Abstract
Alternative splicing (AS) has been extensively studied in mammalian systems but much less in plants. Here we report AS events deduced from EST/cDNA analysis in two model plants: Arabidopsis and rice. In Arabidopsis, 4,707 (21.8%) of the genes with EST/cDNA evidence show 8,264 AS events. Approximately 56% of these events are intron retention (IntronR), and only 8% are exon skipping. In rice, 6,568 (21.2%) of the expressed genes display 14,542 AS events, of which 53.5% are IntronR and 13.8% are exon skipping. The consistent high frequency of IntronR suggests prevalence of splice site recognition by intron definition in plants. Different AS events within a given gene occur, for the most part, independently. In total, 36–43% of the AS events produce transcripts that would be targets of the non-sense-mediated decay pathway, if that pathway were to operate in plants as in humans. Forty percent of Arabidopsis AS genes are alternatively spliced also in rice, with some examples strongly suggesting a role of the AS event as an evolutionary conserved mechanism of posttranscriptional regulation. We created a comprehensive web-interfaced database to compile and visualize the evidence for alternative splicing in plants (Alternative Splicing in Plants, available at www.plantgdb.org/ASIP).
Keywords: exon skipping, intron retention, non-sense-mediated decay, spliced alignment, conserved alternative splicing
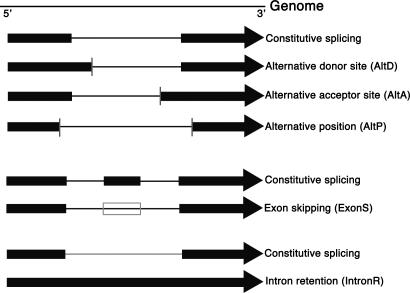
Alternative splicing (AS) is an important posttranscriptional regulatory mechanism that can increase protein diversity and affect mRNA stability (1, 2). Relative to the predominant transcript isoform, different types of AS have been observed, including exon skipping (ExonS), alternative donor (AltD) or acceptor (AltA) site, and intron retention (IntronR) (Fig. 1; reviewed in ref. 3). AS has been extensively studied by EST/cDNA-based analysis in mammalian systems, and 35–60% human genes were suggested to be alternatively spliced (4–9). In humans, ExonS is the most common type (58% of the total number of AS events are ExonS events involving a single exon, and 11% are ExonS events in which multiple exons are skipped in tandem) (4). IntronR is the least common type of AS in humans (5%) (4); ≈70–88% of the AS events occur in protein coding regions (reviewed in ref. 10), and approximately one-third produce premature termination codons (PTCs) (11). These PTC-containing transcripts are apparent targets for non sense-mediated mRNA decay (NMD) (11). Not all of the predicted AS are real and functional, because many possible sources of false positives exist (10). An AS event has been defined as functional “if it is required during the life cycle of the organism and activated in a regulated manner” (12). To identify functional AS events, conserved AS events between human and mouse were studied (12–17), with the assumption that conservation indicates function. Twenty-five percent of a set of 980 ExonS events in human were found to be conserved in mouse (12). Another study reported that ≈10% of human gene loci with mouse orthologs show conserved AS events (15).
Fig. 1.
Visualization of five alternative splicing types. The top black line represents the genome sequence. Filled boxes and arrows denote exons, with the arrow indicating the direction of transcription. Thin lines connecting the boxes indicate introns. The open box represents a skipped exon. Vertical bars represent AltD/AltA.
The splicing mechanism in plants is generally conserved compared with mammals (18, 19). However, introns in plant are usually short in length and U-rich (18–20), with a much less apparent polypyrimidine tract near the 3′ splice site than in mammals (21). Our recent genomewide survey on Arabidopsis splicing-related genes revealed variations in SR proteins and hnRNP proteins between plants and mammals, suggesting plant-specific differences in splicing-regulation mechanisms (22). A few AS events were identified experimentally in plants, including genes involved in splicing (23, 24), transcription (25), flowering regulation (26), disease resistance (27), enzyme activities (28, 29), and many other physiological processes and functions (19). A database (PASDB) collecting known alternatively spliced genes in plants is available at http://pasdb.genomics.org.cn (30). Although the prevalence of AS is not yet clear in plants, it is now recognized as playing an important role in the generation of plant proteome diversity (31).
Computational studies of AS in plants have recently been published. Ner-Gaon et al. (32) identified 436 alternatively spliced genes in Arabidopsis by EST-pair alignment. The fraction of IntronR observed in their study was as high as 64%. A sampling of the IntronR events were confirmed by RT-PCR with polyribosome RNA, demonstrating that these IntronR events are not the byproduct of incomplete splicing (32). Iida et al. (33) aligned 248,514 RIKEN Arabidopsis full-length cDNA/EST sequences to the Arabidopsis genome by using a blast-based method. They identified 15,214 transcription units (TUs) containing at least two sequences each and observed alternative splicing for 11.6% of these TUs (33). Three other studies with a smaller collection of EST/cDNA data briefly reported fewer AS events in Arabidopsis (9, 34, 35). All these pioneering studies revealed that a low fraction of genes (5–10%) are alternatively spliced, with IntronR the most prevalent AS type in Arabidopsis. However, none of the above studies included detailed analyses on the position and outcome of AS. Two recent studies with full-length cDNAs reported similar fractions of alternatively spliced genes but conflicting predominant AS types and positions of AS events relative to the coding regions (36, 37). Systematic analysis of AS in other plants has not been reported so far.
Compared with human, the lower fraction of alternatively spliced genes detected to date in plants is possibly due, in part, to lower cDNA/EST coverage. Millions of ESTs were used in human AS analyses (9), whereas less than one-10th of that number were available for Arabidopsis (9, 32–35). The number of publicly available plant cDNA/EST sequences has increased dramatically since the original studies, and, thus, it seemed likely that more AS events would be identified by using current data. Because the rice genome sequence has recently become available (38, 39) and differences are known to exist in the splicing mechanisms of monocot and dicot plants (19), it is of great interest to explore the AS events in rice and compare them with Arabidopsis. In this study, we applied the geneseqer spliced-alignment program (40, 41) to map currently available Arabidopsis and rice full-length cDNAs and ESTs to their respective genome sequences and identified thousands of AS events by exhaustive comparison of the deduced transcription units. The alternatively spliced genes were comparatively analyzed, and a small portion of the AS events were found to be conserved in the two plants. These data strongly suggest that, similar to mammals, AS occurs in plants on a large scale as a mechanism of regulation of gene expression. A user-friendly database has been constructed to store and visualize these AS events, which is frequently updated to reflect increases in cDNA/EST collections in both plants.
Results and Discussion
Genomewide EST/cDNA Alignments in Arabidopsis and Rice.
A total of 95.8% and 85.7% of the current Arabidopsis and rice EST/cDNA collections could be unambiguously aligned to their respective genomes by using the geneseqer spliced alignment program (40). The unaligned ESTs/cDNAs are either from the organelle (chloroplast and mitochondrion) genomes or different subspecies or are short and low-quality sequences. In total, 369,218 Arabidopsis ESTs/cDNAs were matched to the genome and producing 372,772 cognate alignments (see Materials and Methods). As shown in Table 3, which is published as supporting information on the PNAS web site, <1% of the EST/cDNAs have multiple-cognate alignments. A total of 25,231 TUs were identified, 23,856 (94.6%) of which correspond to annotated gene regions and 1,375 (5.4%) to previously uncharacterized gene regions. The average number of ESTs/cDNAs per TU is 14.8. In rice, 283,816 ESTs/cDNAs produced 319,391 cognate alignments, with ≈3% of the aligned EST/cDNAs showing multiple cognate alignments. The high proportion of multiple cognate alignments relative to Arabidopsis presumably reflects recent gene duplications in rice (42). We defined a total of 36,270 rice TUs, 87.7% of which overlap with annotated genes and 12.3% are in previously uncharacterized regions. The average number of ESTs/cDNAs per rice TU is ≈8.8. Rice introns are generally longer and have higher GC-content compared with Arabidopsis introns (see Supporting Text, Table 4, and Fig. 3, which are published as supporting information on the PNAS web site).
Approximately One-Fifth of Expressed Genes Are Alternatively Spliced in both Plants.
As described in Table 3, a total of 21,641 Arabidopsis genes (including 1,375 previously uncharacterized genes that did not overlap any annotated gene) and 30,917 of rice genes (4,466 previously uncharacterized genes) were defined as “expressed genes” by comparing the GenBank and The Institute for Genomic Research (TIGR)-annotated genes with our TUs. In Arabidopsis, 4,707 (21.8%) of the expressed genes display a total of 8,264 AS events (Table 1). Compared with recent estimates of 11.6% based on RIKEN Arabidopsis full-length cDNA sequences (33) and <5% indicated in a TIGR study (35) and other previous estimates (9, 34), our AS ratio is much higher. This increase may be because of the use of (i) more recent, larger EST/cDNA collections and/or (ii) a more sensitive AS detection method. Our Arabidopsis EST/cDNA collection includes with few exceptions all of TIGR's collection and ≈176,000 new sequences not included in the TIGR analysis. As shown in Fig. 2, our list includes 844 of the 909 (92.8%) TIGR-annotated alternatively spliced genes (excluding 279 genes with AS types involving terminal exons not discussed here). Sixty-five genes from the TIGR list are absent in our collection. Among these genes, three AS events actually are presented in our study under different gene names (because of annotation changes), 21 genes are really missed by our method, and the remaining 41 genes are likely false annotations given the lack of cognate EST/cDNA evidence (see Table 5, which is published as supporting information on the PNAS web site). In addition, 10 ExonS cases identified by TIGR lack EST/cDNA evidence, although other types of AS events were identified in Alternative Splicing in Plants (ASIP) for these genes (Table 5). Our study identified 3,863 alternatively spliced genes not identified previously in the TIGR analysis, 2,712 (70%) of which can be attributed to the previously uncharacterized transcript sequences that have become available since TIGR's study, whereas the remaining additional cases are method-specific (detailed data is available upon request).
Table 1.
AS events and alternatively spliced genes in Arabidopsis and rice
| AS type |
Arabidopsis |
Rice |
||
|---|---|---|---|---|
| Events (%) | Genes (%) | Events (%) | Genes (%) | |
| AltD | 845 (10.2) | 724 (3.3) | 1,642 (11.3) | 990 (3.2) |
| AltA | 1,810 (21.9) | 1,452 (6.7) | 2,201 (15.1) | 1,698 (5.5) |
| AltP | 308 (3.7) | 200 (0.9) | 921 (6.3) | 562 (1.8) |
| ExonS | 666 (8.1) | 379 (1.8) | 2,004 (13.8) | 999 (3.2) |
| IntronR | 4,635 (56.1) | 3,094 (14.3) | 7,774 (53.5) | 4,513 (14.6) |
| Total | 8,264 | 4,707 (21.8) | 14,542 | 6,568 (21.2) |
Percentages in the events columns represent the proportion of certain AS types relative to the total number of AS events. Percentages in the genes columns indicate the frequency of the AS type in all expressed genes studied (21,641 for Arabidopsis and 30,917 for rice). The total numbers of alternatively spliced genes are smaller than the sum of genes with different AS types, because some genes have multiple AS events.
Fig. 2.
Comparison among the TIGR, RIKEN, and our (ASIP) data sets of alternatively spliced genes in Arabidopsis. Numbers represent sizes of the indicated gene sets. TIGR represents data from ref. 35, and RIKEN represents data from ref. 33. ASIP data are from this study.
As for the 1,431 RIKEN-annotated alternatively spliced genes showing AltD/AltA, ExonS, or IntronR, 798 of them (55.8%) were included in our list. We did not perform a detailed comparison between RIKEN′s and our AS list, because only approximately two-thirds of RIKEN′s data set (as available at National Center for Biotechnology Information at the time of this study) were included in our EST/cDNA collection, and ≈200,000 EST/cDNA sequences now available were not included in the RIKEN study (further details available upon request). Another study by using pairwise EST comparisons detected 436 alternatively spliced Arabidopsis genes (32), 418 of which (95.9%) are included in our database.
In rice, the overall AS ratio is very similar to that of Arabidopsis. Approximately 21.2% (6,568 of 30,917) of the expressed genes showed a total of 14,542 AS events (Table 1). The TIGR rice genome annotation project identified 2,538 alternatively spliced genes (43) (http://rice.tigr.org/tdb/e2k1/osa1/expression/alt_spliced.info.shtml) showing all five of the AS types discussed here or alternative initiation or termination sites, which were not considered here (see Materials and Methods). The TIGR gene list is not broken up by AS type, and, therefore, we can only estimate overlap with our determination. Because ≈2,014 (80%) of the TIGR-annotated alternatively spliced genes (including other splicing variation types, roughly 23% as estimated from the TIGR Arabidopsis study by using the same program) are included in our database, we are confident that the vast majority of the TIGR-annotated five AS types considered here also were detected by our method. Thus, in summary, we estimate the occurrence of AS in both Arabidopsis and rice at ≈20% of all expressed genes.
Intron Retention Is the Most Prevalent AS Type in both Arabidopsis and Rice.
Among the five AS types in Arabidopsis and rice, Table 1 indicates IntronR as the most prevalent type (>50% of AS events), followed by AltA, AltD, and ExonS. Alternative position (AltP) is the least prevalent type. IntronR in Arabidopsis recently was demonstrated to be a bona fide AS event instead of merely the product of incomplete splicing in a study by using ribosome-associated mRNAs as template for RT-PCR (32). Although it is possible that some partially processed pre-mRNA are exported to the cytoplasm and associate with ribosomes, it is difficult to distinguish these events from regulated AS events. For both Arabidopsis and rice, ≈10% of IntronR cases in our database derive from transcripts with multiple introns that are all retained. These transcripts may result from sampling unprocessed pre-mRNAs or genomic contaminations in the cDNA library construction. We did not remove these events because they also may be real AS events, but in any case, the general conclusions regarding frequencies of AS types remain the same. IntronR is prevalent in introns of short size and high GC content (see Supporting Text; see also Fig. 4, which is published as supporting information on the PNAS web site).
Compared with AS events in human, where ExonS is the most abundant (4), the high frequency of IntronR in plants may reflect distinct features of plant pre-mRNA splicing. Mammalian genes with long introns and small exons are likely spliced by using the exon definition mechanism (44). This hypothesis is supported by the observation that mutations of mammalian splice sites most frequently lead to ExonS (45). In contrast, the intron definition mechanism may be prevalent in species in which genes typically have small introns (44). As plant introns are generally short and likely recognized by intron definition, failure of splice-site recognition would mostly lead to IntronR. The observed AS frequency differences between plants and human are consistent with this model. As observed AS events are likely a mixture of regulated events and unregulated events (splicing errors), it is not surprising to see a high frequency of ExonS in species in which the exon definition mechanism may be prevalent (human) and a high frequency of IntronR in species in which the intron definition mechanism may be prevalent (Arabidopsis and rice). Our result that short introns and introns with high GC content are more likely to be retained further supports the intron definition mechanism in plants (see Supporting Text). If an intron is too small or has a high GC content, steric hindrance or lack of typical intron-recognition signals may lead to inefficient splice-site recognition and, concomitantly, to a high frequency of IntronR. On the other hand, the exon definition mechanism appears to apply to some plant introns, because the mutation of certain plant splice sites lead to ExonS (46, 47). Comparing Arabidopsis and rice, we note a prevalence of relatively long introns in rice and an ≈5% higher incidence of ExonS in rice. We propose that in both plants, the intron definition mechanism is the predominant mode of intron recognition and exon definition is a more prominent mechanism of intron recognition in rice than it is in Arabidopsis.
Most AS Events Are Mutually Independent.
For most of the identified alternatively spliced genes (3,140 or 67% in Arabidopsis and 3,968 or 60% in rice), only a single AS event has been observed, and for the genes with multiple AS events, most transcripts contain only a single AS event. Because our analysis is based mostly on partial cDNA sequences, the latter observation includes transcripts not long enough to cover multiple AS events in a gene. However, detailed analysis showed that ≈56% of alternatively spliced transcripts covering the locations of multiple alternatively spliced introns in a gene actually only showed a single AS event relative to the constitutive transcript. Of 1,088 Arabidopsis genes and 1,813 rice genes with multiple AS events not limited to IntronR, a small fraction, 115 (10.6%) and 134 (7.4%), respectively, have multiple events within the same transcripts and no single AS event transcripts, suggesting that in these genes, multiple AS events are coordinated (see www.plantgdb.org/ASIP/EnterDB.php for a listing of these genes in ASIP). However, most AS events appear to be mutually independent.
More than One-Third of AS Events May Be Coupled with NMD.
More than half of the observed AS events alter the reading frame and possibly generate an early stop codon (Supporting Text; see also Table 6, which is published as supporting information on the PNAS web site), which renders the alternative mRNA isoforms possible candidates for NMD. PTCs in human were defined as in-frame stop codons residing >50 bp upstream of the 3′ most exon–exon junction (48). Arabidopsis homologs of human proteins involved in NMD and the exon junction complex have been identified (22). It seems that plants also have an NMD-like surveillance system, as suggested by the observation of increased PTC+/PTC− mRNA isoform ratios in a mutant of Arabidopsis UPF3, a homolog of a factor required for NMD in mammals and yeast (49). To investigate whether the 50-bp PTC rule applies in Arabidopsis, we screened a set of 4,868 Arabidopsis genes, for which the annotated longest ORF is fully supported by cDNA evidence and, therefore, is presumably within the constitutively spliced isoform. Fig. 5, which is published as supporting information on the PNAS web site, shows a histogram of the distances between the stop codon and last exon–exon junction in these genes. In 4,675 of the transcripts (96%), the stop codon occurs within the last exon. For 117 of the remaining 193 genes, the distance between the stop codon and the last exon–exon junction is <50 bp. Thus, overall >98% of Arabidopsis genes fit the human pattern of stop codon positioning close to or downstream of the last exon–exon junction. Therefore, we treat as potential substrates for NMD all AS isoforms producing PTCs defined as in-frame stop codons >50 bp upstream of the 3′ most exon–exon junction.
As summarized in Table 6, ≈43% Arabidopsis AS events and 36% rice events produce NMD candidates. These frequencies are similar to the frequency observed in human (35%) (11). Among the five types of AS events, AltP has the lowest incidence of NMD (18–22% of all AltP events), IntronR has the highest (40–48%), and AltA, AltD, and ExonS have frequencies at ≈34–43%. Because NMD is a surveillance mechanism that removes erroneous mRNAs containing PTCs (50), the one-third of AS events that could generate NMD candidates will produce much less protein diversity than might be expected. The NMD mechanism also can be routinely used by the cell to regulate gene expression (51), therefore coupling of AS and NMD could be an important posttranscriptional regulation to adjust the level of transcript isoforms (2).
Conserved Alternatively Spliced Genes.
Gene ontology annotation analysis on the alternatively spliced genes showed consistency between Arabidopsis and rice but no strong biases with respect to occurrence of AS events (see Supporting Text). By further searching Arabidopsis and rice close homolog pairs generated from reciprocal blast best hits (see Materials and Methods), 1,988 (41.7%) of the 4,707 alternatively spliced genes in Arabidopsis were found to have close homologs in rice that are also alternatively spliced and 1,154 of these gene pairs conserved the same AS type (Table 2). Among the five AS types, IntronR is most likely to be conserved, with ≈30% of the intron-retaining genes in Arabidopsis also showing IntronR in rice (Table 2). It should be noted, however, that the conserved alternatively spliced genes do not necessarily conserve the same AS event. For instance, Arabidopsis gene At1g28570 and rice gene LOC_Os01g46120 (GDSL-motif lipases) have similar gene structure and both have ExonS in the ORFs. In the Arabidopsis gene, the second exon is skipped, whereas in the rice gene, the fourth exon is skipped. Details regarding these conserved genes are available from the ASIP database (www.plantgdb.org/ASIP/EnterDB.php).
Table 2.
Conserved alternatively spliced genes in Arabidopsis and rice
| AS type | Conserved genes* (%) | Conserved introns† | Conserved events‡ |
|---|---|---|---|
| AltD | 108 (14.9) | 61 | 5 |
| AltA | 259 (17.8) | 117 | 5 |
| AltP | 9 (4.5) | 21 | 5 |
| ExonS | 47 (12.4) | 4 | 1 |
| IntronR | 951 (30.7) | 263 | 29 |
| Total | 1154 (24.5) | 445 | 41 |
*Conserved genes indicate the number of Arabidopsis genes showing any type of AS that have rice homologs also showing the same AS type. The percentages in parentheses indicate the fractions of the conserved genes relative to the total number of alternatively spliced genes with the specific AS type. The total numbers are smaller than the sums of entries by AS type, because some genes have multiple AS events.
†The number of introns that conserved between rice and Arabidopsis and are alternatively spliced with the indicated AS type in Arabidopsis (but not necessarily in rice).
‡The number of conserved introns showing the same AS type in both Arabidopsis and rice.
To identify conserved AS events, we first searched for conserved intron pairs as described in Materials and Methods. A total of 4,142 genes containing 7,640 conserved introns were identified in Arabidopsis, including 352 genes (445 introns) that are alternatively spliced. The frequency of genes containing conserved introns being alternatively spliced is ≈8.5%, much lower than the overall AS frequency (≈22%). As shown in Table 2 (and Table 7, which is published as supporting information on the PNAS web site), within this set of genes we could identify 41 Arabidopsis genes with AS events that are conserved in rice (same type of AS event for the conserved intron pair). Eleven of these events (26.8%) maintained the constitutive reading frame, and 22 (53.6%) produce NMD candidates.
The intron-pair method can identify only conserved introns in coding regions where the flanking exons are well conserved between Arabidopsis and rice. Therefore, we also visually compared a large number of homologous gene structures in Arabidopsis and rice and selected those with the same intron alternatively spliced. A small portion of additional conserved AS events were identified in this way, including U1-70K (U1 snRNP-70K, At3g50670, and LOC_Os10g02630), Tra2/SFRS10 (Transformer-2-β, At1g07350, and LOC_Os03g15890), and polypyrimidine tract binding protein (PTB; At3g01150, At5g53180, and LOC_Os01g43170). For most of the homologous gene pairs with conserved AS events, the AS event has similar effects on translation. For example, both Arabidopsis and rice IDH2 genes have three introns and can use an alternative acceptor site 12 bases downstream for splicing the first intron. A protein with four amino acid residues (VITK) removed will be generated from the alternative mRNA isoform in both Arabidopsis and rice. The PTB genes in both rice and Arabidopsis show transcripts, which include an additional exon in their third introns and produce PTC-containing mRNA isoforms. The mammalian PTB gene can autoregulate its splicing by promoting the skipping of exon 11, which introduces a PTC and leads to NMD (52). This regulation pathway may be conserved in plants. As shown in Supporting Text (and Fig. 6, which is published as supporting information on the PNAS web site), RT-PCR by using mixed Arabidopsis tissues validated the AS events in the atIDH2 and atPTB2a genes.
Interestingly, 30 conserved intron pairs and many more conserved gene pairs have different AS types in Arabidopsis and rice. For instance, At3g60370 and LOC_Os07g30800 (FKBP-type peptidyl-prolyl cis-trans isomerase) are orthologs with similar gene structure. Splicing of the third intron in At3g60370 can use an alternative acceptor site that generates a PTC-containing isoform. Splicing of the third intron in LOC_Os07g30800 alternatively includes an additional exon, which also produces a NMD candidate. Although the two genes have different AS types, it may be that the function of their AS events is conserved.
Conclusions
Based on our large-scale EST/cDNA alignments to the genomes of Arabidopsis and rice, we estimate that more than one-fifth of plant genes are alternatively spliced. Clearly, not all of the thousands of AS events will be biologically functional. Aberrant splicing will inevitably occur during the complicated dynamic splicing process that is happening continuously and under varied physiological conditions. Most aberrant splicing (splicing errors) will be removed by mRNA surveillance mechanisms such as NMD and, thus, are neutral to the organism. In the course of evolution, some splicing variants may be selectively beneficial and, thus, can be fixed as functional AS events.
How to distinguish splicing errors from biologically functional AS events remains an unresolved question. Interruption of protein coding sequences and/or production of PTC are not necessarily good landmarks for splicing errors, because coupling of AS and NMD may be regulated and add another level of regulation to gene expression. Conservation of AS events seems to be a good indication of functional AS events. Several papers addressed conserved cassette exons between human and mouse (12, 15, 17). We have shown that approximately one-quarter of the alternatively spliced Arabidopsis genes undergo the same type of AS in rice, and we identified a small number of conserved events among those. We also experimentally validated a few AS events in Arabidopsis by RT-PCR (see Supporting Text and Fig. 6). The web-interfaced ASIP database can serve as a starting point for the community to perform additional experiments, including high-throughput methods such as oligonucleotide microarrays studies, to identify functional AS events in plants.
Materials and Methods
Data Sources.
Our initial analysis was based on a total of 323,340 Arabidopsis ESTs that were downloaded from GenBank by using the ENTREZ query “Arabidopsis[ORGN] AND gbdiv_est[PROP].” Arabidopsis full-length cDNAs were retrieved from GenBank (query “Arabidopsis[ORGN] AND (FLI_CDNA[KYWD] OR GSLT_cDNA [KYWD]),” with some additional sequences obtained from AtGDB (www.plantgdb.org/AtGDB/resource.php) for a total of 62,009 sequences. For rice (Oryza sativa), sets of 298,857 ESTs and 32,136 cDNAs were obtained from GenBank by using similar queries. Updates to our database of plant AS events include more recent sequence depositions, but the analysis results reported here reflect only the sequences available at the time. The five Arabidopsis chromosome sequences were obtained from GenBank (accession nos. NC_003070, NC_003071, NC_003074, NC_003075, and NC_003076). For rice, our analysis was based on TIGR 3.0 of the 12 pseudochromosome sequences (downloaded from http://rice.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml).
Identification of AS in Arabidopsis and Rice.
Arabidopsis and rice full-length cDNAs and ESTs were aligned against their corresponding genomic origins by using the geneseqer (40) with species-specific splice site models (41) and the minimum allowed exon and intron lengths set conservatively to 2 (53) and 20 (54, 55), respectively. More information regarding the minimum intron size is available upon request. unix shell scripts and perl scripts were written to automate the following steps: (i) Loading of geneseqer alignment data was performed, including the predicted exon and intron coordinates and similarity and splice site scores, into a mysql database. (ii) Filtering of the alignment data was performed by using only high-quality alignments (overall similarity and coverage scores at least 0.8). For ESTs/cDNAs matching multiple genomic loci, only the best match (presumed cognate) was used, as described in ref. 34. For highly conserved duplicated genes, there may be multiple locations with equal score. In such cases, all these alignments were used. Exons with individual similarity scores <0.8 and their flanking introns were removed from the remaining alignments. Introns with a local similarity score <0.8 for either the 50 bp upstream or downstream flanking exon sequences also were removed. Redundancy in the sets of qualified exons and introns was eliminated. For any pair of terminal exons sharing the same exon/intron border only the longer exon was retained. (iii) For each intron, its coordinates were compared with each overlapping exon and intron to identify AS events. Two overlapping introns with different 5′ and/or 3′ ends, and all overlapping intron/exon pairs were considered as candidate AS events. For overlapping introns, the intron with the most cDNA/EST evidence was taken to be the constitutive intron. Several cases of AS were distinguished as follows (cf. Fig. 1). Use of an AltD/AltA is defined as an intron differing from the constitutive intron only in the donor or acceptor site. AltP refers to an intron overlapping the constitutive intron but differing in both donor and acceptor site position. ExonS occurs when an exon in one transcript isoform is completely contained in an intron of another isoform. IntronR occurs in a transcript isoform that contains a sequence segment that is precisely spliced out in another transcript isoform (i.e., an intron is fully contained in an exon in our database). (iv) Other transcript variants, including alternative terminal exons and alternative initial exons, were not considered. These events may involve the coupling of other mechanisms such as alternative transcription initiation sites with splicing (56). Candidate exon skipping events involving first or last exons also were removed, because these events also constitute examples of alternative initial or terminal exons, respectively.
Web Interface and Visualization of AS.
We created a web interface for the MySQL database to access all of the alignment data and graphically display the AS events by using perl-cgi, php, and javascript. The site is referred to as ASIP and is accessible at www.plantgdb.org/ASIP. All scripts were written in modular fashion such that the database can be easily updated and expanded to other species. All displays are integrated with the PlantGDB (57) Arabidopsis and rice genome browsers (www.plantgdb.org/AtGDB and www.plantgdb.org/OsGDB, respectively), allowing retrieval of sequence records and viewing of the AS events in an expanded genome context.
Diagrams of different AS types at ASIP are illustrated in Fig. 1. For AltD, AltA, and AltP events, a vertical color bar at the exon/intron border denotes the alternative sites. Multiple alternative sites are distinguished by different colors. For ExonS events, a green box within an intron indicates the position of the skipped exon. To visualize IntronR events, the intron is drawn in a light green color in transcripts from which it is spliced.
Derivation of Putative Transcript Isoforms (PTIs).
To classify AS events with respect to their impact on the translation of the alternatively spliced transcript, it is necessary to deduce the likely full-length alternative transcripts from overlapping ESTs (in addition to experimentally obtained full-length cDNA isoforms). For the purpose of this study, we make the assumption that all AS events within a TU (as defined in ref. 58, with the caveat that because of incomplete coverage, there may be multiple TUs per annotated gene; see Table 3) are mutually independent, and, thus, we derived all possible combinations of alternative exons to generate the set of PTIs for each transcription unit. To do so, we first clustered all exons and introns from our database into sets corresponding to distinct transcription units and then assembled complete gene structures by concatenating compatible exons and introns in the 5′ to 3′direction. This approach is different from TAP (8), which relies on a reference sequence structure, and also from PASA (35), which assembles only observed combinations of exons. With tightly spaced genes, assignment of some ESTs to a particular transcription unit may be ambiguous. However, this ambiguity, did not affect our evaluation of AS, because events involving terminal exons were excluded. The positions of the start and stop codons for each PTI were determined by searching for the longest ORF in the forward direction for PTIs with introns and in both directions for PTIs without introns for which the direction of transcription is ambiguous.
Position and Outcome of AS Events.
To classify AS events relative to their predicted effect on mRNA translation and stability, the constitutive ORF was defined as the longest ORF with the most abundant EST/cDNA evidence in all of the PTIs from the same gene. Any two PTIs differing in only one AS event were then compared. If multiple PTI pairs contain the same AS event, only the pair with the longest ORF was considered for classification of that AS event. If both ORFs from the PTI pair differ from the defined constitutive ORF, then the longer ORF from the PTI pair was redefined as the constitutive ORF for this comparison. To check whether an AS event will generate a transcript that contains premature stop codons and could be degraded by NMD, the distance between the stop codon and the last exon junction was calculated for both PTIs. If one isoform had a distance >50 nt, whereas the other isoform had a distance <50, then the AS event was regarded to produce a NMD candidate. If the AS event produced a PTC in one PTI relative to the other, this stop codon was used to calculate the NMD distance.
Conserved Alternatively Spliced Genes in Arabidopsis and Rice.
To identify conserved AS events between Arabidopsis and rice, all annotated Arabidopsis proteins were matched against all annotated rice proteins, and vice versa, by using blast (59). We used a 1 × 10−20 as the cutoff for blast and labeled the highest scoring hit as “uniquely best” if its value was at least 1 × 10−20 times lower than that of the next highest scoring hit. Reciprocal uniquely best hits were selected as close homolog pairs (potential orthologs). If there was evidence in both genes for the same type of AS event, this gene pair was characterized as conserving AS, although not necessarily the same position and outcome of AS. To further identify conserved AS position, we first derived the set of conserved introns between Arabidopsis and rice by matching all Arabidopsis introns plus 30-bp flanking sequences against rice intron and flanking sequences by using tblastx (value <1 × 10−4 and requiring both flanking exons to have at least 10 bases as part of the blast hits). The subset of these introns that occur in close homolog pairs were defined as conserved introns between Arabidopsis and rice. Conserved AS events were defined as AS events of the same type occurring in conserved introns.
Supporting Information.
For more information, see Figs. 7–9 and Table 8, which are published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Wei Huang for help with statistical analysis and Robert Fluhr, Qunfeng Dong, Yuanbin Ru, and Michael Sparks for critical reading of the manuscript. This work was supported in part by National Science Foundation Grants DBI-0110189 and DBI-0321600 and Research Grant IS-3454-03 from the United States–Israel Binational Agricultural Research and Development Fund.
Abbreviations
- AltA
alternative acceptor site
- AltD
alternative donor site
- AltP
alternative position
- AS
alternative splicing
- ASIP
alternative splicing in plants
- ExonS
exon skipping
- IntronR
intron retention
- NMD
non-sense-mediated decay
- PTC
premature termination codon
- PTI
putative transcript isoform
- TIGR
The Institute for Genomic Research
- TU
transcription unit.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T. A., Soreq H. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Lareau L. F., Green R. E., Bhatnagar R. S., Brenner S. E. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Black D. L. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S., Zink D., Korn B., Vingron M., Haas S. A. Bioinformatics. 2004;20:2579–2585. doi: 10.1093/bioinformatics/bth288. [DOI] [PubMed] [Google Scholar]
- 5.Modrek B., Resch A., Grasso C., Lee C. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brett D., Hanke J., Lehmann G., Haase S., Delbruck S., Krueger S., Reich J., Bork P. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 7.Mironov A. A., Fickett J. W., Gelfand M. S. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kan Z., Rouchka E. C., Gish W. R., States D. J. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brett D., Pospisil H., Valcarcel J., Reich J., Bork P. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 10.Modrek B., Lee C. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 11.Lewis B. P., Green R. E., Brenner S. E. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorek R., Shamir R., Ast G. Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Pan Q., Bakowski M. A., Morris Q., Zhang W., Frey B. J., Hughes T. R., Blencowe B. J. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Thanaraj T. A., Clark F., Muilu J. Nucleic Acids Res. 2003;31:2544–2552. doi: 10.1093/nar/gkg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugnet C. W., Kent W. J., Ares M., Jr., Haussler D. Pac. Symp. Biocomput. 2004;9:66–77. doi: 10.1142/9789812704856_0007. [DOI] [PubMed] [Google Scholar]
- 16.Nurtdinov R. N., Artamonova I. I., Mironov A. A., Gelfand M. S. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- 17.Yeo G. W., Van Nostrand E., Holste D., Poggio T., Burge C. B. Proc. Natl. Acad. Sci. USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorkovic Z. J., Wieczorek Kirk D. A., Lambermon M. H., Filipowicz W. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 19.Reddy A. S. N. Crit. Rev. Plant Sci. 2001;20:523–571. [Google Scholar]
- 20.Ko C. H., Brendel V., Taylor R. D., Walbot V. Plant Mol. Biol. 1998;36:573–583. doi: 10.1023/a:1005932620374. [DOI] [PubMed] [Google Scholar]
- 21.Brown J. W., Smith P., Simpson C. G. Plant Mol. Biol. 1996;32:531–535. doi: 10.1007/BF00019105. [DOI] [PubMed] [Google Scholar]
- 22.Wang B. B., Brendel V. Genome Biol. 2004;5:R102. doi: 10.1186/gb-2004-5-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golovkin M., Reddy A. S. Plant Cell. 1996;8:1421–1435. doi: 10.1105/tpc.8.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazar G., Goodman H. M. Plant Mol. Biol. 2000;42:571–581. doi: 10.1023/a:1006394207479. [DOI] [PubMed] [Google Scholar]
- 25.Montag K., Salamini F., Thompson R. D. Nucleic Acids Res. 1995;23:2168–2177. doi: 10.1093/nar/23.12.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., et al. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- 27.Dinesh-Kumar S. P., Baker B. J. Proc. Natl. Acad. Sci. USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werneke J. M., Chatfield J. M., Ogren W. L. Plant Cell. 1989;1:815–825. doi: 10.1105/tpc.1.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baga M., Glaze S., Mallard C. S., Chibbar R. N. Plant Mol. Biol. 1999;40:1019–1030. doi: 10.1023/a:1006286807176. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y., Zhou C., Ye L., Dong J., Xu H., Cai L., Zhang L., Wei L. Genomics. 2003;82:584–595. doi: 10.1016/s0888-7543(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 31.Kazan K. Trends Plant Sci. 2003;8:468–471. doi: 10.1016/j.tplants.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 33.Iida K., Seki M., Sakurai T., Satou M., Akiyama K., Toyoda T., Konagaya A., Shinozaki K. Nucleic Acids Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W., Schlueter S. D., Brendel V. Plant Physiol. 2003;132:469–484. doi: 10.1104/pp.102.018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haas B. J., Delcher A. L., Mount S. M., Wortman J. R., Smith R. K., Jr., Hannick L. I., Maiti R., Ronning C. M., Rusch D. B., Town C. D., et al. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov N. N., Troukhan M. E., Brover V. V., Tatarinova T., Flavell R. B., Feldmann K. A. Plant Mol. Biol. 2006;60:69–85. doi: 10.1007/s11103-005-2564-9. [DOI] [PubMed] [Google Scholar]
- 37.Nagasaki H., Arita M., Nishizawa T., Suwa M., Gotoh O. Gene. 2005;364:53–62. doi: 10.1016/j.gene.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 38.Feng Q., Zhang Y., Hao P., Wang S., Fu G., Huang Y., Li Y., Zhu J., Liu Y., Hu X., et al. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki T., Matsumoto T., Yamamoto K., Sakata K., Baba T., Katayose Y., Wu J., Niimura Y., Cheng Z., Nagamura Y., et al. Nature. 2002;420:312–316. doi: 10.1038/nature01184. [DOI] [PubMed] [Google Scholar]
- 40.Brendel V., Xing L., Zhu W. Bioinformatics. 2004;20:1157–1169. doi: 10.1093/bioinformatics/bth058. [DOI] [PubMed] [Google Scholar]
- 41.Sparks M. E., Brendel V. Bioinformatics. 2005;21(Suppl. 3):iii20–iii30. doi: 10.1093/bioinformatics/bti1205. [DOI] [PubMed] [Google Scholar]
- 42.Yu J., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., et al. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., et al. Plant Physiol. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berget S. M. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 45.Nakai K., Sakamoto H. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 46.Brown J. W. Plant J. 1996;10:771–780. doi: 10.1046/j.1365-313x.1996.10050771.x. [DOI] [PubMed] [Google Scholar]
- 47.Simpson C. G., McQuade C., Lyon J., Brown J. W. Plant J. 1998;15:125–131. doi: 10.1046/j.1365-313x.1998.00184.x. [DOI] [PubMed] [Google Scholar]
- 48.Nagy E., Maquat L. E. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 49.Hori K., Watanabe Y. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 50.Baker K. E., Parker R. Curr. Opin. Cell Biol. 2004;16:293–299. doi: 10.1016/j.ceb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Lejeune F., Maquat L. E. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Wollerton M. C., Gooding C., Wagner E. J., Garcia-Blanco M. A., Smith C. W. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- 53.Haas B. J., Volfovsky N., Town C. D., Troukhan M., Alexandrov N., Feldmann K. A., Flavell R. B., White O., Salzberg S. L. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-research0029. research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deutsch M., Long M. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim L. P., Burge C. B. Proc. Natl. Acad. Sci. USA. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kornblihtt A. R. Curr. Opin. Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Dong Q., Schlueter S. D., Brendel V. Nucleic Acids Res. 2004;32:D354–D359. doi: 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Nature. 2002;420:563–573. [Google Scholar]
- 59.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.