Abstract
The PglB oligosaccharyltransferase (OTase) of Campylobacter jejuni can be functionally expressed in Escherichia coli, and its relaxed oligosaccharide substrate specificity allows the transfer of different glycans from the lipid carrier undecaprenyl pyrophosphate to an acceptor protein. To investigate the substrate specificity of PglB, we tested the transfer of a set of lipid-linked polysaccharides in E. coli and Salmonella enterica serovar Typhimurium. A hexose linked to the C-6 of the monosaccharide at the reducing end did not inhibit the transfer of the O antigen to the acceptor protein. However, PglB required an acetamido group at the C-2. A model for the mechanism of PglB involving this functional group was proposed. Previous experiments have shown that eukaryotic OTases have the same requirement, suggesting that eukaryotic and prokaryotic OTases catalyze the transfer of oligosaccharides by a conserved mechanism. Moreover, we demonstrated the functional transfer of the C. jejuni glycosylation system into S. enterica. The elucidation of the mechanism of action and the substrate specificity of PglB represents the foundation for engineering glycoproteins that will have an impact on biotechnology.
Keywords: glycoengineering, glycoproteins, LPS, PglB, Stt3p
Campylobacter jejuni possesses a general N-linked glycosylation system responsible for the glycosylation of a range of proteins (1–3). In this process, the oligosaccharide is assembled on the lipid carrier undecaprenyl-pyrophosphate (Und-PP) at the cytoplasmic side of the inner membrane and translocated to the periplasm by the ABC transporter homologue PglK (4–6). Next, the oligosaccharyltransferase (OTase) PglB transfers the oligosaccharide from the lipid carrier to the acceptor proteins. Bacterial and eukaryotic N-linked glycosylation pathways are homologous processes. In eukaryotes, the oligosaccharide is assembled on the lipid carrier dolichyl pyrophosphate at the membrane of the endoplasmic reticulum (7). The preassembled oligosaccharide is then transferred to selected Asn residues within the sequence Asn-X-Ser/Thr of nascent polypeptide chains (8). This process is catalyzed by the OTase complex (9). Stt3p is the most conserved protein in this complex and is homologous to PglB (7, 10).
The generation of the lipid-linked oligosaccharide substrate for N-glycosylation has significant similarities to the O antigen biosynthesis pathway in Gram-negative bacteria (11, 12). O antigen is the outer component of the LPS (13). At least two different mechanisms for biosynthesis and assembly of O antigen have been described. One of them, referred as the “Wzy-dependent mechanism,” involves the synthesis of repeating subunits on the lipid carrier Und-PP at the cytoplasmic side of the inner membrane. Once completed, O antigen subunits are flipped across the cytoplasmic membrane, polymerized by the Wzy polymerase in the periplasmic space, and transferred to the lipid A core by the WaaL ligase (13). The alternative “ABC transporter-dependent” pathway involves the formation of a polymeric O antigen by reactions occurring at the cytosolic face of the cytoplasmic membrane (13). The nascent polysaccharide chain is transported across the inner membrane by an ATP-binding cassette transporter and subsequently ligated to the lipid A core (14). In Escherichia coli, the WecA UDP-GlcNAc:Und-P GlcNAc-1-P transferase can initiate either assembly pathway (15, 16).
The C. jejuni N-glycosylation machinery can be functionally transplanted to E. coli (17). PglB expressed in a waaL mutant strain of E. coli can efficiently accept diverse Und-PP-linked glycans as substrates (12). To further study the specificity of PglB, we have chosen to work in E. coli and Salmonella enterica serovar Typhimurium. E. coli E69 cells express the O9a antigen with a high mannose structure as well as the capsular K30 antigen at its surface. The O9a polymannan is linked to the lipid A core, whereas the K30 antigen forms the capsular polysaccharide. The O9a antigen is synthesized by the ABC transporter-dependent mechanism (18–20). The K30 capsular polysaccharide is the established prototype for group 1 capsules (21). The early stages in assembly are similar to the Wzy-dependent mechanism in the O antigen biosynthesis, and it starts with the transfer of Gal-1-P to the Und-P carrier lipid, catalyzed by the WecA homolog WbaP. Then, mannose, galactose (Gal), and glucuronic acid (GlcA) residues are added by different glycosyltransferases, and the lipid-linked oligosaccharide is transferred into the periplasm by Wzx (22), polymerized, and subsequently translocated to the surface (23–25). A small fraction of the repeating unit is also transferred to the lipid A core, forming the KLPS (26). Interestingly, WbaP also catalyzes the reversible transfer of Galp-1-phosphate from the precursor UDP-Galp to Und-P as the initial synthetic reaction for the S. enterica O antigen.
In this work, we show that PglB transferred a polysaccharide that is assembled by a Wzy-independent pathway. We have found that an N-acetyl group in position 2 of the sugar directly linked to the Und-PP-carrier was necessary for recognition and/or catalysis. Based on the experimental data, we present a model for OTase catalysis involving this acetamido group.
Results
A Branching Glc at the Reducing End of the Oligosaccharide Substrate Did Not Prevent Transfer by PglB.
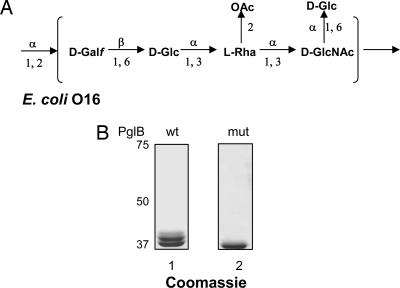
PglB has relaxed specificity toward its lipid-linked glycan substrate. It can transfer its endogenous substrate, the heptasaccharide Glc(GalNAc)5Bac of C. jejuni, as well as different O antigen polysaccharides that were assembled by the Wzy (polymerase)-dependent mechanism on the lipid carrier Und-PP (12). The O16 polysaccharide from the E. coli K-12 strain W3110 was among the O antigens transferred by PglB. Strain W3110 does not normally produce the O16 polysaccharide due to an IS5 insertion interrupting the function of the wbbL gene (27). This gene encodes a rhamnosyltransferase required for the addition of rhamnose to the second position of the O16 repeating unit (Fig. 1A). O16 biosynthesis can be restored by complementation in trans with pMF19, a plasmid encoding a functional rhamnosyltransferase gene (28). O16 also has a branched Glc residue in the C-6 position of the initiating sugar GlcNAc (Fig. 1A). The incorporation of this Glc residue in the repeating unit appears to be a nonstoichiometric modification, and the glucosyltransferase responsible for this modification is encoded outside the O16 operon (29, 30).
Fig. 1.
Transfer of a single O16 antigen subunit to AcrA. (A) Structure of the repetitive O16 subunit. Galf, Galactofuranose; Rha, rhamnose. (B) AcrA was purified from EVV11 (Δwzy) cells carrying the rhamnosyltransferase gene wbbL and expressing PglB (lane 1) or PglBmut (lane 2), separated by SDS/PAGE, and visualized by Coomassie blue staining.
To further study the specificity of PglB, we wanted to check whether the C-6 substitution of the initiating sugar GlcNAc prevented the transfer of the O16 glycan to the acceptor protein AcrA. Thus, we investigated whether the oligosaccharide bound to AcrA had a Glc residue. For this purpose, the rhamnosyltransferase (WbbL) was expressed in the W3110 derivative EVV11. Because this strain contains a deletion of the polymerase gene wzy, only one repeating unit of the O16 antigen was synthesized on the Und-PP carrier. After induction of PglB, the AcrA products were purified and analyzed. Mono- and di-glycosylated AcrA forms were observed in the presence of PglB, whereas only unglycosylated AcrA was found in the presence of PglBmut, an inactive version of PglB (17) (Fig. 1B). Mono- and di-glycosylated AcrA forms were digested with trypsin, and the product mixture was analyzed by MALDI-TOF/TOF MS and tandem MS to determine the structure of the short oligosaccharide. These analyses confirmed that a branching Hexose was present on the peptide-proximal N-acetylhexoseamine (HexNAc) residue of this oligosaccharide (see Fig. 5 and Supporting Text, which are published as supporting information on the PNAS web site). Based on the known structure and genetic data, the only candidate for this hexose residue was the branching Glc at the C-6 position of the reducing GlcNAc residue of the O16 antigen repeating unit. Thus, we concluded that a substitution at the C-6 position in the reducing end of the oligosaccharide did not prevent its PglB-mediated transfer to the protein acceptor AcrA.
PglB Transfers the O9a Antigen to AcrA.
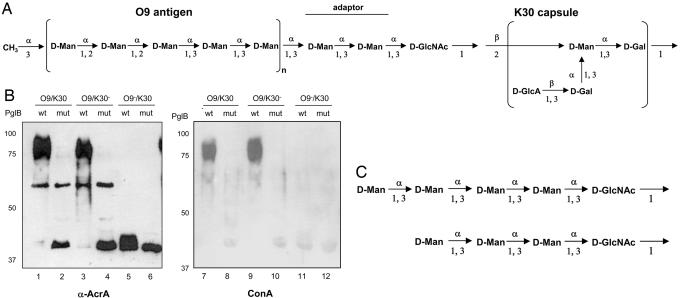
To further study the substrate requirements of PglB, we tested whether PglB transferred the O9a antigen and/or the K30 capsular antigen in E. coli E69. In this strain, both the O9a antigen (an ABC transporter-dependent O antigen) and the K30 capsule (a capsule synthesized by a Wzy-dependent mechanism) are assembled onto Und-PP-glycan intermediates offering PglB OTase two potential substrates (18, 22). The structures of the oligosaccharides are given in Fig. 2A.
Fig. 2.
Glycosylation of AcrA in E. coli E69 (O9a/K30) and its derivatives CWG28 (O9a/K30−) and CWG44 (O9a−/K30). (A) Structure of different polysaccharides in E. coli E69 cells. The O9a polymannan is synthesized by the ABC transporter-dependent mechanism and is linked to the lipid A core, whereas the K30 polysaccharide is the established prototype for group 1 capsules. GlcA, glucuronic acid; Man, mannose. (B) Proteins derived from periplasmic extracts (Left) as well as Ni2+ affinity-purified proteins from the same extracts (Right) derived from E. coli cells expressing AcrA and PglB or PglBmut were separated by SDS/PAGE, transferred to nitrocellulose membranes, and detected with antibodies directed against either AcrA (lanes 1–6) or Con A lectin (lanes 7–12). (C) Structure of the oligosaccharides detected by tandem MS linked to AcrA in strain CWG44 (O9a−/K30). For details on the MS results, see Supporting Text.
PglB was expressed in E. coli E69 by using an arabinose-inducible promoter, and expression of soluble AcrA was controlled by the constitutive tetracycline promoter (12). As a control, we used cells expressing PglBmut (17). After induction of PglB expression, periplasmic extracts were prepared by lysozyme treatment and proteins separated by SDS/PAGE. After transfer to nitrocellulose, AcrA was visualized by immunodetection with anti-AcrA antiserum (Fig. 2B, lanes 1 and 2). Nonglycosylated AcrA (40 kDa) was the only immunoreactive protein present when PglBmut was expressed (lane 2). In contrast, a ladder of bands corresponding to higher-molecular weight proteins, centered at 80 kDa, was detected with anti-AcrA antibodies in the presence of functional PglB (Fig. 2B, lane 1). Based on the following results, these bands represented glycosylated forms of AcrA. In strain CWG28, an E69 derivative that does not produce the K30 capsule (31), the same ladder profile was detected in the presence of a functional PglB (Fig. 2B, lanes 3 and 4). But in CWG44, a strain that does not produce the O9a antigen due to an uncharacterized mutation (32), nonglycosylated and shorter glycosylated AcrA forms were observed in the presence of functional PglB (Fig. 2B, lanes 5 and 6). To prove that protein-bound polysaccharide originated from the O9a antigen pathway, Ni2+ affinity-purified AcrA proteins derived from periplasmic extracts of E. coli E69, CWG28 and CWG44 cells were incubated with Con A, a lectin that recognizes high mannose structures (Fig. 2B, lanes 7–12). The high-molecular weight proteins that were observed in the presence of functional PglB reacted with the lectin (Fig. 2B, lanes 7 and 9) when proteins were extracted from E69 and CWG28 cells. As expected, there was no reactivity with purified proteins extracted from O9a-deficient CWG44 cells (Fig. 2B, lanes 11 and 12). Similar results as with the Con A lectin were obtained with the serotype-specific O9a antiserum (data not shown).
The CWG44 strain does not produce O9a LPS, but it produces the K30 capsule, as confirmed by immunoreactivity with the K30-specific antiserum (data not shown). We detected modifications of AcrA in the presence of functional PglB (Fig. 2B, lane 5). These modified proteins showed a slightly slower mobility than unglycosylated AcrA but did not react with the capsule specific antiserum (data not shown). We did not observe a glycosylation of AcrA in the higher-molecular range in CWG44 (Fig. 2B, lanes 5 and 6). We concluded that AcrA was not glycosylated with the K30 capsule. However, AcrA was modified with short-chain oligosaccharides distinct from K30 in the CWG44 strain.
Glycopeptide Analysis Shows That the K30 Antigen Is Not Transferred to AcrA.
To identify the nature of the AcrA modifications in CWG44 cells, we carried out MS analysis of peptides resulting from trypsin digestion of purified protein. After induction of PglB, periplasmic AcrA was extracted, purified by affinity chromatography by using a NTA-agarose column, separated by SDS/PAGE, and Coomassie-stained. Glycosylated AcrA was digested with trypsin, and the product mixture was analyzed by MALDI-TOF/TOF MS. This analysis showed that AcrA indeed was glycosylated in the CWG44 strain (for details, see Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site). The oligosaccharides covalently linked to protein were most likely incompletely assembled O9a antigen, because they consist of HexNAc and additional Hex residues. Based on the results from tandem MS analysis and the known structure of the O9a antigen, the oligosaccharides linked to AcrA are depicted in Fig. 2C. Importantly, no transfer of K30 capsule to AcrA was detected in CWG44 or in a mutant that accumulated Und-PP-bound K30 capsule in the periplasm (data not shown).
The S. enterica O Antigen Containing Gal at the Reducing End Is Not a Substrate for PglB.
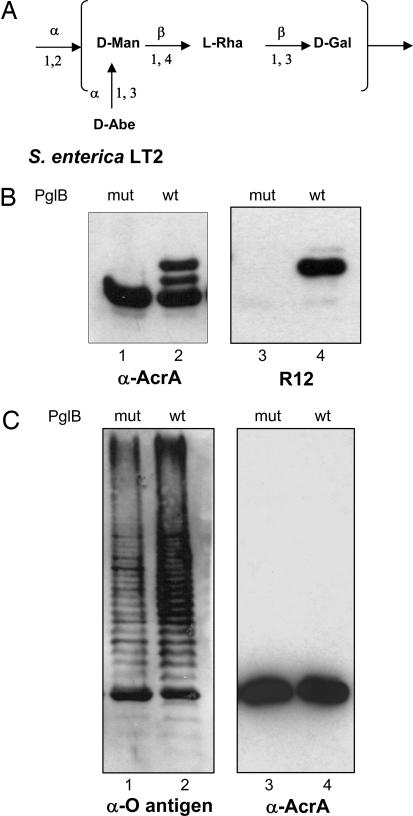
We have shown previously that oligo- or polysaccharides containing bacillosamine (Bac), GlcNAc, GalNAc, or fucosamine (FucNAc) at the reducing end (Table 1) were transferred from the Und-PP carrier to proteins by PglB (12). The capsular K30 antigen was the first of all of the tested polysaccharides that was not a substrate for PglB. In contrast to the other polysaccharides transferred, K30 antigen contains a Gal residue at the reducing end. We hypothesized that the acetamido group at the C-2 position at the reducing-end sugar plays a crucial role in recognition and/or catalysis by PglB. To test our hypothesis and to confirm that the lack of transfer of capsule was not simply due to the different features in the assembly of the polysaccharide in the capsular biosynthesis, we tested whether PglB transferred the O polysaccharide from S. enterica LT2, which also contains a Gal residue as initiating sugar (Fig. 3A). We used the strain SL3749 (33) that lacks the WaaL O antigen ligase to avoid competition between WaaL and PglB for the same lipid-linked oligosaccharide. We first verified whether the C. jejuni N-glycosylation machinery was active in Salmonella cells. Periplasmic extracts of S. enterica SL3749 expressing soluble AcrA and the C. jejuni pgl locus were prepared and visualized by immunodetection. Although only a single band corresponding to unglycosylated AcrA was detected with anti-AcrA antibodies in the presence of PglBmut, three bands were visualized in the presence of functional PglB (Fig. 3B, lanes 1 and 2). The upper band also reacted with the R12 antiserum (Fig. 3B, lane 4). This antiserum recognizes primarily the glycosylated form of AcrA (17). These results were equivalent to those previously obtained under similar experimental conditions in E. coli (17). We concluded that the C. jejuni N-glycosylation machinery can be functionally expressed in Salmonella. We then investigated the ability of PglB to mediate the transfer of the S. enterica LT2 O antigen onto AcrA. SL3749 cells expressing AcrA, together with either functional PglB or PglBmut, were cultured in the presence of arabinose. Periplasmic AcrA was extracted and purified by affinity chromatography by using a NTA-agarose column. Immunodetection of the purified fractions using anti-AcrA antibodies showed only unglycosylated AcrA (Fig. 3C, lanes 3 and 4). Bands that could correspond to AcrA glycosylated with O polysaccharide were absent when an antiserum directed toward the LT2 O polysaccharide was used (data not shown). However, the O polysaccharide-specific antiserum detected a ladder of bands in whole-cell extracts from the strains expressing either PglB or PglBmut (Fig. 3C, lanes 1 and 2). Because the SL3749 strain is devoid of the O polysaccharide ligase, precluding the formation of long-chain O polysaccharide-substituted LPS (33), we assigned these bands to Und-PP-linked O antigen. Our results indicated that the absence of protein glycosylation was not due to lack of production of the potential PglB substrate, but the S. enterica O antigen containing Gal as initiating sugar was not a substrate for PglB.
Table 1.
Different oligo-/polysaccharides transferred by PglB
| Structure | Bacteria | PglB substrate | Hexose at the reducing end | Substitutions of the hexose at the reducing end | Reference (glycan structure, PglB substrate) |
|---|---|---|---|---|---|
| Oligosaccharide | C. jejuni | Yes | Bac | GalNAc (α1→3) | 3, 17 |
| O7 antigen | E. coli | Yes | GlcNAc | Gal (α1→3) | 12, 44 |
| O16 antigen | E. coli | Yes | GlcNAc | Rha(α1→3) and Glc (α1→6) | 12, 29 |
| O11 antigen | Pseudomonas aeruginosa | Yes | D-FucNAc | L-FucNAc (β1→3) | 12, 45 |
| O157 antigen | E. coli | Yes | GalNAc | Glc (α1→3) | 46, and M.W., unpublished work |
| O1 antigen | Shigella dysenteriae | Yes | GlcNAc | Gal (α1→3) | ref. 47, and M.W., unpublished work |
| O9a antigen | E. coli | Yes | GlcNAc | Man (α1→3) | ref. 48, this work |
| K30 capsular antigen | E. coli | No | Gal | Man (α1→3) | 49, this work |
| LT2 antigen | S. enterica | No | Gal | Rha (β1→3) | This work |
Bac, bacillosamine (2,4-diacetamido-2,4,6-trideoxyglucose); Rha, rhamnose; GalNAc, N-acetylgalactosamine; GlcNAc, N-acetylglucosamine; FucNAc, N-acetylfucosamine; Gal, galactose; Glc, glucose; Man, mannose.
Fig. 3.
Glycosylation of AcrA in S. enterica LT2. (A) Structure of the Salmonella LT2 O-antigen subunit. Man, mannose; Rha, rhamnose; Abe, abequose. (B) Western blot analysis of S. enterica SL3749 (ΔWaaL) periplasmic extracts expressing AcrA, and the pgl locus containing either PglBwt or PglBmut. Antisera anti-AcrA (lanes 1 and 2) or R12 (lanes 3 and 4) were used. (C Left) Western blot analysis of S. enterica SL3749 whole-cell extracts expressing AcrA and PglBwt or PglBmut in the absence of the pgl locus, showing production of S. enterica LT2 Und-PP-bound O antigen. Crossreaction of the antisera with AcrA can be detected. (C Right) Western blot of purified AcrA from periplasmic extracts of the cells is shown. Only one band, corresponding to the unglycosylated protein, is detectable in the presence of PglBwt or PglBmut.
Discussion
We have previously shown that PglB can transfer a number of different glycans from the Und-PP carrier to the C. jejuni acceptor protein AcrA (12, 17). The glycans transferred were diverse, and it was not possible to establish the structural restrictions imposed by PglB. We hypothesized that the sugar at the reducing end of the glycan must play an important role in recognition and/or catalysis, because it is directly linked to the Asn residue in the protein. All of the PglB substrates so far identified contain Bac, GlcNAc, GalNAc, or FucNAc at the reducing end (Table 1), all of which have in common an acetamido group at the C-2 position.
In this work, we tested the PglB-mediated transfer of three different polysaccharides: the O9a antigen and K30 capsular polysaccharide from E. coli and the O antigen of S. enterica LT2. The O9a antigen is a polymannan synthesized by an ABC transporter-dependent Wzy-independent pathway. The O9a antigen and the K30 capsular polysaccharide are produced simultaneously in strain E69 used in this study. Furthermore, the K30 antigen exists in two forms. One is the high-molecular weight capsular form, and the other exists as short oligosaccharides linked to the LPS lipid A core, a form known as KLPS (26). Thus, O9a and K30 antigens are both effectively presented to the WaaL ligase enzyme in a format suitable for attachment to the lipid A core. The O9a antigen was efficiently transferred to AcrA, but we did not detect glycosylated AcrA containing the K30 polysaccharide. However, two short oligosaccharides, both containing a HexNAc residue at the reducing end, were transferred to AcrA in E. coli CWG44. This strain was isolated based on its resistance to the coliphage O9-1, which lyses bacterial cells expressing O9 polymannan LPS at the surface (32). CWG44 carries a deletion in the O9a antigen biosynthesis locus, but the extent of the deletion has not been determined. The results presented here are consistent with production of an incompletely assembled O9a Und-PP-linked intermediate due to inactivation of a gene encoding a mannosyltransferase involved in the biosynthesis of the polymannan. Interestingly, these short oligosaccharides detected on AcrA were not previously reported on the lipid A core. It is possible that our immunological detection methods for AcrA are responsible for the increased sensitivity.
The initiating sugars of the O9a antigen and the K30 capsules are GlcNAc and Gal, respectively. The absence of AcrA glycosylated with K30 polysaccharide could be explained if PglB has a specific requirement of HexNAc residues for the oligosaccharyl transfer reaction. Alternatively, the mechanism of group I capsule export and assembly, which involves a protein complex spanning the inner membrane, the periplasm, and the outer membrane, may prevent PglB from accessing the glycan substrate (24). This seems unlikely, because some lipid intermediates in the capsular assembly system become available to the WaaL ligase to form the KLPS structure (26). This material is assembled in the absence of the Wza and Wzc proteins, which form the envelope-spanning complex for capsule assembly (23, 25). Our observation that the S. enterica LT2 polysaccharide, also containing Gal at the reducing end, was not transferred to AcrA by PglB strongly supports the notion that PglB has specificity for HexNAc residues at the reducing end of the glycan substrate. Lack of PglB-mediated transfer in S. enterica was not due to loss of functionality of PglB, because coexpression of the C. jejuni N-glycosylation system and AcrA resulted in glycosylated AcrA containing the C. jejuni glycan in S. enterica cells. Thus, we propose that the acetamido group at C-2 in the sugar located at the reducing end plays a crucial role during recognition and/or catalysis by PglB.
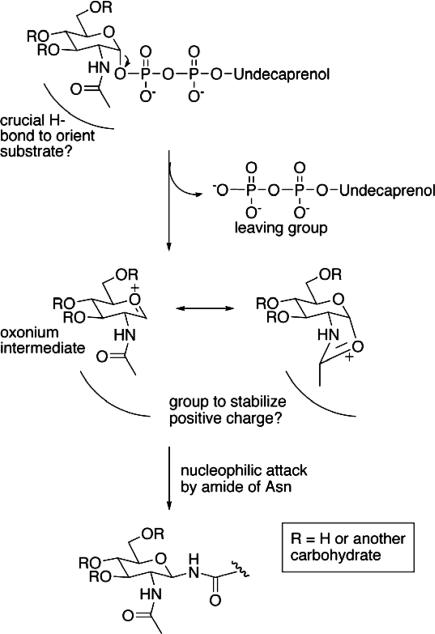
Several explanations for the requirement of a C-2 acetamido group in the reducing end sugar can be envisioned. In previous work addressing the substrate specificity of a eukaryotic OTase, three unnatural dolichol-linked disaccharide analogues were synthesized and evaluated as substrates or inhibitors for OTase from yeast. Minor changes, such as replacement of the hydrogens with isosteric fluorines, in the acetamido group of the proximal GlcNAc residue resulted in significant or total loss of activity (34). The loss of transferase activity could not be explained by a reduction in the binding of the unnatural analogs, because the affinity constants were not dramatically modified. Based on this evidence, the authors proposed that the acetamido group is involved in the catalytic activity and hypothesized that the acetamido group may interact with the OTase enzyme through critical hydrogen bonding with the OTase catalytic site. An alternate explanation that accounts for the yeast OTase results as well as our observations that the transferred sugar must have a 2-acetamido group is that the acetamido group acts as a neighboring group to stabilize an oxonium intermediate (Fig. 4). Even a trifluoracetamido group would not serve as well as an acetamido group as such a neighboring group participant. In contrast to mechanisms of chitinases and N-acetyl-β-glycosaminidase that use the acetamido group for substrate-assisted catalysis with relatively poor leaving groups (35–39), the OTase and PglB proteins act on sugars with good phosphate leaving groups that could allow an SN1- rather than SN2-like reaction. The enzyme active site then serves to exclude water, and stabilizes a full positive charge to create a particularly reactive electrophile to compensate for the relatively poor nucleophilicity of the asparagine amide, and thereby does not require deprotonation of the amide to generate a sufficiently reactive nucleophile. Support for such a positively charged intermediate comes from the report that peptide substrates for the yeast OTase that were modified with an amino group or amino-linked sugar in place of the asparagines served as much better inhibitors of the enzyme than either the unglycosylated or GlcNAc-linked Asn-containing peptides (40). Of course, detailed kinetics studies will be required to ascertain how concerted or stepwise this process is.
Fig. 4.
A mechanism to explain the necessity of the 2-acetamido group for carbohydrate transfer by PglB involves initial enzyme activation of the undecaprenol diphosphate. After cleavage of the anomeric linkage, the acetamido group acts as a neighboring group to stabilize the resulting oxonium intermediate before nucleophilic attack by the amide of an asparagine side chain. The electrophile created is reactive enough not to require deprotonation of the asparagine. The enzyme active site serves to hold the substrates in proximity and exclude water from the reactive intermediates.
If the enzyme needs to bind the carbohydrate-containing intermediate in the absence of the charged isoprenoid, the presence of additional sugars attached to the HexNAc could also serve to increase the binding affinity of the reactive intermediate. Indeed, the yeast OTase reaction is much more efficient with a disaccharide than monosaccharide substrate, and this additional sugar has been proposed to assist in binding the sugar donor (34). In the prokaryotic system, we showed that even an O16 subunit containing a Glc residue α-6 linked to the GlcNAc was efficiently transferred in the Wzy polymerase mutant EVV1 strain. This experiment showed that a substitution at C-6 of the initiating GlcNAc residue did not prevent the transfer of the glycan by PglB. Although detailed mechanistic studies of PglB in vitro are required, our experiments extend the conclusions of the yeast system to the prokaryotic N-glycosylation system with additional evidence for alternate mechanistic possibilities.
Our finding that the C. jejuni N-glycosylation machinery was functional in S. enterica cells may be useful to develop novel live attenuated antibacterial vaccines. S. enterica cells exposing glycoproteins on their surface could be an alternative to the use of conjugate vaccines. Understanding the basis of PglB substrate recognition specificity will permit us to engineer glycoproteins containing a diversity of bacterial glycans. The efficacy of attenuated S. enterica cells as glycoprotein display devices is currently under investigation.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
E. coli strains were grown on LB at 37°C. Trimethoprim (100 μg/ml)/20 μg/ml tetracycline/80 μg/ml spectinomycin/20 μg/ml chloramphenicol/100 g/ml ampicillin were added to the media for selection as needed. E. coli DH5α (Invitrogen) was the host for cloning experiments. Plasmids pACYC184 and pBR322 (NEB, Beverly, MA) were used as cloning vectors. Plasmids pACYC184(pgl), pACYC184(pglmut) (encoding the C. jejuni pgl cluster) (12), and pMF19 (encoding the rhamnosyl transferase wbbL) (28) have been described elsewhere. Strains and plasmids used are listed in Table 2.
Table 2.
Strains and plasmids used in this study
| Strain | Characteristic/description | Serotype | Reference |
|---|---|---|---|
| Bacteria | |||
| DH5α | F-ϕ80lacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rk−, mk+) gal− phoA supE44 λ−thi−1 gyrA96 relA1 | Invitrogen | |
| E69 | Prototroph | O9a:K30:H12 | F.  rskov rskov |
| CWG28 | E69 derivative; trp his lac rpsL cpsK30; Smr | O9a:K−:H12 | 31 |
| CWG44 | E69 derivative; trp his lac rpsL rfbO9 | O−:K30:H12 | 32 |
| EVV11 | W3110, Δwzy | This work | |
| SL3749 | S. enterica sv. Typhimurium LT2; waaL446 | R-LPS | SGSC228* |
| Plasmid | |||
| pACYC(pgl) | Encodes the C. jejuni pgl cluster, CmR | 17 | |
| pACYC(pglmut) | Encodes the C. jejuni pgl containing mutations W458A and D459A in PglB, CmR | 17 | |
| pMAF10 | Hemagglutinin-tagged PglB cloned in pMLBAD, TmpR | 12 | |
| pWA1 | Hemagglutinin-tagged PglBmut cloned in pMLBAD, TmpR | 12 | |
| pWA2 | Soluble periplasmic hexahis-tagged AcrA under control of Tet promoter, in pBR322, AmpR | 12 | |
| pMF19 | Expresses WbbL rhamnosyltransferase; restores O16 antigen biosynthesis, SpR | 28 |
*Salmonella Genetic Stock Centre (maintained by K. E. Sanderson, University of Calgary, Calgary, AL, Canada).
Production and Purification of Glycosylated AcrA.
E. coli cells expressing the O antigen genes, AcrA and PglB, were induced by the addition of l-arabinose to 0.2% (wt/vol) as described (12). After induction at 37°C for 5 h, arabinose was added again to ensure that PglB expression was maintained when the carbon source becomes limiting (as the cells metabolize the arabinose). Cells were harvested by centrifugation after 20 h of induction. Preparations of periplasmic extracts were carried out either by osmotic-shock lysis or by lysozyme treatment. For the former, cells were consecutively incubated first in 20% sucrose/30 mM Tris·Cl, pH 8.0/1 mM EDTA and then in 5 mM MgSO4 at 4°C for 2–4 h (20 OD600 units/ml). The latter consisted of a single incubation of 1 h at 4°C in 30 mM Tris·Cl, pH 8.5/20% (wt/vol) sucrose/1 mM EDTA/1 mg/ml lysozyme (Sigma). A final centrifugation step in both methods yielded periplasmic proteins in the supernatant. For purification of (His-6)-tagged AcrA protein, the extracts were diluted with 1/9th volume 10× buffer A (100 mM imidazole/300 mM Tris·Cl, pH 8.0/3 M NaCl), sterile-filtered, loaded on a HisTrap HP column (Amersham Pharmacia Biosciences) at 1–5 ml/min, washed with at least 25 column volumes of buffer A containing 20 mM imidazole, and finally eluted into 1–3 ml of buffer A containing 0.25 M imidazole.
Immunoblotting.
Immunoblotting was performed as described (41). Antisera specific for AcrA, R12 (17), K30 capsular polysaccharide (42), and O9a polysaccharide (20) have been described. Con A–horseradish peroxidase was from Sigma.
Characterization of Glycosylated Peptides.
Purified AcrA samples were separated by SDS/PAGE and stained using Novex colloidal blue reagent (Invitrogen); then the desired band was excised, lyophilized, and digested with trypsin by an adaptation of the method of ref. 43. Because AcrA does not contain cysteine residues, the reduction step of disulfide bonds and the alkylation of free cysteine thiols could be omitted. The gel pieces were immediately washed with 50 mM NH4HCO3, dehydrated with CH3CN, and dried. Gel pieces were rehydrated for 1 h at 4°C in 50 mM NH4HCO3 containing 0.02 mg/ml trypsin (EC 3.4.21.4; Promega) and digested overnight at 37°C in 50 μl of 50 mM NH4HCO3. The (glyco)peptides were extracted from the gel pieces with a step gradient of 100% CH3CN to 75% CH3CN. Tryptic peptides extracted from in-gel-digested SDS/PAGE bands were desalted over a NuTip 10 Carbon cleanup tip (Glygen, Columbia, MD). The tip was first preconditioned with three washes of 5 μl of 60% acetonitrile, 0.05% trifluoroacetic acid (TFA), followed by three washes of 5 μl with 0.05% TFA. The sample (in 5 μl of 0.05% TFA) was applied by pipetting it up and down 25 times over the tip. Subsequently, the tip was washed 10 times with 10 μl of 0.05% TFA, and the peptides were eluted with 5 × 1.75 μl of 60% acetonitrile containing 0.05% TFA (pipetting up and down the eluent 10 times for each elution and pooling the five eluates). Then the pooled eluate was vacuum-evaporated to dryness and reconstituted in 2 μl of “superDHB” MALDI matrix solution, i.e., a 9:1 mixture of 20 mg/ml 2,5-dihydroxybenzoic acid (2,5-DHB) and 20 mg/ml 5-methoxysalicylic acid in 70% MeOH containing 0.1% TFA (both matrix compounds from Fluka). This final sample (0.8 μl) was applied to a stainless-steel ABI 192-target MALDI plate (Applied Biosystems) and air-dried. The detailed procedure for the MALDI-TOF analysis is presented in Supporting Text.
Supplementary Material
Acknowledgments
N.L.P. acknowledges Prof. J. Siegel and the Organisch-Chemisches Institut for hosting her summer stay at the University of Zurich. This research was supported by Swiss National Foundation Grant 3100A0-105541 and grants from the Union Bank of Switzerland and the Gebert Rüf Foundation (to M.A.); Canadian Institutes of Health Research Grant MT10206 (to M.A.V.); a grant from the Natural Sciences and Engineering Council of Canada (to C.W.); and the GlycoInit Initiative from Eidgenössiche Technische Hochschule Zurich. M.A.V. and C.W. hold Canada Research Chairs in Infectious Diseases and Microbial Pathogenesis and in Molecular Microbiology, respectively.
Abbreviations
- Und-PP
undecaprenyl-pyrophosphate
- OTase
oligosaccharyltransferase
- Gal
galactose
- Glc
glucose
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office. C.R.H.R. is a guest editor invited by the Editorial Board.
References
- 1.Linton D., Allan E., Karlyshev A. V., Cronshaw A. D., Wren B. W. Mol. Microbiol. 2002;43:497–508. doi: 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 2.Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. Mol. Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 3.Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. J. Biol. Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 4.Alaimo C., Catrein I., Morf L., Marolda C. L., Callewaert N., Valvano M. A., Feldman M. F., Aebi M. EMBO J. 2006;25:967–976. doi: 10.1038/sj.emboj.7601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glover K. J., Weerapana E., Imperiali B. Proc. Natl. Acad. Sci. USA. 2005;102:14255–14259. doi: 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linton D., Dorrell N., Hitchen P. G., Amber S., Karlyshev A. V., Morris H. R., Dell A., Valvano M. A., Aebi M., Wren B. W. Mol. Microbiol. 2005;55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- 7.Burda P., Aebi M. Biochim. Biophys. Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld R., Kornfeld S. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 9.Knauer R., Lehle L. Biochim. Biophys. Acta. 1999;1426:259–273. doi: 10.1016/s0304-4165(98)00128-7. [DOI] [PubMed] [Google Scholar]
- 10.Fry B. N., Korolik V., ten Brinke J. A., Pennings M. T., Zalm R., Teunis B. J., Coloe P. J., van der Zeijst B. A. Microbiology. 1998;144:2049–2061. doi: 10.1099/00221287-144-8-2049. [DOI] [PubMed] [Google Scholar]
- 11.Bugg T. D., Brandish P. E. FEMS Microbiol. Lett. 1994;119:255–262. doi: 10.1111/j.1574-6968.1994.tb06898.x. [DOI] [PubMed] [Google Scholar]
- 12.Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., Morris H. R., Dell A., Valvano M. A., Aebi M. Proc. Natl. Acad. Sci. USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raetz C. R., Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronner D., Clarke B. R., Whitfield C. Mol. Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 15.Rick P. D., Hubbard G. L., Barr K. J. Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt G., Mayer H., Makela P. H. J. Bacteriol. 1976;127:755–762. doi: 10.1128/jb.127.2.755-762.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., et al. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 18.Kido N., Torgov V. I., Sugiyama T., Uchiya K., Sugihara H., Komatsu T., Kato N., Jann K. J. Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke B. R., Cuthbertson L., Whitfield C. J. Biol. Chem. 2004;279:35709–35718. doi: 10.1074/jbc.M404738200. [DOI] [PubMed] [Google Scholar]
- 20.Cuthbertson L., Powers J., Whitfield C. J. Biol. Chem. 2005;280:30310–30319. doi: 10.1074/jbc.M504371200. [DOI] [PubMed] [Google Scholar]
- 21.Whitfield C., Roberts I. S. Mol. Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 22.Drummelsmith J., Whitfield C. Mol. Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 23.Drummelsmith J., Whitfield C. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nesper J., Hill C. M., Paiment A., Harauz G., Beis K., Naismith J. H., Whitfield C. J. Biol. Chem. 2003;278:49763–49772. doi: 10.1074/jbc.M308775200. [DOI] [PubMed] [Google Scholar]
- 25.Wugeditsch T., Paiment A., Hocking J., Drummelsmith J., Forrester C., Whitfield C. J. Biol. Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 26.MacLachlan P. R., Keenleyside W. J., Dodgson C., Whitfield C. J. Bacteriol. 1993;175:7515–7522. doi: 10.1128/jb.175.23.7515-7522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D., Reeves P. R. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 28.Feldman M. F., Marolda C. L., Monteiro M. A., Perry M. B., Parodi A. J., Valvano M. A. J. Biol. Chem. 1999;274:35129–35138. doi: 10.1074/jbc.274.49.35129. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson G., Neal B., Liu D., Hobbs M., Packer N. H., Batley M., Redmond J. W., Lindquist L., Reeves P. J. Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Z., Valvano M. A. J. Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitfield C., Schoenhals G., Graham L. J. Gen. Microbiol. 1989;135:2589–2599. doi: 10.1099/00221287-135-10-2589. [DOI] [PubMed] [Google Scholar]
- 32.McCallum K. L., Laakso D. H., Whitfield C. Can. J. Microbiol. 1989;35:994–999. doi: 10.1139/m89-166. [DOI] [PubMed] [Google Scholar]
- 33.Kaniuk N. A., Vinogradov E., Whitfield C. J. Biol. Chem. 2004;279:36470–36480. doi: 10.1074/jbc.M401366200. [DOI] [PubMed] [Google Scholar]
- 34.Tai V. W., Imperiali B. J. Org. Chem. 2001;66:6217–6228. doi: 10.1021/jo0100345. [DOI] [PubMed] [Google Scholar]
- 35.Brameld K. A., Shrader W. D., Imperiali B., Goddard W. A., III J. Mol. Biol. 1998;280:913–923. doi: 10.1006/jmbi.1998.1890. [DOI] [PubMed] [Google Scholar]
- 36.Mark B. L., Vocadlo D. J., Knapp S., Triggs-Raine B. L., Withers S. G., James M. N. J. Biol. Chem. 2001;276:10330–10337. doi: 10.1074/jbc.M011067200. [DOI] [PubMed] [Google Scholar]
- 37.Terwisscha van Scheltinga A. C., Armand S., Kalk K. H., Isogai A., Henrissat B., Dijkstra B. W. Biochemistry. 1995;34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 38.Tews I., Terwisscha van Scheltinga A. C., Perrakis A., Wilson K. S., Dijkstra B. W. J. Am. Chem. Soc. 1997;119:7954–7959. [Google Scholar]
- 39.Williams S. J., Mark B. L., Vocadlo D. J., James M. N., Withers S. G. J. Biol. Chem. 2002;277:40055–40065. doi: 10.1074/jbc.M206481200. [DOI] [PubMed] [Google Scholar]
- 40.Peluso S., de L. U. M., O’Reilly M. K., Imperiali B. Chem. Biol. 2002;9:1323–1328. doi: 10.1016/s1074-5521(02)00281-8. [DOI] [PubMed] [Google Scholar]
- 41.Aebi M., Gassenhuber J., Domdey H., te Heesen S. Glycobiology. 1996;6:439–444. doi: 10.1093/glycob/6.4.439. [DOI] [PubMed] [Google Scholar]
- 42.Dodgson C., Amor P., Whitfield C. J. Bacteriol. 1996;178:1895–1902. doi: 10.1128/jb.178.7.1895-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shevchenko A., Wilm M., Vorm O., Mann M. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 44.L’Vov V L., Shashkov A. S., Dmitriev B. A., Kochetkov N. K., Jann B., Jann K. Carbohydr. Res. 1984;126:249–259. doi: 10.1016/0008-6215(84)85382-3. [DOI] [PubMed] [Google Scholar]
- 45.Knirel Y. A. Crit. Rev. Microbiol. 1990;17:273–304. doi: 10.3109/10408419009105729. [DOI] [PubMed] [Google Scholar]
- 46.Perry M. B., MacLean L., Griffith D. W. Biochem. Cell Biol. 1986;64:21–28. doi: 10.1139/o86-004. [DOI] [PubMed] [Google Scholar]
- 47.Dmitriev B. A., Knirel Y. A., Kochetkov N. K. Eur. J. Biochem. 1976;66:559–566. doi: 10.1111/j.1432-1033.1976.tb10582.x. [DOI] [PubMed] [Google Scholar]
- 48.Prehm P., Jann B., Jann K. Eur. J. Biochem. 1976;67:53–56. doi: 10.1111/j.1432-1033.1976.tb10631.x. [DOI] [PubMed] [Google Scholar]
- 49.Chakraborty A. K., Friebolin H., Stirm S. J. Bacteriol. 1980;141:971–972. doi: 10.1128/jb.141.2.971-972.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.