.1128/mcb.17.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpichev I. V., Small G. M. Mol. Cell. Biol. 1998;18:6560–6570. doi: 10.1128/mcb.18.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith J. J., Marelli M., Christmas R. H., Vizeacoumar F. J., Dilworth D. J., Ideker T., Galitski T., Dimitrov K., Rachubinski R. A., Aitchison J. D. J. Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgartner U., Hamilton B., Piskacek M., Ruis H., Rottensteiner H. J. Biol. Chem. 1999;274:22208–22216. doi: 10.1074/jbc.274.32.22208. [DOI] [PubMed] [Google Scholar]
- 23.Einerhand A. W., Voorn-Brouwer T. M., Erdmann R., Kunau W. H., Tabak H. F. Eur. J. Biochem. 1991;200:113–122. doi: 10.1111/j.1432-1033.1991.tb21056.x. [DOI] [PubMed] [Google Scholar]
- 24.Rottensteiner H., Palmieri L., Hartig A., Hamilton B., Ruis H., Erdmann R., Gurvitz A. Biochem. J. 2002;365:109–117. doi: 10.1042/BJ20011495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sajbidor J., Certik M., Grego J. J. Chromatogr. A. 1994;665:191–195. [Google Scholar]
- 26.Erdmann R., Veenhuis M., Mertens D., Kunau W. H. Proc. Natl. Acad. Sci. USA. 1989;86:5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson D. R., Knoll L. J., Levin D. E., Gordon J. I. J. Cell Biol. 1994;127:751–762. doi: 10.1083/jcb.127.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faergeman N. J., Black P. N., Zhao X. D., Knudsen J., DiRusso C. C. J. Biol. Chem. 2001;276:37051–37059. doi: 10.1074/jbc.M100884200. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Benoit G., Liu J., Prasad S., Aarnisalo P., Liu X., Xu H., Walker N. P., Perlmann T. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 30.Desvergne B., Michalik L., Wahli W. Mol. Endocrinol. 2004;18:1321–1332. doi: 10.1210/me.2004-0088. [DOI] [PubMed] [Google Scholar]
- 31.Forman B. M., Chen J., Evans R. M. Proc. Natl. Acad. Sci. USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helledie T., Jorgensen C., Antonius M., Krogsdam A. M., Kratchmarova I., Kristiansen K., Mandrup S. Mol. Cell. Biochem. 2002;239:157–164. [PubMed] [Google Scholar]
- 33.Poch O. Gene. 1997;184:229–235. doi: 10.1016/s0378-1119(96)00602-6. [DOI] [PubMed] [Google Scholar]
- 34.Baldauf S. L. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 35.Picard D., Schena M., Yamamoto K. R. Gene. 1990;86:257–261. doi: 10.1016/0378-1119(90)90287-2. [DOI] [PubMed] [Google Scholar]
- 36.Leskinen P., Virta M., Karp M. Yeast. 2003;20:1109–1113. doi: 10.1002/yea.1024. [DOI] [PubMed] [Google Scholar]
- 37.Richieri G. V., Ogata R. T., Kleinfeld A. M. Mol. Cell. Biochem. 1999;192:87–94. [PubMed] [Google Scholar]