Abstract
Sustained activation of poly(ADP-ribose) polymerase-1 (PARP-1) and extracellular signal-regulated kinases 1/2 (ERK1/2) both promote neuronal death. Here we identify a direct link between these two cell death pathways. In a rat model of hypoglycemic brain injury, neuronal PARP-1 activation and subsequent neuronal death were blocked by the ERK1/2 inhibitor 2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one (PD98059). In neuron cultures, PARP-1-mediated neuronal death induced by N-methyl-d-aspartate, peroxynitrite, or DNA alkylation was similarly blocked by ERK1/2 pathway inhibitors. These inhibitors also blocked PARP-1 activation and PARP-1-mediated death in astrocytes. siRNA down-regulation of ERK2 expression in astrocytes also blocked PARP-1 activation and cell death. Direct effects of ERK1/2 on PARP-1 were evaluated by using isolated recombinant enzymes. The activity of recombinant human PARP-1 was reduced by incubation with alkaline phosphatase and restored by incubation with active ERK1 or ERK2. Putative ERK1/2 phosphorylation sites on PARP-1 were identified by mass spectrometry. Using site-directed mutagenesis, these sites were replaced with alanine (S372A and T373A) to block phosphorylation, or with glutamate (S372E and T373E) to mimic constitutive phosphorylation. Transfection of PARP-1 deficient mouse embryonic fibroblasts with the mutant PARP-1 species showed that the S372A and T373A mutations impaired PARP-1 activation, whereas the S372E and T373E mutations increased PARP-1 activity and eliminated the effect of ERK1/2 inhibitors on PARP-1 activation. These results suggest that PARP1 phosphorylation by ERK1/2 is required for maximal PARP-1 activation after DNA damage.
Keywords: DNA damage, hypoglycemia, mitogen-activated protein kinase, astrocyte, neuron
Poly(ADP-ribose) polymerase-1 (PARP-1) is an abundant nuclear enzyme that is activated by DNA strand breaks and functionally linked to DNA repair (1). PARP-1 activation in neurons occurs in ischemia, trauma, hypoglycemia, excitotoxicity, multiple sclerosis, and other conditions involving oxidative stress or inflammation (2–5). When activated, PARP-1 consumes NAD to form poly(ADP-ribose) (PAR) on specific acceptor proteins. The poly(ADP-ribosyl)ation modifies protein binding to DNA and other proteins to facilitate DNA repair and prevent chromatid exchange (1, 6). However, extensive activation of PARP-1 leads to NAD depletion and cell death (7–9). PARP inhibitors and PARP-1 gene deletion can markedly improve neuronal survival in ischemia and other disorders (10–12). PARP-1 also interacts with several transcription factors (13), and the coactivation of nuclear factor κB (NF-κB) by PARP-1 promotes the cellular inflammatory response (14–16).
Like PARP-1, the extracellular signal-regulated kinases 1 and 2 (ERK1/2) are activated by ischemia and other conditions that generate oxidative stress (17–22). ERK1/2, p38, and c-Jun-N-terminal kinase (JNK) comprise the major subgroups of the mitogen-activated protein kinases (MAPKs) (23), which are serine/threonine kinases involved in diverse aspects of signal transduction. ERK1/2 are activated when phosphorylated by the upstream MAPK/ERK kinases 1 and 2 (MEK1/2). Early studies suggested that p38 and JNK activation promoted cell death, whereas ERK1/2 activation promoted cell survival, growth, and differentiation (24); however, an increasing body of evidence suggests that ERK1/2 activation can also promote neuronal death, particularly when oxidative stress induces sustained ERK1/2 activation (as reviewed in ref. 22). Inhibitors of ERK1/2 activation can markedly improve neuronal survival under these conditions (17–22), but the relevant downstream phosphorylation targets and the mechanisms by which survival is influenced by ERK1/2 activation remain poorly defined.
Here we demonstrate that pharmacological inhibition of ERK1/2 prevents PARP-1 activation and reduces PARP-1 mediated neuronal death, both in cell culture and in a rat model of hypoglycemia. siRNA down-regulation of ERK2 also blocks PARP-1 activation, and mass spectrometry identified serine and threonine residues phosphorylated by ERK2. Substitution of these residues with either unphosphorylatable or phosphomimetic residues had striking effects on PARP-1 activation and response to ERK1/2 inhibitors. Given the central role of PARP-1 in both cell death and inflammation, these results suggest that effects of ERK1/2 on PARP-1 activity may be a general mechanism by which ERK1/2 influences cell survival.
Results
Inhibition of the MEK1/2–ERK1/2 Cascade Prevents PARP-1-Mediated Cell Death.
The alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) triggers PARP-1 activation and PARP-1-mediated cell death in cultured neurons and astrocytes (8, 25). To determine whether MAPKs influence the PARP-1 cell death program, cortical neuron cultures were incubated with MNNG along with inhibitors of the three main MAPK subgroups. 2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one (PD98059) and 1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene (U0126), which inhibit MEK1/2 activity and thereby block phosphorylation and activation of ERK1/2, prevented neuron death at 10-μM concentrations (Fig. 1A). By contrast, neuron death was not reduced by a 10 μM concentration of 4-[5-(4-fluorophenyl)-2-[4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl]pyridine (SB203580) or anthra[1–9-cd]pyrazol-6(2H)-one (SP600125), which inhibit p38 and JNK, respectively. Neuronal death induced by MNNG was also prevented by the PARP inhibitor 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone (DPQ), confirming the dominant role for PARP-1 activation in this cell death pathway (8, 25).
Fig. 1.
Inhibition of ERK1/2 prevents PARP-1-mediated cell death. (A) Neuron death induced by MNNG is blocked by the PARP inhibitor DPQ (25 μM) and by the MEK1/2 inhibitors PD98059 (P, 10 μM) and U0126 (U, 10 μM), but not by inhibitors of p38 [SB203580 (SB), 10 μM] or JNK [SP600125 (SP), 10 μM]. (B and C) Neuronal death induced by SIN-1 (B) or NMDA (C) was blocked by PD to an extent similar to that achieved with DPQ. (D) Death of astrocytes incubated with MNNG was prevented by the PARP inhibitor DPQ (25 μM) and the MEK1/2 inhibitors PD and U, but not by the p38 inhibitor SB or the JNK inhibitor SP (each at 10 μM). (E) siRNA targeting ERK2 alone or together with ERK1 reduced MNNG-induced astrocyte death, whereas mismatch siRNA or siRNA to ERK1 alone did not. The immunoblot shows representative duplicate assessments of ERK1 and ERK2 protein expression under each of the designated siRNA treatment conditions. n = 3; ∗, P < 0.05 vs. MNNG, SIN-1, or NMDA alone.
PARP-1 activation also mediates neuronal death induced by N-methyl-d-aspartate (NMDA) and peroxynitrite (2, 3). The influence of ERK1/2 activation on PAPR-1-mediated cell death in these settings was evaluated in neuronal cultures treated with NMDA or the peroxynitrite generator SIN-1. As with MNNG, neuron death induced by these agents was blocked by PD98059 with efficacy comparable with that achieved by 25 μM DPQ (Fig. 1 B and C). Higher concentrations of DPQ showed no greater effect (data not shown). PARP-1-mediated astrocyte death was also blocked by the ERK1/2 inhibitors (Fig. 1D).
Although PD98059 and U0126 are considered selective inhibitors of the ERK1/2 pathway at the 10-μM concentration used for these studies, these compounds can have other pharmacological effects (26). The possibility that PD98059, U0126, or the DMSO vehicle might have direct effects on PARP-1, independent of their effects on ERK1/2, was evaluated by using recombinant PARP-1 in a cell-free activity assay. None of these compounds had a significant direct effect on PARP-1 activity (data not shown).
To further confirm the effects of ERK1/2 on PARP-1-mediated cell death, we used siRNA to down-regulate ERK1 and ERK2 expression in the astrocyte cultures. Astrocyte cultures were used for the siRNA studies because these cells can be grown as homogenous monotype cultures, display the same biochemical responses to PARP-1 activation as neurons, are more reproducibly responsive to siRNA, and are less susceptible to nonspecific effects of siRNA delivery. Western blotting showed ERK1 expression was reduced by 72 ± 8% and ERK2 expression was reduced by 72 ± 5% (n = 4) at day 4 after the siRNA incubations. Down-regulation of both ERK1 and ERK2 together reduced MNNG-induced astrocyte death. (Fig. 1E). When siRNA to ERK1 and ERK2 were used separately, down-regulation of ERK2, but not ERK1, reduced cell death.
Effects of ERK1/2 on PARP-1 Enzymatic Activity.
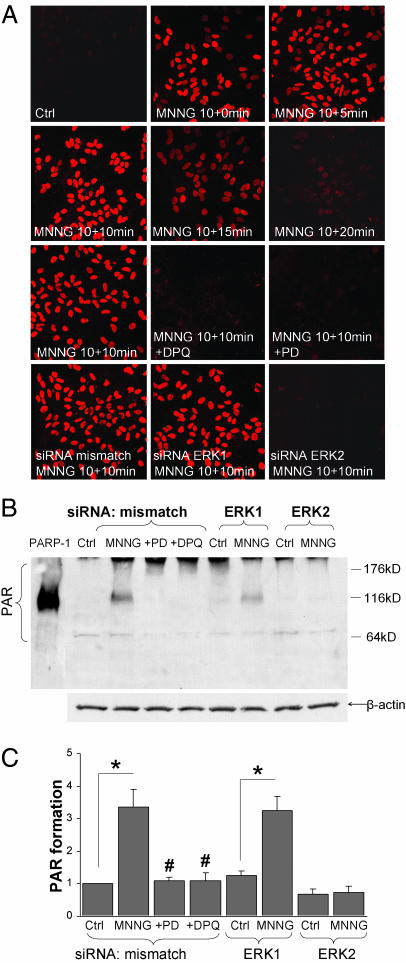
ERK1/2 activation could prevent PARP-1-mediated cell death either by blocking PARP-1 enzymatic activity or by blocking other, downstream events in this cell death pathway (9, 27). We evaluated PARP-1 activity by measuring the enzymatic product of PARP-1, PAR, in the presence and absence of ERK1/2 inhibition. Immunostaining showed PAR formation in astrocyte cultures within 5 min of a 10-min incubation with 100 μM MNNG. PAR accumulation peaked at 10 min and dissipated after 15 min (Fig. 2A). Both the MEK1/2–ERK1/2 inhibitor PD98059 and siRNA down-regulation of ERK2 (but not ERK1) blocked PAR formation at each of the time points examined. Effects of these agents at the peak 10-min time point are shown in Fig. 2A. PAR formation quantified by immunoblotting at the 10-min time point confirmed the pattern observed with immunostaining (Fig. 2 B and C).
Fig. 2.
Inhibition of the MEK–ERK pathway prevents PARP-1 activation. (A) Astrocyte cultures immunostained for poly(ADP-ribose) (PAR) at serial time points after a 10-min incubation with 100 μM MNNG. PAR formation is prevented by DPQ (25 μM) and by the MEK1/2 inhibitor PD98059 (10 μM). siRNA down-regulation of ERK2 also reduced PAR formation, whereas down-regulation of ERK1 had no effect. Representative of n = 4. (B) Western blots were prepared from cultures harvested 10 min after 10-min incubations with MNNG or control. MNNG induced PAR immunoreactivity at 116 kDa, corresponding to PARP-1 automodification, and a more diffuse signal at higher molecular masses corresponding to poly(ADP-ribosyl)ation of many protein targets. Recombinant PARP-1 was loaded in the far left lane as a positive control for autopoly(ADP-ribosyl)ation of PARP-1. Signal at the extreme top of the gel is a loading artifact. (C) PAR immunoreactivity under each condition is quantified. n = 4; ∗, P < 0.01 vs. control (Ctrl); #, P < 0.01 vs. the MNNG/mismatch siRNA condition.
ERK1 and ERK2 Regulate PARP-1 Activity by Direct Phosphorylation.
Dephosphorylation of recombinant human PARP-1 with calf intestinal alkaline phosphatase (CIAP) reduced enzymatic activity by ≈75%, whereas preincubation with heat-inactivated CIAP had no effect (Fig. 3A). These results suggest that phosphorylation is required for maximal PARP-1 activity and, further, that the commercially obtained recombinant PARP-1 is phosphorylated at a site (or sites) that up-regulate enzymatic activity. PARP-1 activity was approximately doubled in the presence of histones, as expected (28), and the effect of CIAP treatment was proportionally the same in the presence or absence of histones (Fig. 3A). Studies performed in the absence of DNA showed no detectable PAR synthesis (data not shown). Consequently, all subsequent PARP-1 activity assays were done in the presence of DNA, but in the absence of histones to exclude possible effects of histone phosphorylation by ERK1/2.
Fig. 3.
Recombinant PARP-1 is phosphorylated and regulated by ERK1 and ERK2. (A) PARP-1 activity was reduced by preincubation with CIAP but not by heat-inactivated CIAP (HI-CIAP). Similar results were obtained in the presence and absence of histones. (B) After CIAP dephosphorylation, enzymatic activity was restored by incubation with 100 ng of ERK1 or ERK2. (n = 3–5; ∗, P < 0.05 vs. PARP-1 + CIAP). (C) Phosphorylation of PARP-1 was assessed after incubation with [32P]ATP substrate and 100 ng of ERK1 or ERK2. Lanes 1–3 of the radiography gel show controls for nonspecific ATP binding; lanes 4 and 5 show ERK1 and ERK2 autophosphorylation; lanes 6 and 7 are positive controls for kinase activity, showing ERK1 and ERK2 phosphorylation of myelin basic protein (MBP); and lanes 8 and 9 show phosphorylation of CIAP-treated PARP-1 by both ERK1 and ERK2. Similar results were obtained in three independent experiments. Equal loading of PARP-1 in the designated lanes was confirmed with Coomassie blue staining.
To determine whether ERK1/2 can phosphorylate and regulate PARP-1 directly, recombinant PARP-1 activity was measured after incubation with active ERK1 or ERK2 in the presence of 500 μM ATP. As shown in Fig. 3B, the loss of enzymatic activity resulting from CIAP dephosphorylation was reversed by subsequent incubation with ERK1 or ERK2 but not by incubation with heat-inactivated ERK1 or ERK2. PARP-1 that had not been treated with CIAP did not increase enzymatic activity after incubation with ERK1 or ERK2 (data not shown). This same sequence of studies was performed by using [32P]ATP substrate to further confirm direct phosphorylation of PARP-1 by ERK1/2. Both ERK1 and ERK2 were found to transfer 32P to PARP-1 (Fig. 3C). 32P labeling was not observed in the absence of ERK1/2, and, as expected, ERK1 and ERK2 produced both autophosphorylation and robust phosphorylation of myelin basic protein (29).
To determine whether the phosphorylation status of PARP-1 is altered by ERK1/2 during activation in situ, PARP-1 was immunoprecipitated from astrocytes after treatment with MNNG, MNNG plus PD98059, or washes only. The recovered PARP-1 was probed with antibodies recognizing phosphothreonine and phosphoserine. Changes in phosphothreonine content were not significantly different among the three treatment groups (data not shown); however, phosphoserine content was significantly increased in cultures treated with MNNG, and this increase was prevented by coincubation with the MEK1/2 inhibitor, PD98059 (Fig. 5, which is published as supporting information on the PNAS web site). Immunoblots prepared from astrocytes at sequential time points after MNNG incubation confirmed a rapid up-regulation of ERK1/2 activity, as evidenced by a rapid increase in the phosphorylated (active) fraction. Inhibition of PARP-1 with DPQ did not affect ERK1/2 activity, whereas PD98059 did, as expected, prevent phosphorylation of ERK1/2 (Fig. 6, which is published as supporting information on the PNAS web site).
Sites of ERK2 Phosphorylation on PARP-1.
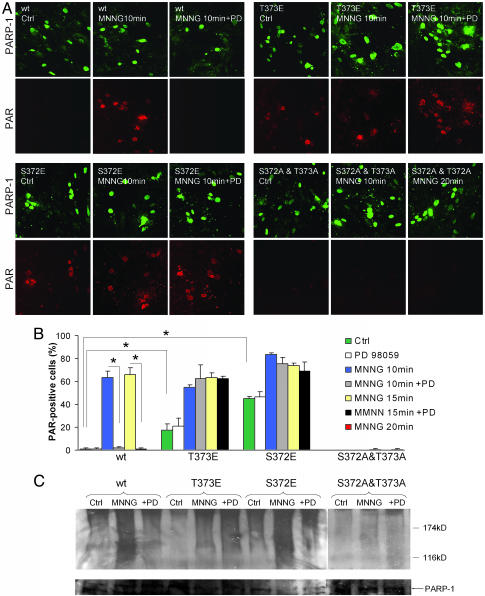
Tandem mass spectrometry was used to analyze phosphorylation sites on human PARP-1 after incubation with ERK1 and ERK2. No phosphorylation was detected with ERK1, but treatment with ERK2 produced a change in the mass of a single peptide fragment consistent with the addition of two phosphate groups. This peptide contained the potential phosphorylation sites T368, S372, and T373. To further evaluate these putative sites, we transfected PARP-1−/− mouse embryonic fibroblast (MEF) cells with PARP-1 constructs in which these residues were replaced by alanine to prevent phosphorylation. These constructs were expressed in PARP-1−/− MEF cells, and PARP-1 activity was assessed by PAR immunostaining (Fig. 4A). PAR formation with the T368A substitution was not discernibly different from wild-type PARP-1; however, the S372A and T373A substitutions each produced a partial reduction in MNNG-induced PAR formation (data not shown), and substitution of both S372 and T373 with alanine completely prevented PAR formation (Fig. 4 A and B). We additionally transfected the PARP-1−/− MEF cells with T368E, S372E, and T373E PARP-1 constructs to mimic continuous phosphorylation at these sites. Again, substitution at the T368 residue had no effect, but cells transfected with either the S372E or the T373E substitutions showed increased basal PARP-1 activity (Fig. 4 A and B). Most importantly, PARP-1 activation in these cells was not blocked by the MEK–ERK1/2 pathway inhibitor PD98059, implicating these sites as targets for ERK1/2 phosphorylation. Western blots prepared under these same conditions corroborated the results obtained by immunostaining (Fig. 4C).
Fig. 4.
The effect of ERK1/2 blockade on PARP-1 activity is abrogated by mutations at S372 and T373 in human PARP-1. (A) Immunostaining shows PARP-1 expression (green) and PAR formation (red) in PARP-1−/− MEF cells transfected with wild-type human PARP-1 or with mutant PARP-1 constructs prepared by site-directed mutagenesis. In all cases, PAR accumulation peaked at 10–20 min after a 10-min incubation with 75 μM MNNG. Cells transfected with the S372E or T373E mutants showed enhanced basal PAR formation and, in contrast to wild-type PARP-1, no inhibition of PAR formation in the presence of PD98059 (PD). Conversely, the double S372A and T373A mutation prevented PARP-1 activation. (B) Quantified data show the percent of PARP-1 expressing cells that had detectable PAR formation at 10, 15, or 20 min after the MNNG incubation, with and without the presence of the PD98059. The 20-min time point was analyzed only for cells transfected with the S372A and T373A construct (far right-hand bar). ∗, P < 0.01; n = 3. (C) Western blots of PAR formation corroborated the pattern seen with immunostaining. Poly(ADP-ribosyl)ated proteins form a smear in the high-molecular-mass regions of the blots. The panel is a composite of lanes run on two separate gels. PARP-1 expression in each of the treatment conditions was assessed by reprobing the membranes with antibody to PARP-1 (Lower).
Inhibition of ERK1/2 Activation Blocks PARP-1 Activation and Neuronal Death After Severe Hypoglycemia.
Hypoglycemia causes PARP-1 activation and PARP-1-mediated neuronal death in selectively vulnerable neuron populations (4, 30). The selectivity of this insult, and the near-complete abrogation of hypoglycemic neuronal death with PARP inhibitors (4), make hypoglycemia a useful in vivo model of PARP-1-mediated neuron death. To determine whether the MEK1/2–ERK1/2 pathway contributes to PARP-1 activation and neuron death in vivo, rats were given an intracerebroventricular injection of 1 nmol of PD90859 (17) immediately before onset of hypoglycemia. Peak brain concentrations were estimated to be <5 μM PD98059 and 0.01% DMSO vehicle. Activation of PARP-1 and ERK1/2 were assessed in brains harvested 3 h after hypoglycemia by immunostaining for PAR and for phosphoERK1/2. PAR and phosphoERK1/2 accumulated in the neuronal populations most vulnerable to hypoglycemic injury, and both PAR and phosphoERK1/2 formation were blocked in rats receiving PD98059 (Fig. 7, which is published as supporting information on the PNAS web site). PhosphoERK1/2 immunoreactivity was evident in neuronal cytoplasm but was most prominent in the nuclei. Hematoxylin-eosin staining of brains harvested 7 days after hypoglycemia showed a large decrease in neuronal death in the rats treated with the MEK1/2 inhibitor, whereas treatment with vehicle alone (DMSO) had no effect (Table 1, which is published as supporting information on the PNAS web site).
Discussion
Inhibition of ERK1/2 can reduce neuronal death in several settings known to cause extensive PARP-1 activation (17, 19, 31). Here we show that ERK1/2 activation is required both for PARP-1 activation and for PARP-1-induced neuron and astrocyte death. In vivo, ERK1/2 inhibition during severe hypoglycemia blocked PAR formation and neuronal death under conditions in which the neuronal death is almost entirely PARP-1 dependent. In cultured cells, PARP-1-mediated death of neurons induced by MNNG, SIN-1, or NMDA was completely blocked by two chemically distinct inhibitors of the MEK1/2–ERK1/2 pathway. PARP-1-mediated death of astrocytes was also blocked by pharmacological ERK1/2 inhibition, and siRNA-mediated down-regulation of astrocyte ERK2 protein expression had a cytoprotective effect comparable with that of the pharmacological inhibitors. These studies are germane to the many prior reports that identify neuroprotective effects of ERK1/2 inhibition in ischemia and other conditions (22).
More direct evidence for ERK1/2 regulation of PARP-1 activity was provided by immunoblot and immunohistochemistry studies showing reduced PAR formation with either pharmacological ERK1/2 inhibition or siRNA down-regulation of ERK2 expression. A direct effect of both ERK1 and ERK2 on PARP-1 activity was confirmed by studies using recombinant enzymes. These studies showed that ERK1 and ERK2 can directly phosphorylate PARP-1 and increase PARP-1 activity relative to its dephosphorylated state. In addition, immunoprecipitation of activated PARP-1 from astrocytes showed an increase in phosphoserine content that was blocked by ERK1/2 inhibition. Last, amino acid substitutions at putative ERK2 phosphorylation sites on PARP-1 produced dramatic changes in PARP-1 activity.
Substitution of S372 and T373 with alanine to prevent phosphorylation generated PARP-1 species with reduced activation after DNA damage. This finding suggests that phosphorylation at these sites is needed for maximal PARP-1 activation; however, it is also possible that the reduced PARP-1 activity results from conformational changes induced by the amino acid substitutions. Stronger support for ERK1/2 phosphorylation at these sites is provided by the finding that replacing either of these residues with glutamate, to mimic constitutive phosphorylation, generated PARP-1 species that were no longer responsive to ERK1/2 inhibition. Of note, these residues are located near the beginning of the BRCT portion of the automodification domain, which is known to modulate PARP-1 activity and binding to DNA strand breaks (6). The S372E and T373E mutations also increased the basal level of PAR formation in transfected MEF cells. This increase may result from an amplified PARP-1 response to basal levels of DNA damage or, alternatively, may reflect an increase in PARP-1 activity that occurs independent of DNA damage (32, 33).
Although ERK1 and ERK2 were both shown to phosphorylate and activate recombinant PARP-1, only ERK2 down-regulation led to reduced PAR formation and PARP-1-mediated cell death in the siRNA studies, despite a comparable, 70–75% reduction in both ERK1 and ERK2 protein expression. Similarly, ERK2, but not ERK1, produced detectable phosphorylation of PARP-1 when assessed by mass spectrometry. Prior studies have also suggested that ERK2 has a greater influence than ERK1 on cell survival under stress conditions (18, 34). Our findings provide evidence for a direct interaction between ERK2 and PARP-1 but are not conclusive regarding interactions between ERK1 and PARP-1 in situ.
Regulation of PARP-1 activity by phosphorylation has been described. Aoufouchi and Shall (35) reported that activity of PARP-1 in developing Xenopus oocytes required phosphorylation of PARP-1 at serine residues. Although the kinase was not identified, ERK1/2 is a plausible candidate given that ERK1/2 exhibits sustained activation during the oocyte maturation process (36). Evidence also suggests that PARP-1 may be phosphorylated by protein kinase C, with a resultant down-regulation of PARP-1 activity by that modification (37, 38). The present results suggest that ERK1/2 activation is a prerequisite for maximal PARP-1 activation after DNA damage. The ERK1/2 signaling pathway is itself activated during DNA damage, through a p53-independent mechanism (39). The ERK1/2 pathway can also be activated at multiple steps by reactive oxygen species (22). Whether ERK1/2 activation is always required for maximal PARP-1 activation remains uncertain, however, because PARP-1 activation is reported in settings that may not involve concomitant ERK1/2 activation (32, 33). Our observation that recombinant human PARP-1 prepared in Escherichia coli is active, but loses activity when treated with alkaline phosphatase, indicates that kinases other than ERK1/2 (which are not expressed in bacteria) can activate PARP-1. PARP-1 purified from mammalian cells is generally active, suggesting that basal ERK1/2 activity is sufficient for measurable PARP-1 activity or that other pathways for PARP-1 regulation exist.
Hypoglycemia produces PARP-1-mediated neuronal death in selectively vulnerable neuron populations (4). Here we showed that hypoglycemia also produces neuronal ERK1/2 phosphorylation (activation). The MEK1/2 inhibitor PD98059 blocked ERK1/2 phosphorylation during hypoglycemia and also blocked PARP-1 activation and subsequent cell death in these neuronal populations. These findings, together with the cell culture and cell-free enzyme studies, suggest that the neuroprotective effects of ERK1/2 inhibition in hypoglycemia are largely attributable to reduced PARP-1 activation. Given that PARP-1 has a critical influence on neuronal survival in ischemia, excitotoxicity, inflammation, and many other conditions, ERK1/2 regulation of PARP-1 activity may be a common and important pathway by which the MEK1/2–ERK1/2 signal cascade influences neuronal survival.
Methods
Reagents.
DPQ was obtained from Calbiochem. PD98059, U0126SB, SB203580, and SP600125 were from Tocris Cookson (Ellisville, MO); rabbit polyclonal and mouse monoclonal anti-PAR (clone 10H), mouse monoclonal anti-PARP-1 (clone C2–10), and recombinant human PARP-1 were from Trevigen (Gaithersburg, MD). Rabbit polyclonal anti-ERK1/2 and anti-phosphoERK1/2 polyclonal antibodies were from Cell Signaling Technology (Beverly, MA). Rabbit anti-phosphoserine and anti-phosphothreonine were from Zymed. Cell culture reagents were obtained from Mediatech (Herndon, VA), and all other reagents were from Sigma/Aldrich except where stated.
Cell Culture Procedures.
Astrocyte and astrocyte–neuron cocultures were prepared as described (40, 41). The cocultures were used on days 12–14 in vitro. PARP-1-deficient MEFs (PARP-1−/− MEF), kindly provided by Dr. Z. Q. Wang [International Agency for Research on Cancer (IACR), Lyon, France] and Dr. Y. Wang (Thomas Jefferson University, Philadelphia), were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FBS. Studies were initiated by placing the cultures in a physiological balanced salt solution (BSS) containing 3.1 mM KCl, 134 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgSO4, 0.25 mM KH2PO4, 15.7 mM NaHCO3, and 2 mM glucose, preequilibrated to pH 7.2 in a 5% CO2 atmosphere. Incubations were terminated by complete medium exchange and replacement with BSS for astrocyte or MEF cultures or with glial-conditioned medium for the astrocyte–neuron cocultures. Control wells received medium exchanges only. Viability of the astrocyte monocultures was evaluated 24 h after drug treatments by the lactate dehydrogenase (LDH) method (40, 42). Neuronal viability was assessed by the propidium iodide (PI) exclusion method (25), in which the number of PI-stained (dead) and live cells in five randomly picked optical fields of each well were counted to calculate percent neuron survival in each well. Fixation and immunostaining of cell cultures was performed as described (16).
siRNA Down-Regulation of ERK1/2.
Astrocytes were incubated with siRNA transfection complexes at day 10 in vitro, at which time they were 95% confluent. siRNA incubations were performed with RNAiFect reagents (Qiagen, Valencia, CA) according to the manufacturer's instructions. The siRNA sense sequence for ERK1 was 5′-ACAAGCGCAUCACAGUAGAtt-3′ and for ERK2 was 5′-CAAAGUUCGAGUUGCUAUCtt-3′ (Ambion, Austin, TX). Controls were prepared with a mismatch sequence lacking significant homology to any known mouse gene sequences. The siRNA complexes were removed after 6 h, and cultures were used for experiments 4 days later.
Western Blotting and Immunoprecipitation.
Western blots were prepared and quantified as described (16). Membranes were reprobed with anti β-actin to quantify protein loading, and band densities were quantified with the National Institutes of Health image j program. Controls performed in the absence of primary or secondary antibodies showed no signal. For immunoprecipitation, cells were lysed in the presence of protease inhibitors, and the lysates were precleaned by incubation with protein G plus agarose beads (Calbiochem). The target protein, PARP-1, was captured by using mouse monoclonal anti-PARP-1 antibody complexed to protein G-agarose beads. Phosphorylation status of the recovered PARP-1 was analyzed by Western blotting. Recombinant PARP-1 was applied as a control for correct molecular weight, and Coomassie brilliant blue was used to quantify protein loading.
Statistics.
For cell culture studies, each “n” denotes an independent experiment composed of three to four parallel treatments per condition. For the in vivo studies, each “n” denotes the summed measurements from an individual animal. Results are presented as a means ± standard error. Statistical significance was evaluated by one-way ANOVA followed by the Student–Neuman–Keuls' test for comparisons between multiple treatment groups or Dunnett's test for comparisons of multiple treatment groups against a common control group.
Additional methods for the PARP-1 phosphorylation and activity assays and rat hypoglycemia studies are in Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Susana Castro-Obregon for assistance with the siRNA studies, Aaron Hamby and Andreu Viader Valls for technical assistance, and Stephen Massa for critical suggestions and reading of the manuscript. The work was supported by National Institutes of Health Grant NS41421 and the Department of Veterans Affairs (both to R.A.S.) and the Finnish Cultural Foundation, Saastamoinen Foundation, and Sigrid Juselius Foundation (all to T.M.K.).
Abbreviations
- CIAP
calf intestinal alkaline phosphatase
- DPQ
3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone
- ERK1/2
extracellular signal-regulated kinases 1 and 2
- JNK
c-Jun-N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MEF
mouse embryonic fibroblast
- MEK1/2
MAPK/ERK kinases 1 and 2
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidine
- NMDA
N-methyl-d-aspartate
- PAR
poly(ADP-ribose)
- PARP-1
poly(ADP-ribose) polymerase-1
- PD98059
2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one
- SB203580
4-[5-(4-fluorophenyl)-2-[4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl]pyridine
- SP600125
anthra[1–9-cd]pyrazol-6(2H)-one
- U0126
1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Malanga M., Althaus F. R. Biochem. Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 2.Mandir A. S., Poitras M. F., Berliner A. R., Herring W. J., Guastella D. B., Feldman A., Poirier G. G., Wang Z. Q., Dawson T. M., Dawson V. L. J. Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo C., Dawson V. L. Trends Pharmacol. Sci. 1998;19:287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 4.Suh S. W., Aoyama K., Chen Y., Garnier P., Matsumori Y., Gum E., Liu J., Swanson R. A. J. Neurosci. 2003;23:10681–10690. doi: 10.1523/JNEUROSCI.23-33-10681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kauppinen T. M., Suh S. W., Genain C. P., Swanson R. A. J. Neurosci. Res. 2005;81:190–198. doi: 10.1002/jnr.20525. [DOI] [PubMed] [Google Scholar]
- 6.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 7.Berger N. A. Radiat. Res. 1985;101:4–15. [PubMed] [Google Scholar]
- 8.Ha H. C., Snyder S. H. Proc. Natl. Acad. Sci. USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 10.Endres M., Wang Z. Q., Namura S., Waeber C., Moskowitz M. A. J. Cereb. Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Eliasson M. J., Sampei K., Mandir A. S., Hurn P. D., Traystman R. J., Bao J., Pieper A., Wang Z. Q., Dawson T. M., Snyder S. H., Dawson V. L. Nat. Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 12.Virag L., Szabo C. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 13.Ha H. C., Hester L. D., Snyder S. H. Proc. Natl. Acad. Sci. USA. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ullrich O., Diestel A., Eyupoglu I. Y., Nitsch R. Nat. Cell Biol. 2001;3:1035–1042. doi: 10.1038/ncb1201-1035. [DOI] [PubMed] [Google Scholar]
- 15.Chiarugi A., Moskowitz M. A. J. Neurochem. 2003;85:306–317. doi: 10.1046/j.1471-4159.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 16.Kauppinen T. M., Swanson R. A. J. Immunol. 2005;174:2288–2296. doi: 10.4049/jimmunol.174.4.2288. [DOI] [PubMed] [Google Scholar]
- 17.Alessandrini A., Namura S., Moskowitz M. A., Bonventre J. V. Proc. Natl. Acad. Sci. USA. 1999;96:12866–12869. doi: 10.1073/pnas.96.22.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Namura S., Iihara K., Takami S., Nagata I., Kikuchi H., Matsushita K., Moskowitz M. A., Bonventre J. V., Alessandrini A. Proc. Natl. Acad. Sci. USA. 2001;98:11569–11574. doi: 10.1073/pnas.181213498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh-hashi K., Maruyama W., Yi H., Takahashi T., Naoi M., Isobe K. Biochem. Biophys. Res. Commun. 1999;263:504–509. doi: 10.1006/bbrc.1999.1237. [DOI] [PubMed] [Google Scholar]
- 20.Stanciu M., Wang Y., Kentor R., Burke N., Watkins S., Kress G., Reynolds I., Klann E., Angiolieri M. R., Johnson J. W., DeFranco D. B. J. Biol. Chem. 2000;275:12200–12206. doi: 10.1074/jbc.275.16.12200. [DOI] [PubMed] [Google Scholar]
- 21.Hu B. R., Wieloch T. J. Neurochem. 1994;62:1357–1367. doi: 10.1046/j.1471-4159.1994.62041357.x. [DOI] [PubMed] [Google Scholar]
- 22.Chu C. T., Levinthal D. J., Kulich S. M., Chalovich E. M., DeFranco D. B. Eur. J. Biochem. 2004;271:2060–2066. doi: 10.1111/j.1432-1033.2004.04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H. Chem. Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 24.Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 25.Ying W., Sevigny M. B., Chen Y., Swanson R. A. Proc. Natl. Acad. Sci. USA. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.English J. M., Cobb M. H. Trends Pharmacol. Sci. 2002;23:40–45. doi: 10.1016/s0165-6147(00)01865-4. [DOI] [PubMed] [Google Scholar]
- 27.Alano C. C., Ying W., Swanson R. A. J. Biol. Chem. 2004;279:18895–18902. doi: 10.1074/jbc.M313329200. [DOI] [PubMed] [Google Scholar]
- 28.Kun E., Kirsten E., Mendeleyev J., Ordahl C. P. Biochemistry. 2004;43:210–216. doi: 10.1021/bi0301791. [DOI] [PubMed] [Google Scholar]
- 29.Alessandrini A., Crews C. M., Erikson R. L. Proc. Natl. Acad. Sci. USA. 1992;89:8200–8204. doi: 10.1073/pnas.89.17.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auer R. N., Siesjo B. K. Baillieres Clin. Endocrinol. Metab. 1993;7:611–625. doi: 10.1016/s0950-351x(05)80210-1. [DOI] [PubMed] [Google Scholar]
- 31.Mori T., Wang X., Aoki T., Lo E. H. J. Neurotrauma. 2002;19:1411–1419. doi: 10.1089/089771502320914642. [DOI] [PubMed] [Google Scholar]
- 32.Cohen-Armon M., Visochek L., Katzoff A., Levitan D., Susswein A. J., Klein R., Valbrun M., Schwartz J. H. Science. 2004;304:1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- 33.Tulin A., Spradling A. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 34.Castro-Obregon S., Del Rio G., Chen S. F., Swanson R. A., Frankowski H., Rao R. V., Stoka V., Vesce S., Nicholls D. G., Bredesen D. E. Cell Death Differ. 2002;9:807–817. doi: 10.1038/sj.cdd.4401035. [DOI] [PubMed] [Google Scholar]
- 35.Aoufouchi S., Shall S. Biochem. J. 1997;325:543–551. doi: 10.1042/bj3250543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer A., Nebreda A. R. Prog. Cell Cycle Res. 2000;4:131–143. doi: 10.1007/978-1-4615-4253-7_12. [DOI] [PubMed] [Google Scholar]
- 37.Bauer P. I., Farkas G., Buday L., Mikala G., Meszaros G., Kun E., Farago A. Biochem. Biophys. Res. Commun. 1992;187:730–736. doi: 10.1016/0006-291x(92)91256-p. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y., Koide S. S., Yoshihara K., Kamiya T. Biochem. Biophys. Res. Commun. 1987;148:709–717. doi: 10.1016/0006-291x(87)90934-x. [DOI] [PubMed] [Google Scholar]
- 39.Tang D., Wu D., Hirao A., Lahti J. M., Liu L., Mazza B., Kidd V. J., Mak T. W., Ingram A. J. J. Biol. Chem. 2002;277:12710–12717. doi: 10.1074/jbc.M111598200. [DOI] [PubMed] [Google Scholar]
- 40.Swanson R. A., Farrell K., Stein B. A. Glia. 1997;21:142–153. doi: 10.1002/(sici)1098-1136(199709)21:1<142::aid-glia16>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Swanson R. A. J. Neurochem. 2003;84:1332–1339. doi: 10.1046/j.1471-4159.2003.01630.x. [DOI] [PubMed] [Google Scholar]
- 42.Koh J. Y., Choi D. W. J. Neurosci. Methods. 1987;20:83–90. doi: 10.1016/0165-0270(87)90041-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.