Abstract
The NS1 protein of influenza A virus (NS1A protein) is a multifunctional protein that counters cellular antiviral activities and is a virulence factor. Its N-terminal RNA-binding domain binds dsRNA. The only amino acid absolutely required for dsRNA binding is the R at position 38. To identify the role of this dsRNA-binding activity during influenza A virus infection, we generated a recombinant influenza A/Udorn/72 virus expressing an NS1A protein containing an RNA-binding domain in which R38 is mutated to A. This R38A mutant virus is highly attenuated, and the mutant NS1A protein, like the WT protein, is localized in the nucleus. Using the R38A mutant virus, we establish that dsRNA binding by the NS1A protein does not inhibit production of IFN-β mRNA. Rather, we demonstrate that the primary role of this dsRNA-binding activity is to protect the virus against the antiviral state induced by IFN-β. Pretreatment of A549 cells with IFN-β for 6 h did not inhibit replication of WT Udorn virus, whereas replication of R38A mutant virus was inhibited 1,000-fold. Using both RNA interference in A549 cells and mouse knockout cells, we show that this enhanced sensitivity to IFN-β-induced antiviral activity is due predominantly to the activation of RNase L. Because activation of RNase L is totally dependent on dsRNA activation of 2′-5′ oligo (A) synthetase (OAS), it is likely that the primary role of dsRNA binding by the NS1A protein in virus-infected cells is to sequester dsRNA away from 2′-5′ OAS.
Keywords: dsRNA-binding, IFN-β
Influenza A viruses cause a highly contagious respiratory disease in humans and are responsible for the periodic wide-spread epidemics, or pandemics, that have caused high mortality rates (1). The most devastating pandemic occurred in 1918, resulting in ≈30 million deaths worldwide (2). The avian influenza A viruses (H5N1 viruses), which are currently circulating primarily in Asia, are strong candidates for causing the next pandemic if they acquire the ability for efficient human-to-human transmission (3, 4). These viruses, which have so far been transmitted largely from chickens to humans, are highly virulent, resulting in death in ≈50% of infected humans. It is not known why these viruses are so virulent, and only a few of the molecular determinants of their virulence have been identified.
The NS1 protein of influenza A virus (NS1A protein) plays a large role in countering host cell antiviral defenses (5, 6) and is expected to play important roles in viral virulence (3, 4). The NS1A protein is a multifunctional protein that participates in both protein–RNA and protein–protein interactions (7). Its RNA target is A-form dsRNA (8–11), and it binds and inhibits the function of two cellular proteins that are required for the 3′ end processing of cellular pre-mRNAs: the 30-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF) and poly(A)-binding protein II (PABII) (13, 14). The dsRNA-binding domain comprises the 73 amino-terminal amino acids of the NS1A protein and forms a symmetric homodimer with a unique six-helical chain fold (15–17). This structure differs from that of the predominant class of dsRNA-binding domains, termed dsRBDs, which are found in a large number of cellular proteins (18–20). Because the NS1A dsRBD has a much lower affinity for dsRNA than cellular dsRBDs (10, 11, 21, 22), it has not been clear that the NS1A protein would effectively compete for dsRNA with dsRBD-containing cellular proteins during virus infection.
In fact, the role of the dsRNA-binding activity of the NS1A protein during influenza A virus infection has not been identified. We have shown that the latent protein kinase PKR is not activated in cells infected with a recombinant influenza A/Udorn/72 virus expressing an NS1A protein lacking dsRNA-binding activity (23), demonstrating that sequestering of dsRNA by the NS1A protein is not the mechanism by which PKR activation is inhibited during influenza A virus infection. A previous study (24) reported that high levels of IFN-β and its mRNA were produced in cells infected with a recombinant influenza A/WSN/33 virus expressing an NS1A protein with a mutated RNA-binding domain (24). However, in the present study, we show that this mutant WSN NS1A protein is located in the cytoplasm, rather than the nucleus of infected cells, and our results reveal that the phenotype of this mutant WSN virus is due to the mislocalization of the mutant NS1A protein rather than to the loss of NS1A dsRNA-binding activity. This conclusion is based on experiments using a recombinant A/Udorn/72 virus expressing an NS1A protein lacking RNA-binding activity. Because this mutant NS1A protein is localized in the nucleus of infected cells, the attenuation of the Udorn mutant virus can be attributed solely to the loss of the dsRNA-binding activity of the NS1A protein. Using this mutant Udorn virus, we establish that the dsRNA-binding activity of the NS1A protein does not play a role in inhibiting the production of IFN-β mRNA but rather is required for the protection of influenza A virus against the antiviral state induced by IFN-β. Further, we demonstrate that the NS1A dsRNA-binding activity provides this protection primarily by inhibiting the IFN-α/β-induced 2′-5′-oligo (A) synthetase (OAS)/RNase L pathway.
Results
The Udorn NS1A Protein Lacking dsRNA-Binding Activity Is Localized in the Nucleus of Virus-Infected Cells.
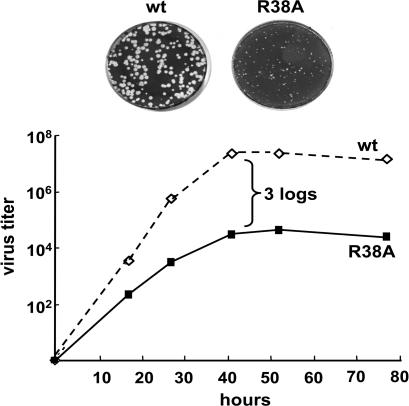
The only amino acid that is absolutely required for dsRNA binding by the NS1A protein is the R at position 38, so that mutation of R38 to A eliminates dsRNA binding (10). To determine the role of dsRNA binding by the NS1A protein during influenza A virus infection, we generated a recombinant influenza A/Udorn/72 expressing a mutant R38A mutant NS1A protein. This recombinant virus is highly attenuated: it formed pin-point plaques in Madin–Darby canine kidney (MDCK) cells, and the virus yield at low multiplicity of infection (moi) of 0.001 plaque-forming units (pfu) per cell was ≈1,000-fold lower than with WT virus (Fig. 1).
Fig. 1.
The R38A influenza A/Udorn/72 mutant virus is attenuated. (Upper) Size of the plaques of the WT and R38A mutant viruses on MDCK cells. (Lower) Growth curve of the WT and R38A mutant viruses after low moi of MDCK cells.
Determining how the loss of the dsRNA-binding activity of the NS1A protein causes this high level of virus attenuation is complicated by the fact that mutation of R38 to A also inactivates the nuclear localization signal (NLS1) that is coincident with the RNA-binding domain (25). However, the NS1A protein of influenza A/Udorn/72 virus contains another NLS (NLS2), a bipartite NLS that extends from amino acid 219 to the C-terminal 237 aa (K. Melen, L. Kinnunen, R.M.K., and I. Julkunen, unpublished results). As shown in Fig. 2, in cells infected with the Udorn R38A mutant virus, the NS1A protein was localized in the nucleus, as was the case for the NS1A protein in WT virus-infected cells. In contrast, the NS1A protein of influenza A/WSN/33 has only the NLS1 that is coincident with the RNA-binding domain, because a stop codon at position 230 disrupts the NLS2 sequence. Consequently, in cells infected with the WSN R38A, K41A mutant virus, the NS1A protein was localized in the cytoplasm, in contrast to the situation in cells infected with WT WSN virus (Fig. 2). The mislocalization of this WSN NS1A mutant protein likely accounts for the high levels of IFN-β and its mRNA that were reported to be induced in cells infected by the WSN R38, K41 mutant virus (24), because this phenotype was not exhibited by the Udorn R38A mutant virus, as documented below.
Fig. 2.
Intracellular localization of the NS1A protein encoded by WT and R38A mutant influenza A/Udorn/72 viruses (Upper) and by WT and R38A, K41A mutant influenza A/WSN/33 viruses (Lower).
The Udorn R38A Mutant Virus Exhibits No Detectable Defect During Single-Cycle Virus Growth and Does Not Induce Higher Levels of IFN-β mRNA.
To determine whether the loss of RNA-binding activity of the NS1A protein affects one or more steps in virus replication, we measured the time course of virus-specific protein synthesis and of infectious virus production during single-cycle growth of the Udorn WT and R38A mutant viruses. MDCK cells were infected with either virus at an moi of 5, and the time course of virus-specific protein synthesis was determined by labeling the cells for 30 min with [35S]methionine and cysteine at various times after infection (Fig. 3A). No difference in the time course and rate of virus-specific protein synthesis between WT and R38A virus-infected cells was detected, demonstrating that virus-specific protein synthesis was not affected by the loss of the RNA-binding activity of the NS1A protein. In addition, the time course of production of infectious virus was the same in cells infected by either virus (Fig. 3B), demonstrating that all of the essential steps in virus replication were not affected by the loss of the RNA-binding activity of the NS1A protein. The same results were observed in single-cycle experiments in human A549 cells (see Fig. 4 below).
Fig. 3.
The Udorn R38A mutant virus exhibits no detectable defect during single-cycle virus growth and does not induce higher levels of IFN-β mRNA. (A) Viral protein synthesis after infecting MDCK cells with either WT or R38A Udorn virus at an moi of 5. Infected cells were labeled with [35S]methionine and [35S]cysteine at the indicated times after infection, and the radiolabeled proteins were analyzed by SDS/PAGE. The mobility of the R38A mutant NS1A protein is reduced relative to that of the WT NS1A protein. As a consequence, the R38A NS1A protein and the M1 protein have similar, but not identical, mobilities. These two proteins can be resolved by electrophoresis on higher concentration gels, which reveals that the relative amounts of M1 and NS1 are the same in WT and R38A virus-infected cells. (B) Growth curve of the WT and R38A mutant viruses after infecting MDCK cells at an moi of 5. (C) The relative amount of IFN-β mRNA produced during single-cycle growth in MDCK cells of WT, R38A mutant, and CPSF mutant viruses. At the times after infection indicated, cells were collected, and the total RNA was extracted. The amount of IFN-β mRNA was determined by using quantitative RT-PCR.
Fig. 4.
The Udorn R38A mutant virus has enhanced sensitivity to the antiviral state induced by IFN-β. A549 cells were treated or mock-treated with 100 units/ml IFN-β for 6 h. The cells were then infected with WT or R38A mutant virus at an moi of 5, and virus production was assayed as described in Materials and Methods.
We also determined whether the loss of the dsRNA-binding activity of the NS1A protein affected the production of IFN-β mRNA during single-cycle virus growth. Human A549 cells were used for these experiments. As shown previously, WT Udorn virus activates IFN regulatory factor 3 (IRF-3), but the production of mature IFN-β mRNA is low because the NS1A protein binds and inhibits the function of two cellular proteins that are essential for the 3′ end processing of cellular pre-mRNAs, the 30-kDa subunit of CPSF (CPSF30) and poly(A)-binding protein II (7, 26). Mutational inactivation of the NS1A site for binding CPSF30 was shown to result in a large increase in IFN-β mRNA production and substantial (1,000-fold) attenuation of the virus (CPSF mutant virus) (7, 27). This increase in IFN-β mRNA production serves as a benchmark for the levels of IFN-β mRNA that result in substantial virus attenuation. We used quantitative RT-PCR to measure the amount of IFN-β mRNA produced after infection with the WT, R38A mutant and a CPSF mutant virus (Fig. 3C). Whereas the CPSF mutant virus induced ≈70 times more IFN-β mRNA than WT virus, the R38A mutant virus induced the same amount of IFN-β mRNA as WT virus. Approximately equal amounts of the NS1A protein were synthesized in cells infected at an moi of 5 with the three viruses (Fig. 3A and ref. 27). These results show that the loss of RNA-binding activity of the NS1A protein does not affect IFN-β mRNA production.
The Udorn R38A Mutant Virus Has Enhanced Sensitivity to the Antiviral State Induced by IFN-β.
Because the R38A mutation did not affect either the production of IFN-β mRNA or virus replication during single-cycle growth, we reasoned that the attenuation of the Udorn R38A virus was likely due to an enhanced sensitivity to one or more antiviral proteins induced by IFN-β. To determine whether this is the case, human A549 cells were treated with 100 units/ml IFN-β for 6 h and were then infected with either Udorn WT or R38A mutant virus at an moi of 5 (Fig. 4). We measured the time course of virus replication relative to that seen in cells that were mock-treated for 6 h before infection. In the absence of IFN-β pretreatment, the time course of virus production was the same in cells infected by either virus, as was the case in MDCK cells. In contrast, after IFN-β pretreatment for 6 h, there was a dramatic difference in the subsequent replication of the two viruses. Whereas this IFN-β pretreatment had no detectable effect on the growth of WT Udorn virus, it caused a substantial inhibition of the growth of the R38A mutant virus. The rate of virus replication and virus yield of the R38A mutant virus was reduced ≈1,000-fold. We conclude that the loss of the RNA-binding activity of the NS1A protein rendered the R38A mutant virus susceptible to the actions of one or more antiviral proteins induced by the 6-h pretreatment with IFN-β.
Activation of the 2′-5′ OAS/RNase L Pathway Is the Primary Cause of the Enhanced Sensitivity of the R38A Mutant Virus to IFN-β.
IFN-β has been shown to induce the synthesis of two antiviral proteins that are activated by dsRNA: PKR and 2′-5′ OAS (28). We have shown previously that the RNA-binding domain of the NS1A protein is not responsible for blocking the activation of PKR in virus-infected cells (23). Consequently, we focused on the possibility that activation of 2′-5′ OAS causes the inhibition of the replication of the R38A mutant virus after IFN-β treatment. Once activated, 2′-5′ OAS polymerizes ATP into 2′-5′ oligo (A) chains, which in turn activates the latent RNase L that degrades mRNAs and ribosomal RNAs and as a result inhibits virus replication (28, 29).
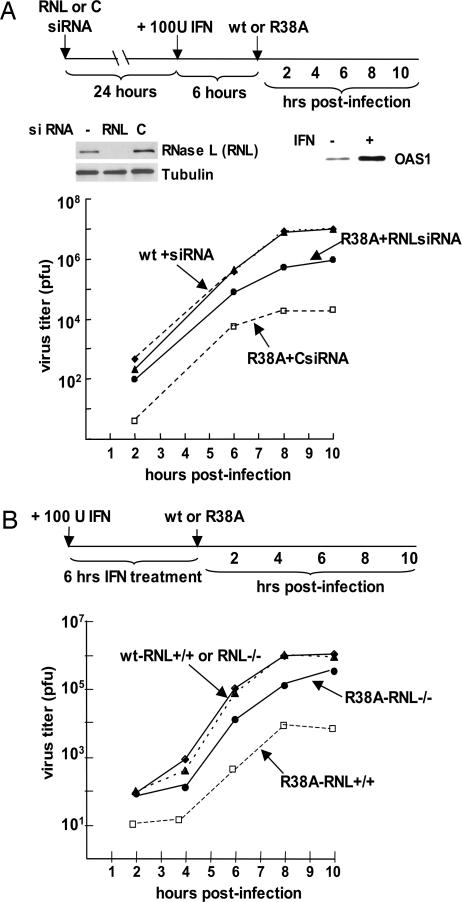
Because there are multiple isoforms of 2′-5′ OAS (28), we used small interfering RNAs (siRNAs) to deplete RNase L rather than 2′-5′ OAS and determined whether this depletion relieved the inhibition of the replication of the R38A mutant virus after IFN-β treatment. As shown in Fig. 5A, transfection of A549 cells with these specific siRNAs reduced RNase L to a level that was not detectable in an immunoblot. We also verified that 2′-5′ OAS was synthesized as a result of IFN-β treatment. The replication of WT virus was not affected by depletion of RNase L, whereas the IFN-β-mediated inhibition of the replication of the R38A mutant virus was substantially relieved. The rate of replication and yield of R38A virus was increased ≈100-fold relative to that produced in cells treated with a control siRNA. Consequently, RNase L, whose activation is mediated by activated 2′-5′ OAS, plays a large role in the IFN-β-induced inhibition of the replication of the R38A mutant virus.
Fig. 5.
Activation of the 2′-5′ OAS/RNase L pathway is the primary cause of the enhanced sensitivity of the R38A mutant virus to IFN-β. (A) siRNA knockdown of RNase L. A549 cells were transfected with either RNase L (RNL)-specific siRNAs or a control siRNA for 24 h. The cells were then treated with 100 units/ml IFN-β for 6 h, followed by infection with WT or R38A virus at an moi of 5. Virus production was assayed as described in Materials and Methods. The depletion of RNase L was assayed by an immunoblot of extracts of cells collected after IFN-β treatment. The production of OAS resulting from IFN-β treatment was monitored by immunoblotting using antibody against the OAS1 isomer (12). (B) Mouse RNase L knockout cells. Mouse RNase L−/− and RNase L+/+ cells were treated with 100 units/ml IFN-β for 6 h. The cells were then infected with WT or R38A mutant virus at an moi of 5, and virus production was assayed as described in Materials and Methods.
Although RNase L was not detected by an immunoblot after transfecting human A549 cells with RNase L-specific siRNAs, it was possible that some residual RNase L enzyme was still present and therefore might be activated to inhibit the replication of the R38A mutant virus, although inefficiently. To test this possibility, we carried out experiments using mouse cells lacking the RNase L gene (30). These mutant mouse cells (RNase L−/−) and WT mouse cells (RNase L+/+) were treated with mouse IFN-β for 6 h and were then infected with either Udorn WT or R38A mutant virus at an moi of 5 (Fig. 5B). The replication of WT virus was the same in the WT and mutant mouse cells after IFN-β treatment, verifying that the 2′-5′ OAS/RNase L pathway does not affect WT virus. In contrast, the replication of the R38A virus differed substantially between the two mouse cell lines after IFN-β treatment. In the WT mouse cells, the replication of the R38A virus relative to that of Udorn WT virus was severely inhibited (≈1,000-fold), similar to the situation in IFN-β-treated human A549 cells. In the mouse RNase L−/− cells, this inhibition was substantially relieved: the rate of replication and yield of the R38A virus was 100- to 200-fold greater than in the WT mouse cells. Thus, the 2′-5′ OAS/RNase L pathway is responsible for most, but not all, of the sensitivity of the R38A mutant virus to the 6-h treatment with IFN-β.
Discussion
The N-terminal RNA-binding domain of the NS1A protein of influenza A virus binds a wide range of dsRNA sequences, with no sequence specificity (8–11). Recent structural data have shown that dsRNA is recognized via two tracks of basic amino acids that align with the polyphosphate backbone of canonical A-form dsRNA (C. Yin, J. A. Khan, G. V. T. Swapna, R.M.K., L. Tong, and G. T. Montelione, unpublished results). These tracks include R38 in both chains of the dimeric structure of the RNA-binding domain. Several of the amino acids, including R38, that participate in the recognition of dsRNA are also required for the binding of importin-α, which mediates the nuclear import of the NS1A protein (K. Melen, L. Kinnunen, R.M.K., and I. Julkunen, unpublished results). As is the case for other nuclear import pathways (31), importin-α would be dissociated from its NS1A protein cargo in the nucleus and exported to the cytoplasm to mediate the nuclear import of other NS1A protein molecules. Consequently, in the cytoplasm, but not in the nucleus, importin-α would be expected to compete with dsRNA for binding to the RNA-binding domain of the WT NS1A protein.
Our results demonstrate that the primary role of the dsRNA-binding activity of the NS1A protein in infected cells is to protect the virus against the antiviral state induced by IFN-α/β. We were able to establish this role of the NS1A protein in the context of influenza A virus-infected cells by using a recombinant influenza A/Udorn/72 virus that expresses an NS1A protein with a point mutation, R38A, that eliminates its dsRNA-binding activity (9). This mutated NS1A protein, like the WT NS1A protein, is localized in the nucleus. Although the R38A mutation also inactivates the NS1A NLS that is coincident with the RNA-binding domain, the Udorn NS1A protein has a second NLS at its C terminus that is sufficient for importing the R38A mutant NS1A protein into the nucleus. Therefore, mutational elimination of the binding of importin-α to the RNA-binding domain has no apparent effect on the function of the Udorn NS1A protein during infection, and the attenuation of the R38A mutant virus can be attributed to the loss of the dsRNA-binding activity of the NS1A protein. We found that the R38A mutant virus differed from WT Udorn virus in only one property, sensitivity to the antiviral state induced by IFN-β. Thus, pretreatment of human A549 cells with IFN-β for 6 h had no detectable effect on the replication of WT Udorn virus, whereas the replication of the R38A mutant virus was inhibited 1,000-fold. Using both RNA interference in A549 cells and mouse knockout cells, we showed that this enhanced sensitivity to IFN-β-induced antiviral activity was due largely, but not totally, to the activation of RNase L. In the absence of RNase L, the sensitivity of the R38A mutant virus to IFN-β-induced antiviral activity was relieved 100- to 200-fold.
These results show that the dsRNA-binding activity of the NS1A protein is responsible for rendering influenza A virus resistant to the antiviral activity of the IFN-α/β-induced 2′-5′ OAS/RNase L pathway. Because the activation of RNase L is totally dependent on the dsRNA activation of 2′-5′ OAS (29), it is likely that the primary role of dsRNA binding by the NS1A protein is to sequester dsRNA away from 2′-5′ OAS. The NS1A protein would be expected to effectively compete with 2′-5′ OAS for dsRNA based on the relative affinities of these two proteins for dsRNA. Unlike the majority of cellular proteins, 2′-5′ OAS does not bind dsRNA via a dsRBD domain (32). Because 2′-5′ OAS has a very low affinity for dsRNA, investigators have found it necessary to use a crosslinking method to stabilize the 2′-5′ OAS–dsRNA complex (32). In contrast, the NS1A protein–dsRNA complex remains intact during gel shift and other assays (10, 11), indicating that the NS1A protein likely has a higher affinity for dsRNA than 2′-5′ OAS.
Activation of the 2′-5′ OAS/RNase L pathway does not account for the totality of the enhanced sensitivity of the R38A virus to the antiviral state induced by IFN-β. In the absence of RNase L, the sensitivity was reduced by 100- to 200-fold, indicating that the remaining 5- to 10-fold increased sensitivity is likely due to another dsRNA-dependent, IFN-β-induced activity. Other than PKR and 2′-5′ OAS, the only known IFN-β-induced enzymes that bind dsRNA are dsRNA-specific adenosine deaminases (ADARs), which catalyze A-to-I editing in dsRNAs (28). ADARs have the potential for antiviral activity, as shown by the inhibition of the replication of hepatitis D viral RNA replication resulting from ADAR-mediated RNA editing (33). It is not known whether ADAR RNA editing plays a similar role in the replication of influenza A viral RNA, or whether the NS1A RNA-binding domain would effectively compete with the high-affinity dsRBD domains of ADAR for dsRNA. Another possibility is that there are one or more dsRNA-dependent, IFN-β-induced antiviral proteins that have not yet been identified.
Our results show that the functions attributed to dsRNA binding by the NS1A protein, as assayed by overexpression of the NS1A protein in transient transfection experiments (34–38), do not correspond to the actual function of NS1A protein-mediated dsRNA binding in influenza A virus-infected cells, as identified here. Further, we demonstrate here that the RNA-binding activity of the NS1A protein does not have a role in inhibiting the influenza A virus-induced synthesis of IFN-β mRNA, disproving the hypothesis that NS1A dsRNA binding has such a role (5, 24, 34, 35, 39). In contrast, it has been definitively established that the NS1A protein effectively inhibits the production of IFN-β mRNA by another mechanism: it binds CPSF30, thereby inhibiting the production of mature cellular mRNAs, including IFN-β mRNA (6, 7, 13). Recombinant viruses that express an NS1A protein containing a mutated binding site for CPSF30 are highly attenuated and induce the production of a substantially increased amount of IFN-β mRNA (7, 27). In addition, we have shown that this NS1A-binding site is a potential target for the development of antivirals, because blocking the access of CPSF30 for this NS1A site results in increased production of IFN-β mRNA and the inhibition of virus replication (27). Because inhibition of IFN-β mRNA production occurs in the nucleus of influenza A virus-infected cells, a failure of the NS1A protein to enter the nucleus would be expected to result in substantially increased production of IFN-β mRNA. In fact, this is the phenotype of the WSN virus expressing an NS1A R38A, K41A mutant protein that remains in the cytoplasm (24). The phenotype of another recombinant virus expressing a mutant NS1A protein that is largely cytoplasmic (36) can likely be attributed at least in part to the inability of the mutated NS1A protein to carry out its essential nuclear functions.
The RNA-binding domain of the NS1A protein, which is conserved among influenza A virus strains, has several properties that make it a promising target for the development of antivirals: it is required for efficient replication of influenza A virus, as documented here; and its six-helical chain fold is unique and differs from the consensus dsRBDs found in cellular proteins (16–20). Antivirals directed against this viral target would be expected to be effective against all influenza A viruses, including the highly virulent H5N1 viruses.
Materials and Methods
Generation of Recombinant Influenza A/Udorn/72 Virus from Cloned DNA.
Position 38 in the NS1A protein of influenza A/Udorn/72 virus was changed from R to A by PCR mutagenesis, and the resulting DNA was cloned into pHH21. This plasmid, plus the seven pHH21 plasmids encoding the other Udorn genomic RNAs, was cotransfected into 293T cells, along with the four plasmids encoding the polymerase (PB1, PB2, PA) proteins and nucleocapsid protein (NP). Culture supernatants were collected when a positive hemagglutinin (HA) assay titer was observed. Virus was titered by plaque assay on MDCK cells, and individual plaques were amplified in 10-day-old embryonic chicken eggs at 34°C. Amplified virus was titered by plaque assay, and all of the genomic RNA segments were sequenced. The generation of the CPSF mutant virus (L at position 144 of the NS1A protein mutated to A) has been described (27). The influenza A/WSN/33 virus expressing an NS1A protein with a mutated RNA-binding domain (R38A, K41A) was kindly provided by A. Garcia-Sastre (24).
Virus Infection and IFN Treatment.
For multiple cycle growth, MDCK cells were infected at an moi of 0.001 pfu per cell with either WT or R38A mutant influenza A/Udorn/72 and were incubated at 37°C in serum-free DMEM supplemented with 2.5 μg/ml N-acetylated trypsin (NAT). An aliquot of the medium was harvested every 12 h, and virus production was measured by plaque assay in MDCK cells. For single-cycle infection, A549 cells or mouse cells [RNase L+/+ or RNase L−/−, kindly provided by R. Silverman (30)] were infected at an moi of 5 pfu per cell with either WT or R38A mutant influenza A/Udorn/72. After 1 h of incubation, the inoculum was removed, and the cells were washed twice with DMEM and then overlaid with DMEM. The medium was assayed for virus production by plaque assay in MDCK cells. Where indicated, the cells were pretreated with 100 units/ml IFN-β (Berlex Biosciences, Richmond, CA) for 6 h before virus infection.
Immunofluorescence.
HEL 299 cells were infected with the viruses indicated in Fig. 2 at an moi of 3 pfu per cell. At 7 h after infection, cycloheximide (100 μg/ml) was added for 1 h to eliminate the background of newly synthesized NS1A proteins in the cytoplasm. Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.1% Triton X-100 (PBST), and incubated with rabbit anti-NS1A antibody for 1 h at 25°C. The cells were washed with PBST and incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit antibody for 1 h at room temperature. The cells were then examined by confocal microscopy as described (14).
Assay for Viral Protein Synthesis.
A549 cells were infected with either WT or R38A mutant influenza A/Udorn/72 at an moi of 5 pfu per cell. At various times after infection, cells were washed twice with methionine-free DMEM, and 5 μl of a mixture of [35S]methionine and [35S]cysteine (Promix; Amersham Pharmacia) was added in a final volume of 1 ml of serum-free DMEM, followed by incubation for 30 min at 37°C. After incubation, cells were washed twice with PBS and lysed in 200 μl of Laemmli sample buffer. An aliquot was loaded onto 15% SDS-polyacrylamide gels for analysis by autoradiography.
Measurement of IFN-β mRNA by Real-Time Quantitative RT-PCR.
A549 cells were infected with WT, R38A mutant, or CPSF mutant influenza A/Udorn/72 at an moi of 5 pfu per cell. At the times indicated in Fig. 3C, the cells were collected, and total RNA was extracted by using TRIzol (Invitrogen). For each sample, 1 μg of total RNA, which corresponds to equal cell equivalents, was reverse transcribed by using an oligo(dT) primer to generate the DNA complementary to all mRNAs. The amount of IFN-β mRNA was then determined by using the TaqMan Gene Expression Assay (Applied Biosystems) as described (27).
siRNA Interference.
A549 cells were transfected with either a pool of siRNAs specific for human RNase L (SMART pool; Dharmacon Research, Lafayette, CO) or a control siRNA directed against E6AP. The final concentration of siRNA was 100 nM, and the transfection reagent was Oligofectamine (Invitrogen). At 24 h after transfection, the cells were treated with IFN-β for 6 h and then infected with WT or R38A mutant virus. Depletion of RNase L was verified by immunoblotting by using anti-RNase L antibody (Invitrogen), with an immunoblot using anti-tubulin antibody (Santa Cruz Biotechnology) serving as the loading control.
Acknowledgments
We thank Robert H. Silverman for providing RNase L knockout mouse cells. This investigation was supported by National Institutes of Health Grant AI-11772 (to R.M.K.).
Abbreviations
- OAS
oligo (A) synthetase
- CPSF
cleavage and polyadenylation specificity factor
- dsRBD
dsRNA-binding domain
- MDCK
Madin–Darby canine kidney
- moi
multiplicity of infection
- pfu
plaque-forming unit
- NLS
nuclear localization signal
- siRNA
small interfering RNA
- ADAR
dsRNA-specific adenosine deaminase
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wright P. F., Webster R. G. In: Fields Virology. 4th Ed. Knipe D. M., Howley P. M., editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1533–1579. [Google Scholar]
- 2.Reid A. H., Taubenberger J. K., Fanning T. G. Microbes Infect. 2001;3:81–87. doi: 10.1016/s1286-4579(00)01351-4. [DOI] [PubMed] [Google Scholar]
- 3.Horimoto T., Kawaoka Y. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 4.Noah D. L., Krug R. M. In: Advances in Virus Research. Maramorsch K., Shatkin A. J., editors. Vol. 65. Amsterdam: Elsevier; 2005. pp. 121–145. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Sastre A. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 6.Krug R. M., Yuan W., Noah D. L., Latham A. G. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 7.Noah D. L., Twu K. Y., Krug R. M. Virology. 2003;307:386–395. doi: 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 8.Hatada E., Fukuda R. J. Gen. Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y., Wambach M., Katze M. G., Krug R. M. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Riedel K., Lynch P., Chien C. Y., Montelione G. T., Krug R. M. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien C. Y., Xu Y., Xiao R., Aramini J. M., Sahasrabudhe P. V., Krug R. M., Montelione G. T. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- 12.Asada-Kubota M., Ueda T., Shimada M., Takeda K., Sokawa Y. J. Interferon Cytokine Res. 1995;15:863–867. doi: 10.1089/jir.1995.15.863. [DOI] [PubMed] [Google Scholar]
- 13.Nemeroff M. E., Barabino S. M., Li Y., Keller W., Krug R. M. Mol. Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 14.Chen Z., Li Y., Krug R. M. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian X. Y., Chien C. Y., Lu Y., Montelione G. T., Krug R. M. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- 16.Chien C. Y., Tejero R., Huang Y., Zimmerman D. E., Rios C. B., Krug R. M., Montelione G. T. Nat. Struct. Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 17.Liu J., Lynch P. A., Chien C. Y., Montelione G. T., Krug R. M., Berman H. M. Nat. Struct. Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- 18.Nanduri S., Carpick B. W., Yang Y., Williams B. R., Qin J. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryter J. M., Schultz S. C. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos A., Grunert S., Adams J., Micklem D. R., Proctor M. R., Freund S., Bycroft M., St Johnston D., Varani G. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormack S. J., Samuel C. E. Virology. 1995;206:511–519. doi: 10.1016/s0042-6822(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 22.Tian B., Mathews M. B. J. Biol. Chem. 2001;276:9936–9944. doi: 10.1074/jbc.M007328200. [DOI] [PubMed] [Google Scholar]
- 23.Li S., Min J.-Y., Krug R. M., Sen G. C. Virology. 2006 in press. [Google Scholar]
- 24.Donelan N. R., Basler C. F., Garcia-Sastre A. J. Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenspan D., Palese P., Krystal M. J. Virol. 1988;62:3020–3026. doi: 10.1128/jvi.62.8.3020-3026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M. J., Latham A. G., Krug R. M. Proc. Natl. Acad. Sci. USA. 2002;99:10096–10101. doi: 10.1073/pnas.152327499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twu K. Y., Noah D. L., Rao P., Kuo R.-L., Krug R. M. J. Virol. 2006;80:3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen G. C. Semin. Cancer Biol. 2000;10:93–101. doi: 10.1006/scbi.2000.0312. [DOI] [PubMed] [Google Scholar]
- 29.Silverman R. H. In: Ribonucleases: Structure and Functions. D'Alessio G., Riordan J. F., editors. New York: Academic; 1997. pp. 515–551. [Google Scholar]
- 30.Zhou A., Paranjape J., Brown T. L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., Silverman R. H. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hood J. K., Silver P. A. Curr. Opin. Cell Biol. 1999;11:241–247. doi: 10.1016/s0955-0674(99)80032-5. [DOI] [PubMed] [Google Scholar]
- 32.Hartmann R., Justesen J., Sarkar S. N., Sen G. C., Yee V. C. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 33.Wong S. K., Lazinski D. W. Proc. Natl. Acad. Sci. USA. 2002;99:15118–15123. doi: 10.1073/pnas.232416799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talon J., Horvath C. M., Polley R., Basler C. F., Muster T., Palese P., Garcia-Sastre A. J. Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Li M., Zheng H., Muster T., Palese P., Beg A. A., Garcia-Sastre A. J. Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig S., Wang X., Ehrhardt C., Zheng H., Donelan N., Planz O., Pleschka S., Garcia-Sastre A., Heins G., Wolff T. J. Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvatore M., Basler C. F., Parisien J. P., Horvath C. M., Bourmakina S., Zheng H., Muster T., Palese P., Garcia-Sastre A. J. Virol. 2002;76:1206–1212. doi: 10.1128/JVI.76.3.1206-1212.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W. X., Li H., Lu R., Li F., Dus M., Atkinson P., Brydon E. W., Johnson K. L., Garcia-Sastre A., Ball L. A., et al. Proc. Natl. Acad. Sci. USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Basler C. F., Williams B. R., Silverman R. H., Palese P., Garcia-Sastre A. J. Virol. 2002;76:12951–12962. doi: 10.1128/JVI.76.24.12951-12962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]