Abstract
PURPOSE. To identify proteinases and growth factors abnormally expressed in human corneas of donors with diabetic retinopathy (DR), additional to previously described matrix metalloproteinase (MMP)-10 and -3 and insulin-like growth factor (IGF)-I.
METHODS. RNA was isolated from 35 normal, diabetic, and DR autopsy human corneas ex vivo or after organ culture. Amplified cRNA was analyzed using 22,000-gene microarrays (Agi-lent Technologies, Palo Alto, CA). Gene expression in each diabetic corneal cRNA was assessed against pooled cRNA from 7 to 9 normal corneas. Select differentially expressed genes were validated by quantitative real-time RT-PCR (QPCR) and immunohistochemistry. Organ cultures were treated with a cathepsin inhibitor, cystatin C, or MMP-10.
RESULTS. More than 100 genes were upregulated and 2200 were downregulated in DR corneas. Expression of cathepsin F and hepatocyte growth factor (HGF) genes was increased in ex vivo and organ-cultured DR corneas compared with normal corneas. HGF receptor c-met, fibroblast growth factor (FGF)-3, its receptor FGFR3, tissue inhibitor of metalloproteinase (TIMP)-4, laminin α4 chain, and thymosin β4 genes were down-regulated. The data were corroborated by QPCR and immuno-histochemistry analyses; main changes of these components occurred in corneal epithelium. In organ-cultured DR corneas, cystatin C increased laminin-10 and integrin α3β1, whereas in normal corneas MMP-10 decreased laminin-10 and integrin α3β1 expression.
CONCLUSIONS. Elevated cathepsin F and the ability of its inhibitor to produce a more normal phenotype in diabetic corneas suggest increased proteolysis in these corneas. Proteinase changes may result from abnormalities of growth factors, such as HGF and FGF-3, in DR corneas. Specific modulation of proteinases and growth factors could reduce diabetic corneal epitheliopathy.
Diabetic retinopathy (DR) is the most severe ocular complication of diabetes mellitus and leads to vision loss in millions of patients worldwide.1,2 DR is a consequence of both insulin-dependent (IDDM) and noninsulin-dependent (NIDDM) diabetes mellitus. Diabetes also affects other parts of the eye, including the cornea. Corneal abnormalities are found in > 70% of patients with diabetes3 and mostly concern corneal nerves (neuropathy) and the epithelium (epitheliopathy). A major review article dealing with standards of care for diabetic eye disease states that eye examination in patients with diabetes must include the assessment of cornea1 Some diabetic corneal alterations can be diagnosed during routine eye examinations, but others are detected only with the use of special tests, after cataract or glaucoma surgery, or after vitrectomy.4 Thus the percentage of patients with diabetes who have corneal problems may be underestimated; these problems are also increased in patients in whom DR develops.4-6 Clinically, the disorders observed are tear and epithelial barrier dysfunction, recurrent epithelial defects and erosions, epithelial debridement after vitrectomy, delayed epithelial wound healing, superficial punctate keratitis, edema, corneal nerve tortuosity and decreased density, loss of corneal sensation, increased autofluorescence, and endothelial dysfunction.4-11
At the molecular level, decreased numbers of epithelial hemidesmosomes, increased expression of glycosyltransferases and advanced glycation end (AGE) products, and altered epithelial basement membrane (BM) have been described in human and animal diabetic corneas.4,12-18 Our previous work identified more specific molecular corneal defects associated with diabetes. Human diabetic and, especially, DR corneas had decreased expression of major corneal epithelial BM components, laminin-10, nidogen-1, and the lamininbinding integrin α3β1.19-21 The expression of respective genes was not reduced in DR corneas, ruling out a possible decrease of synthesis.21 Another explanation for the observed changes−ie, increased degradation−was corroborated by a finding of elevated expression and activity of matrix metalloproteinase (MMP)-10 and MMP-3 in DR corneas.21 These enzymes are able to degrade various laminin isoforms and nidogen-1.22-24 Changes observed in ex vivo corneas were fully reproduced in a corneal organ culture model.25
Based on these data, it may be suggested that diabetic corneas have increased activity of specific proteinases and that proteolytic degradation of epithelial BM may alter cell-BM adhesion and migration, leading to the clinically observed epithelial abnormalities. Because proteinases, including MMPs, are known to be regulated by growth factors/cytokines 23,26,27 elevated proteolysis may result from abnormal expression of these factors (such as insulin-like growth factor [IGF]-I)28 in diabetic corneas. We hypothesized that alterations of additional proteinases, growth factors/cytokines, and BM components may occur in diabetic and DR corneas. To identify these additional factors in diabetic corneas, we took advantage of gene microarray technology. This report describes abnormalities in the expression of cathepsin F, laminin α4 chain, MMP inhibitor TIMP-4, and several growth factors and their receptors in corneas from patients with diabetes and DR, fully supporting the above hypothesis. Functional significance of the observed expression changes was tested using treatment of corneal organ cultures with a cathepsin inhibitor and MMP-10.
MATERIALS AND METHODS
Tissues
Age-matched human autopsy normal, diabetic, and DR corneas were obtained from the National Disease Research Interchange (NDRI, Philadelphia, PA); donor identity was withheld by the supplier. NDRI has a human subject protection protocol that is enforced by the managerial committee operating under National Institutes of Health oversight. The Cedars-Sinai Medical Center Institutional Review Board (IRB) granted our study exemption 4 from IRB review in May 2000. For subsequent RNA isolation, corneas obtained in storage medium (Optisol; Chiron Vision, Claremont, CA) within 24 hours of donor death were trephined, immediately frozen in liquid nitrogen, and stored at -80°C. Other corneas obtained within 24 to 48 hours of donor death were embedded in OCT compound for immunohistochemistry or were processed for organ culture.
In preliminary experiments, antibodies were tested on cryostat sections of normal brain tissue from three patients who sustained trauma and brain tissue from three patients with glioblastoma multiforme to verify their positive reaction in immunostaining. Tissues were collected under Cedars-Sinai Medical Center IRB protocol 3637, approved in August 2002 and renewed in September 2003.
Corneal Organ Culture and Treatments
For organ culture, corneal concavity was filled with agar-collagen gel, and corneas were subjected to organ culture in serum-free medium for 10 to 35 days at the air-liquid interface, as described in detail previously.25 At the end of culture, the gel was removed, and corneas were trephined, immediately snap-frozen, and stored in liquid nitrogen or at -80°C until RNA isolation.
Organ-cultured DR corneas were treated with the cathepsin inhibitor cystatin C (SCIPAC, Sittingbourne, United Kingdom). Although it is not entirely specific for cathepsin F, cystatin C produces stronger inhibition of this enzyme than of other cathepsins.29 Cystatin C (50μg/mL) or vehicle (BSA in PBS) was added twice daily to the airexposed top of wounded organ-cultured DR corneas until the wounds fully closed. After the completion of wound healing, corneas were cultured for another week and were processed for immunofluorescence analysis of diabetic markers,20,25 laminin α5 chain, and integrin α3β1. Ten corneas were analyzed. Wound healing rates were determined as described.25
Purified recombinant MMP-10 (R&D Systems, Minneapolis, MN) was added in medium to the air-exposed tops of normal wounded organ-cultured corneas at 1 γg/mL in 20 γL, twice a day until wound closure. Control cultures received BSA. Wound healing rates were determined as described.25 Corneas were processed for immunohisto-chemistry after another week in culture. Heat activation of MMP-10 (1 hour at 55°C) did not change wound healing rates or its effect on laminin and α3β1 integrin, possibly because MMPs may not need full activation for cleaving their extracellular matrix (ECM) substrates.30 Thus, most experiments were conducted without enzyme activation. Twenty-six corneas were analyzed.
Isolation of Total RNA
RNA was isolated from ex vivo or organ-cultured corneas with Trizol reagent (Invitrogen, Carlsbad, CA). All ex vivo corneas used for RNA isolation were received and snap-frozen within 24 hours of donor death. Isolated RNA was further purified using a purification kit (NucleoSpin RNA and Virus Purification; BD Biosciences, Palo Alto, CA) according to the manufacturer's instructions and were stored at -80°C. Total RNA quality and yield were assessed using a bioanalyzer system (Agilent 2100; Agilent Technologies, Palo Alto, CA) and a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE).
Microarray Probe Synthesis and Hybridization
For gene microarray analysis, 16 normal corneas (9 ex vivo and 7 organ-cultured; mean patient age, 71.1 ± 12.8 [mean ± SD] years), 8 diabetic corneas without DR (5 ex vivo and 3 organ-cultured; 2 with IDDM, 6 with NIDDM; mean patient age, 67.3 ± 10.2 years), and 11 DR corneas (6 ex vivo and 5 organ-cultured; all with IDDM; mean patient age, 65.8 ± 9.8 years) were used. Labeled normal cRNA samples were prepared from pooled total RNA of 9 normal ex vivo or of 5 normal organ-cultured corneas. This approach minimizes differences between healthy persons and was extensively published.31,32 Diabetic and DR corneas were analyzed individually without pooling.31 Amplified cRNA probes were generated and labeled by cyanine 3-CTP (Cy3-CTP) or cyanine 5-CTP (Cy5-CTP; both Amersham Biosciences, Piscataway, NJ) using an amplification kit (Low RNA Input Fluorescent Linear Amplification Kit; Agilent) as recommended by the manufacturer. For cDNA synthesis, 1 μg total RNA in 10.3 μL was added to 1.2 μL T7 promoter primers. The mixture was incubated first at 65°C for 10 minutes and then on ice for 5 minutes before adding 8.5 μL of a cDNA synthesis master mix (4 μLof 5× first-strand buffer, 2 μL of 0.1 M dithiothreitol (DTT), 1 μL of 10 mM dNTP mix, 1 μL MMLV reverse transcriptase, and 0.5 μL RNase Out; all in Agilent kit) to each sample and incubating the mixture at 40°C for 2 hours, followed by heating to 65°C for 15 minutes for enzyme inactivation. cRNA was generated from cDNA and labeled as follows: samples were put on ice and 2.4 μL of either 10 mM Cy3-CTP or 10 mM Cy5-CTP (Amersham) and 57.6 μL transcription master mix (15.3 μL nuclease-free water, 20 μL of 4× transcription buffer, 6 μL of 0.1 M DTT, 8 μL of NTP mix, 6.4 μL of 50% polyethylene glycol, 0.5 μL RNase Out, 0.6 μL inorganic phosphatase, and 0.8μL T7 RNA polymerase; all in Agilent kit) were successively added to each sample. All reactions were performed in a thermal cycler (DNA Engine Tetrad; MJ Research, Waltham, MA) according to Agilent's protocol. cRNA synthesis and labeling were performed at 40°C for 2 hours. Amplified labeled cRNA was purified using mini-spin columns (RNeasy; Qiagen, Valencia, CA) according to the manufacturer's instructions and were quantitated using a spectrophotometer (ND-1000; NanoDrop Technologies).
For every hybridization, a mixture consisting of 0.5 μg Cy3-labeled cRNA from a diabetic or a DR cornea, 0.5 μg Cy5-labeled pooled normal corneal cRNA, 50 μL 10× control targets (Agilent), and nuclease-free water (added up to 250 μL) was fragmented by adding 10 μL of 25χ fragmentation buffer (Agilent) and incubating at 60°C for 30 minutes. Then, 250 μL of 2× hybridization buffer (Agilent) were added to the fragmented mixture, and the resultant 500-μL probe was injected into a hybridization chamber holding a human 1A oligo microarray (both from Agilent). Hybridization was performed by gently rotating the slide chamber at 60°C for 16 hours in an oven (Robbins Scientific, Sunnyvale, CA). Slides were washed once with 6× SSC/0.005% Triton X-102 (Agilent) at room temperature for 10 minutes and once with 0.1× SSC/0.005% Triton X-102 on ice for 5 minutes. Washed slides were blow-dried with nitrogen before scanning.
Data Acquisition and Analysis
Microarray slides were scanned at 10-μm resolution using an Agilent G2565BA fluorescence dual laser scanner for Cy3 and Cy5 excitation. Image intensity data were recorded and transferred into text files (Feature Extraction software, version A.4.0.45; Agilent). Spots that did not pass quality control procedures in this software were flagged and removed from further analysis. A possible dye bias was eliminated using an algorithm for the same extraction software (Feature Extraction software DyeNorm algorithm; Agilent) that applies normalization factors. Generated files were imported into spreadsheets (Excel; Microsoft Corp., Redmond, WA) for downstream data analysis and statistical evaluation. Raw Cy3 and Cy5 values were extracted from the data sets and treated as individual measurements. These values were then normalized directly to the median brightness data set across all arrays to render them intercomparable. The normalized values were fed into the dChip 1.3 software33 as an external data source. Values were clustered using Euclidean distance with centroid linkage for gene targets of interest (growth factors/cytokines and proteinases), as shown in Figure 1.
FIGURE 1.
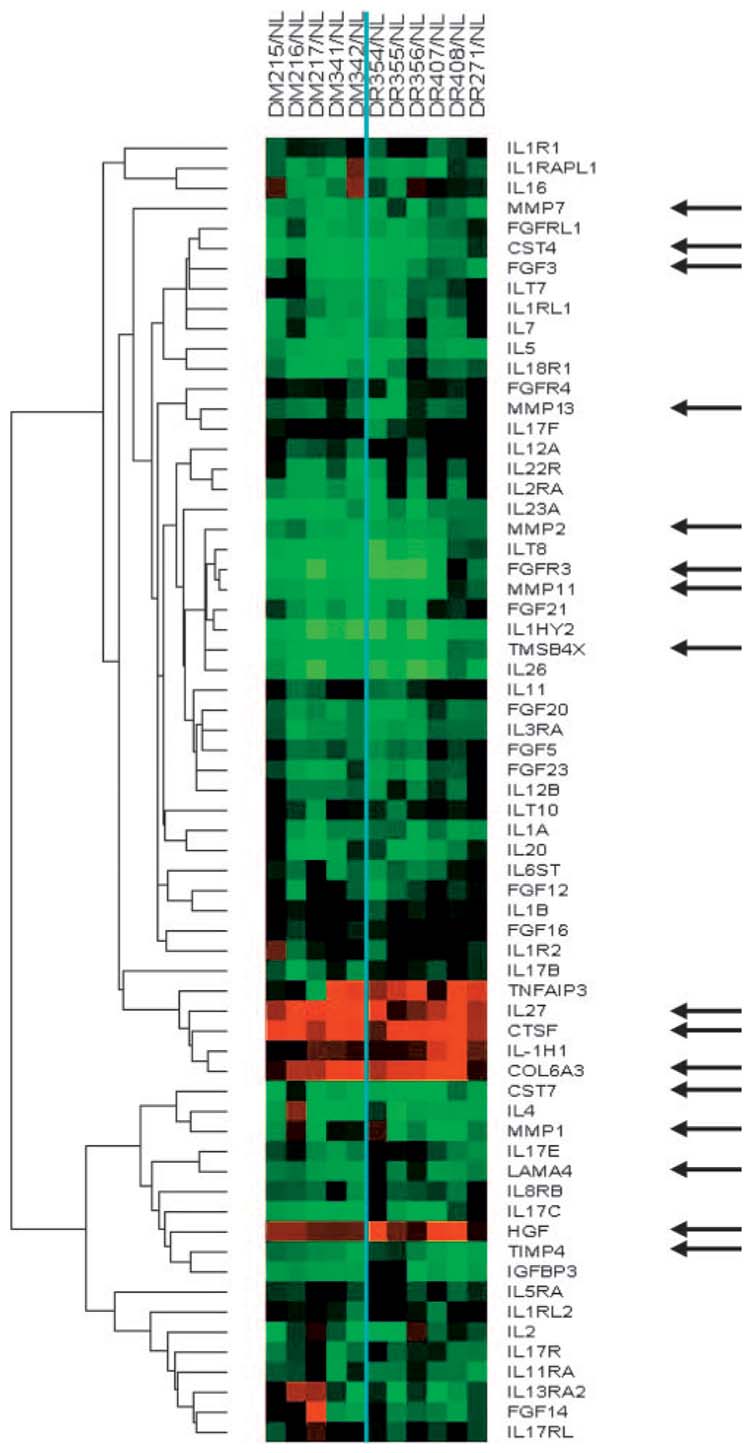
Cluster analysis of gene microarray data using ex vivo corneas. Each column shows proteinase and growth factor/cytokine-related genes in an individual diabetic (DM) or DR cornea that are significantly changed compared with normal (NL) corneas. The vertical line divides the DM and DR groups. For each gene in a DM or DR sample, the signal intensity is presented as a ratio to normal (NL) signal at the same spot on the same array. (green) Gene downregulation in diabetic sample compared with normal. (black) No change. (red) Gene upregulation in diabetic sample compared with normal sample. Arrows point to individual genes of interest. For comparison, type VI collagen α3 chain (COL6A3) and laminin α4 chain (LAMA4) genes are also shown. Note the consistent changes of selected genes across the DM and DR groups. CST4, cystatin S; CST7, cystatin F; CTSF, cathepsin F; IL27, interleukin-27; TMSB4X, X-linked T β4.
Quantitative RT-PCR
Gene microarray results were validated using quantitative real-time RT-PCR (QPCR) on the same samples used for microarray experiments and on additional samples. Each normal or DR cornea (with 5 ex vivo and 5 organ-cultured corneas in each group) was analyzed individually. Total RNA (2 μgin20-μL reaction) was treated with Turbo DNA-free (Ambion, Austin, TX) to remove possible genomic DNA contamination. Of this mix, 5 μL (0.5 μg) was reverse transcribed into first-strand cDNA using the TaqMan reverse-transcription (RT) reagents (Applied Biosystems, Foster City, CA) and oligo (dT) primers. The RT reaction mixture contained the following: 1× TaqMan RT buffer, 1.25 U/μL MultiScribe reverse transcriptase, 5.5 mM MgCl2, 0.4 U/μL RNase inhibitor, 500 μM dNTP, and 2.5 μM oligo (dT) primers in a total volume of 25 μL. Reactions were cycled on a PTC-200 Peltier Thermal Cycler (MJ Research) as follows: 25°C for 10 minutes, 45°C for 45 minutes, and inactivation at 95°C for 5 minutes. Primers for selected genes (gene sequences were from GenBank, National Center for Biotechnology Information) were designed using Primer Express version1.5 software (Applied Biosystems). The selected primers had melting temperatures of 59°C or 60°C and not more than two G-C pairs in the final 5 bp of the 3′ end. All QPCR primers (Table 1) were synthesized by Invitrogen and were confirmed to amplify the predicted products by testing under QPCR conditions.
TABLE 1.
Primers Used for QPCR
| Gene, GenBank Accession no. | Forward Primer | Reverse Primer | Size (bp) |
|---|---|---|---|
| Cathepsin F, AF088886 | CATTCCCACCCTGAAGTTCTG | TGCCAAGATACCACCCTTCAC | 87 |
| Cathepsin F, AF088886 | GCCTGTCCGTCTTTGTCAAT | TTGTTGCCAGGCTCTTTTCT | 160 |
| Cathepsin B, M14221 | CCCTGCATCTATCGAGTTTGC | CATGATTAGCAGAGTGCACGAA | 101 |
| Cathepsin H, X07549 | CAGCCTGGAAACCTACAGACAAG | GGCACATGGCTGGTGATCTT | 81 |
| Cathepsin L, M20496 | GTGTCGGATACACACTCGAATCA | AGAATTCAAATCACACTCGGATCTT | 50 |
| HGF, M60718 | AGAAATGCAGCCAGCATCATC | CACATGGTCCTGATCCAATCTTT | 90 |
| c-met, J02958 | TGCAAAGCTGCCAGTGAAGT | GCCAAAGGACCACACATCTGA | 85 |
| FGF-3, NM_005247 | CTTGACTCTGACTCTCCTCCTTGAG | AGTTCCCACTGGACCCTGATT | 99 |
| FGFR3, M58051 | AGTGGAGCCTGGTCATGGAA | GGATGCTGCCAAACTTGTTCTC | 83 |
| TIMP-4, NM_003256 | CAGACCCTGCTGACACTGAA | GTCCAGAGGCACTCGTTAGG | 352 |
| Thymosin β4, NM_021109 | GGCTGAGATCGAGAAATTCG | GAAGGCAATGCTTGTGGAAT | 165 |
| Laminin α4 chain, S78569 | CTCCATCTCACTGGATAATGGTACTG | GACACTCATAAAGAGAAGTGTGGACC | 362 |
| β2-MG, NM_004048 | ATAATTCTACTTTGAGTGCTGTCTCCAT | TCCTAGAGCTACCTGTGGAGCAA | 71 |
| β2-MG, NM_004048 | CTCGCGCCTACTCTCTCTTTCTG | GCTTACATGTCTCGATCCCACTT | 335 |
All primers are shown from 5′ to 3′. Both primers for cathepsin F and β2-MG gave identical results.
Real-time QPCR was carried out in a MicroAmp Optical 96-well plate using SYBR Green PCR master mix reagents (Applied Biosystems). Each well contained 25 to 100 ng (depending on genes) reversetranscribed cDNA; SYBR Green PCR buffer, which includes SYBR Green 1 dye and passive reference 1; 5.5 mM MgCl2; 200 μM each of dATP, dCTP, and dGTP; 400 μM dUTP; 300 nM each of forward and reverse primers; 0.01 U/μL AmpErase UNG; and 0.03 U/μL AmpliTaq Gold DNA polymerase in a total volume of 25 μL The thermal cycling conditions for real-time PCR were 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of melting (95°C, 15 seconds) and annealing/extension (60°C, 60 seconds). PCR reactions were monitored in real time using the ABI PRISM 7700 Sequence Detector (Applied Biosystems). Signals from each sample were normalized to values obtained for housekeeping gene β2-microglobulin (β2-MG), which was run simultaneously with experimental samples. A comparative threshold cycle (Ct) method (ΔΔCt) was used to calculate the relative gene expression (fold change) between test and reference sample for genes whose primers passed an efficiency test. Otherwise, a standard curve for each target gene as well as for β2-MG was generated with reference RNA, and relative quantitation of gene expression was performed. Briefly, five serial 10-fold dilutions of one of the normal cDNA samples were defined as the standard for all the reactions. Dilutions ranged from 0.015 to 150 ng normal cDNA and were analyzed in duplicate within each run. Controls without the RT step or without template were included for each primer pair to check for any contaminants. None of the primer pairs made within the same exon generated products when the amplification was performed without the RT step, confirming lack of genomic DNA contamination. Such primers gave the same results as those that spanned an intron (not shown). As another quality control measure, melting (dissociation) curves of PCR reactions were monitored to ensure that there was only a single PCR product and no primer dimers. Further testing by agarose gel electro phoresis confirmed that there was only one PCR product of the expected size.
Indirect Immunofluorescence Staining
In total, 36 normal corneas (15 ex vivo and 21 organ-cultured; mean age, 67.9 ± 13.4 years) and 29 DR corneas (11 ex vivo and 18 organ-cultured; 23 with IDDM and 6 with NIDDM; mean age, 66.8 ± 11.2 years) were used. Diabetic corneas without DR were not analyzed because marker changes were not as pronounced as in DR corneas.20,21 Ex vivo and organ-cultured corneas were embedded in OCT and stored at -80°C. General procedures, secondary antibodies, and routine controls were as described previously.20 Primary antibodies are listed in Table 2. Because cryostat section immunolocalization of cathepsins and select growth factors was described only in a limited number of publications, many antibodies were evaluated using several fixation protocols and human gliomas as positive controls.21 Recommendations for immunohistochemical use of each antibody are shown in Table 2.
TABLE 2.
Antibodies Used for Immunostaining
| Antigen | Antibody and Name | Immunostaining | Source/Reference |
|---|---|---|---|
| Cathepsin B | Mouse mAb CA10 | Positive, weak | Oncogene Research Products |
| Cathepsin B | Goat pAb AF953 | Nonspecific | R&D Systems |
| Cathepsin B | Mouse mAb 3E1 | Negative | Serotec |
| Cathepsin B | Rabbit pAb PC41 | Nonspecific | Oncogene Research Products |
| Cathepsin B | Mouse mAb CB131 | Negative | Novocastra Laboratories |
| Cathepsin B | Goat pAb S-12 | Negative | Santa Cruz Biotechnology |
| Cathepsin B | Goat pAb N-19 | Positive | Santa Cruz Biotechnology |
| Cathepsin B | Rabbit pAb FL-339 | Positive | Santa Cruz Biotechnology |
| Cathepsin F | Mouse mAb CTF12 | Unusual staining | Novocastra Laboratories |
| Cathepsin F | Goat pAb C-20 | Positive | Santa Cruz Biotechnology |
| Cathepsin F | Goat pAb V-20 | Positive | Santa Cruz Biotechnology |
| Cathepsin F | Rabbit pAb H-110 | Weak | Santa Cruz Biotechnology |
| Cathepsin H | Goat pAb K-18 | Negative | Santa Cruz Biotechnology |
| Cathepsin H | Goat pAb N-18 | Negative | Santa Cruz Biotechnology |
| Cathepsin H | Goat pAb C-20 | Negative | Santa Cruz Biotechnology |
| Cathepsin H | Rabbit pAb H-130 | Positive | Santa Cruz Biotechnology |
| Cathepsin H | Mouse mAb 1D10 | Negative | Serotec |
| Cathepsin L | Mouse mAb N135 | Negative | Serotec |
| Cathepsin L | Rabbit pAb H-80 | Weak | Santa Cruz Biotechnology |
| Cathepsin L | Goat pAb S-20 | Positive | Santa Cruz Biotechnology |
| Cathepsin L | Goat pAb N-20 | Unusual staining | Santa Cruz Biotechnology |
| Cathepsin L | Goat pAb C-18 | No blocking with peptide | Santa Cruz Biotechnology |
| Cathepsin L | Goat pAb AF952 | Positive | R&D Systems |
| Cathepsin L | Mouse mAb 13C2 | Negative | Novocastra Laboratories |
| c-Met | Rabbit pAb C-12 | Positive | Santa Cruz Biotechnology |
| c-Met | Rabbit pAb C-28 | Positive | Santa Cruz Biotechnology |
| c-Met | Mouse mAb B-2 | Negative | Santa Cruz Biotechnology |
| HGF | Goat pAb AF294 | Positive | R&D Systems |
| HGFα | Rabbit pAb H-145 | Positive | Santa Cruz Biotechnology |
| HGFβ | Rabbit pAb H-170 | Negative | Santa Cruz Biotechnology |
| Thymosin β4 | Rabbit pAb 609270 | Positive | Calbiochem |
| Thymosin β4 | Rabbit pAb A9550.2 | Positive | ALPCO Diagnostics |
| Laminin α4 | chain Rabbit pAb 1129 | Positive | Ref. 34 |
| Laminin α4 | chain Rabbit pAb 1100 | Variable and weak | Ref. 35 |
| Laminin α5 chain | Mouse mAb 4C7 | Positive | Chemicon |
| Integrin α3β1 | Mouse mAb M-KID 2 | Positive | Chemicon |
| TIMP-4 | Mouse mAb 153934 | Positive | R&D Systems |
| TIMP-4 Rabbit | pAb AB19087 | Positive, with some nuclearstaining | Chemicon |
| TIMP-4 | Rabbit pAb AB19168 | Nonspecific | Chemicon |
| TIMP-4 | Rabbit pAb AB19086 | Negative | Chemicon |
| TIMP-4 | Rabbit pAb AB816 | Positive, with somenonspecific staining | Chemicon |
| TIMP-4 | Rabbit pAb AB19081 | Positive | Chemicon |
| FGF-3 | Goat pAb AF1206 | Positive | R&D Systems |
| FGF-3 | Goat pAb N-19 | Negative | Santa Cruz Biotechnology |
| FGF-3 | Mouse mAb MSD 1 | Negative | Santa Cruz Biotechnology |
| FGFR3 | Rabbit pAb C-15 | Positive | Santa Cruz Biotechnology |
| FGFR3 | Rabbit pAb H-100 | Positive, weak | Santa Cruz Biotechnology |
| FGFR3 | Mouse mAb B-9 | Negative | Santa Cruz Biotechnology |
mAb, monoclonal antibody; pAb, polyclonal antibody.
Fixation protocols included 100% acetone at -20°C for 15 minutes, 100% methanol at -20°C for 15 minutes, 1% formalin (0.37% formaldehyde) in PBS at room temperature for 5 minutes, and 2% formalin in PBS at room temperature for 5 minutes After acetone or methanol fixation, only weak to no specific fluorescence was observed. Therefore, for most experiments, 1% formalin fixation was used.
Routine controls without primary antibodies were included with each experiment and were negative. Each antibody was analyzed at least twice in most cases, with identical results. All anti-peptide anti-bodies were neutralized by a corresponding peptide and then were used to stain tissues. In all these cases, no staining was observed attesting to the specificity of the respective antibody.
Statistical Analysis
Gene microarray results were statistically analyzed by two-tailed t-test using dChip software, with P < 0.01 considered significant. This statistical analysis method has been validated before.36 QPCR data (target gene relative to β2-MG in a healthy group compared with a DR group) were analyzed by two-tailed t-test using InStat software program (GraphPad Software, San Diego, CA). Immunostaining results were analyzed by two-sided Fisher exact test (InStat; GraphPad). The number of cases with a specific staining pattern (e.g., strong, weak) in one experimental group (e.g., normal) was compared with the number of cases with the same staining pattern in another experimental group(e.g., DR).20,21 P < 0.05 was considered significant in QPCR and immunostaining analyses.
RESULTS
Gene Microarray Analysis
More than 100 genes were upregulated and 2200 were downregulated in DR corneas, with a twofold cutoff and P < 0.01. Changes were observed in genes coding for proteinases, growth factors/cytokines, apoptosis mediators, and structural proteins and in those involved in metabolic processes. Generally, results obtained with diabetic and DR corneas were similar (Fig. 1). Therefore, QPCR and immunohistochemical analysis was conducted on DR corneas compared with normal corneas. Within the same group (normal or diabetic), similarities were also observed between gene expression profiles of ex vivo and organ-cultured corneas.
For in-depth analysis of differentially expressed genes, the list has been shortened according to our working hypothesis of the importance of alterations of proteinases and growth factors/cytokines in diabetic and DR corneas. This hypothesis was based on the identification of specific proteinases and growth factors (possible proteinase regulators) with abnormal expression levels in diabetic corneas.20,28 We have now attempted to identify additional proteinases and growth factors with significant up-or downregulation in these corneas using a global gene expression approach. The respective differentially expressed genes were clustered (Fig. 1) and analyzed further. The choice of individual components to be looked at in more detail was dictated by significant changes on gene array and detectability by immunostaining (some MMPs were excluded from further testing because no staining could be seen). For growth factors, it was important to take into account a simultaneous change in a factor and its receptor, which suggests the functional significance of a change, and a known function of the growth factor or receptor in wound healing and cell migration that are impaired in diabetic corneas.
Some proteinase and growth factor genes (e.g., cathepsin F and HGF) were upregulated in diabetic and DR corneas, but a number of them (including several MMPs) showed decreased expression compared with normal corneas (Fig. 1; Table 3). Table 3 lists these genes, some of which were selected for further analysis. Of the tissue inhibitors of metalloproteinase (TIMP) genes, only TIMP-4, which was recently found in the cornea,37 was differentially decreased in DR corneas; the other TIMPs were unchanged (Table 3; Ref. 24). A member of the cathepsin family of cysteine proteinases,38 cathepsin F was upregulated in DR corneas and was studied in more detail. The same was true for HGF (Table 3), important for cell migration and wound healing.39,40 Although it was a little less than twofold higher in DR corneas, its change was statistically significant and it was thus selected for further analysis. Because HGF signals through its tyrosine kinase receptor, c-met,41,42 its post-microarray analysis was also carried out even though it was decreased less than twofold. Another potent cell migration mediator, thymosin β4 (X-linked),43-45 was downregulated in DR corneas and was also evaluated further. A similar decrease was observed for one member of fibroblast growth factor (FGF) family, FGF-3/Int-2 (Fig. 1; Table 3). It was selected for further analysis because of its importance for corneal epithelial cell differentiation46 and the fact that one of its receptors, FGFR3, was concomitantly downregulated. A marked increase in diabetic corneas of recently described interleukin (IL)-27 was not further verified because of the lack of antibodies reacting with human tissues. Many other interleukins and their receptors were downregulated in diabetic corneas (Fig. 1). A decreased expression for laminin α4 gene was observed in DR corneas (Table 3). This chain has not been documented in cornea before, and it was of interest to identify its distribution patterns.
TABLE 3.
Changes in Select Proteinase, Growth Factor, and Structural Genes in Diabetic Corneas as Revealed by Gene Microarray Analysis
| Gene | Diabetic*/Normal (P) | Possible Role |
|---|---|---|
| MMP-1⇓ | 0.50 (0.0001) | Proteolysis |
| MMP-2⇓ | 0.40 (<0.0001) | Proteolysis |
| MMP-7⇓ | 0.42 (<0.0001) | Proteolysis |
| MMP-11⇓ | 0.35 (<0.0001) | Proteolysis |
| MMP-13⇓ | 0.43 (0.0002) | Proteolysis |
| TIMP-4⇓ | 0.44 (<0.0001) | Proteolysis |
| Cathepsin F⇑ | 3.70 (<0.0001) | Proteolysis |
| Cystatin F⇓ | 0.37 (<0.0001) | Proteolysis |
| Cystatin S⇓ | 0.35 (<0.0001) | Proteolysis |
| HGF⇑ | 1.92 (0.006) | Growth factor |
| c-met proto-oncogene ⇓ | 0.76 (0.014) | Growth factor receptor |
| FGF-3⇓ | 0.38 (<0.0001) | Growth factor |
| FGFR3⇓ | 0.28 (<0.0001) | Growth factor receptor |
| Thymosin β4⇓ | 0.35 (<0.0001) | Apoptosis, cell migration |
| IL27⇑ | 4.57 (<0.0001) | Inflammatory cytokine |
| Type VI collagen α3 chain⇑ | 3.50 (0.0004) | Structural |
| Laminin α4 chain⇓ | 0.47 (<0.0001) | Structural |
Because the expression levels of all select genes data in diabetic and DR groups were similar, combined data are presented. Arrows show changes in diabetic and normal corneas (⇑, increase; ⇓, decrease).
QPCR
Quantitative RT-PCR was used to verify gene microarray expression data on select genes coding for proteinases, their inhibitors, and growth factors/cytokines. As shown on Figure 2, cathepsin F and HGF gene expression was significantly elevated in DR compared with normal corneas, in accordance with microarray data. The expression of c-met, FGF-3, FGFR3, Tβ4, and TIMP-4 genes was decreased in DR corneas, again in line with microarray results. Microarray analysis of cathepsin B gene expression showed no significant change in DR corneas (not shown), and the same result was obtained by QPCR (Fig. 2). Overall, there was a good correlation between the results obtained by gene microarray analysis and QPCR.
FIGURE 2.
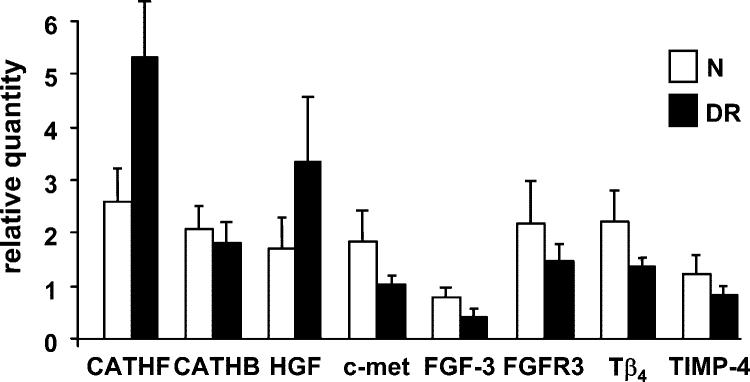
QPCR data for select genes of interest. Data for ex vivo corneas are shown. Results with organ-cultured corneas were generally similar. N, normal; cath, cathepsin. Bars represent SEM.
Immunohistochemistry
Gene microarray and QPCR results were further verified by immunostaining for select gene products, as previously published by us47 and by others48 (Table 4). Weak epithelial staining was observed for cathepsin B, and it was slightly increased in DR corneas ex vivo and after organ culture (Fig. 3). This was consistent with no difference in mRNA expression identified by QPCR (Fig. 2) and microarray analysis. Similarly, no significant changes in DR compared with normal corneas were observed for cathepsins H and L by gene microarray, QPCR (not shown), and immunohistochemistry (Table 4). In contrast, cathepsin F, with elevated expression by gene microarray and QPCR, was significantly increased by immunohistochemistry in DR corneal epithelium compared with normal corneal epithelium (Table 4). This result applied for ex vivo and organ-cultured corneas (Fig. 3).
TABLE 4.
Marker Distribution in Human Corneas by Immunostaining
| Protein | Ex Vivo Normal | Ex Vivo DR | OC Normal | OC DR |
|---|---|---|---|---|
| Cathepsin F⇑ | 2/15 positive* | 6/11 positive | 0/11 positive* | 4/8 positive |
| Cathepsin B | 5/12 positive | 8/11 positive | 2/4 positive | 5/5 positive |
| Cathepsin H | 7/10 positive | 11/11 positive | 5/11 positive | 6/8 positive |
| Cathepsin L | 2/12 positive | 6/11 positive | 2/4 positive | 4/5 positive |
| Laminin α4⇓ | 10/10 intact* | 2/10 intact | 5/7 intact* | 1/8 intact |
| TIMP-4⇓ | 7/10 positive* | 1/10 positive | 7/10 positive* | 0/8 positive |
| HGF⇑ | 4/15 positive* | 8/11 positive | 0/11 positive* | 7/8 positive |
| c-met⇓ | 15/15 strong* | 6/11 strong | 8/11 strong | 3/8 strong |
| FGF-3⇓ | 11/11 positive* | 7/12 positive | 11/11 positive* | 3/7 positive |
| FGFR3⇓ | 11/11 positive* | 6/11 positive | 9/11 positive* | 2/7 positive |
| Thymosin β4⇓ | 5/11 strong* | 0/11 strong | 8/11 positive* | 0/7 positive |
The first number in each pair is the number of cases with a specific pattern (eg, positive, strong); the second number represents the total number of cases analyzed. OC, organ culture; intact, continuous staining of epithelial BM;
P < .05 vs DR; arrows show changes in DR and normal corneas (a, increase; s, decrease).
FIGURE 3.
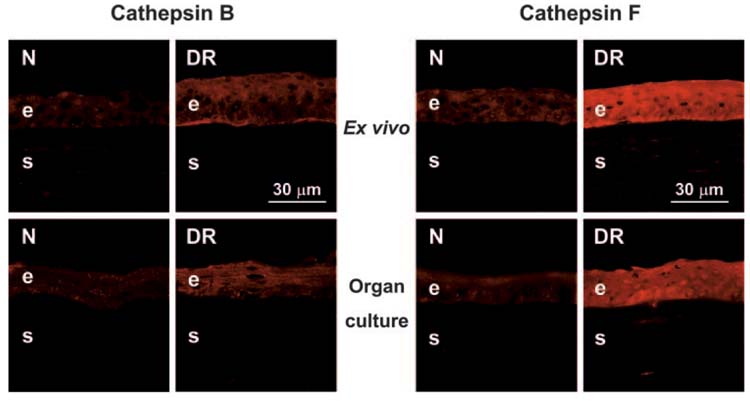
Distribution of cathepsins B and F in ex vivo and organ-cultured normal (N) and DR corneas. Note a slight increase of cathepsin B (left panels, polyclonal antibody [pAb] N-19) and a significant increase of cathepsin F (right panels, pAb V-20) in DR corneal epithelium. Data for ex vivo and organ-cultured corneas are similar. Indirect immunofluorescence. e, epithelium; s, stroma.
When analyzed by immunostaining, HGF and its receptor, c-met, showed patterns in normal corneas that were similar to those described previously.49 HGF was seen mostly in epithelial cells, with some endothelial staining, using two different antibodies. c-met was found in the epithelium, keratocytes, and endothelium (Fig. 4). In ex vivo and organ-cultured DR corneas, epithelial HGF staining increased, whereas epithelial c-met staining decreased (Fig. 4; Table 4). These changes were significant and consistent with gene microarray and QPCR results (Figs. 1, 2, 4).
FIGURE 4.
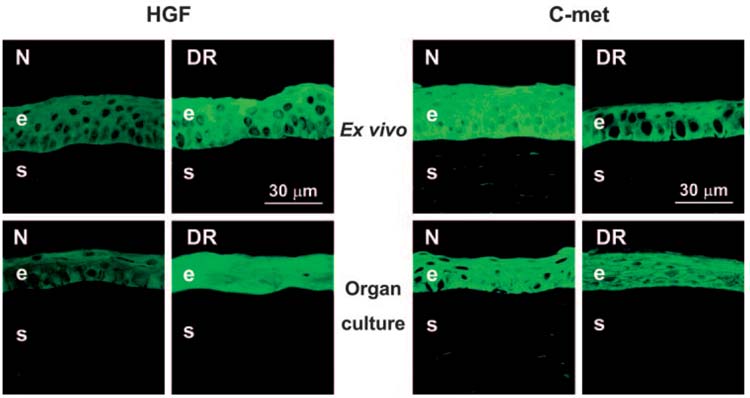
Distribution of HGF and c-met in ex vivo and organ-cultured normal (N) and DR corneas. HGF is increased in DR corneal epithelium (left panels, pAb AF294), but c-met shows a decrease (right panels, pAb C-28). Ex vivo and organ-cultured corneas have similar distributions of both components. Indirect immunofluorescence. e, epithelium; s, stroma.
Staining for another changed growth factor, FGF-3, was observed in normal corneal epithelium and keratocytes. In agreement with gene microarray and QPCR data, the epithelial staining was decreased in ex vivo and organ-cultured DR corneas (Fig. 5; Table 4). One of the FGF-3 receptors, FGFR3, had a similar distribution50 but distinct nuclear staining in the corneal epithelium. FGFR3 was also decreased in DR corneas by all three methods used (Figs. 1, 2, 5; Table 4).
FIGURE 5.
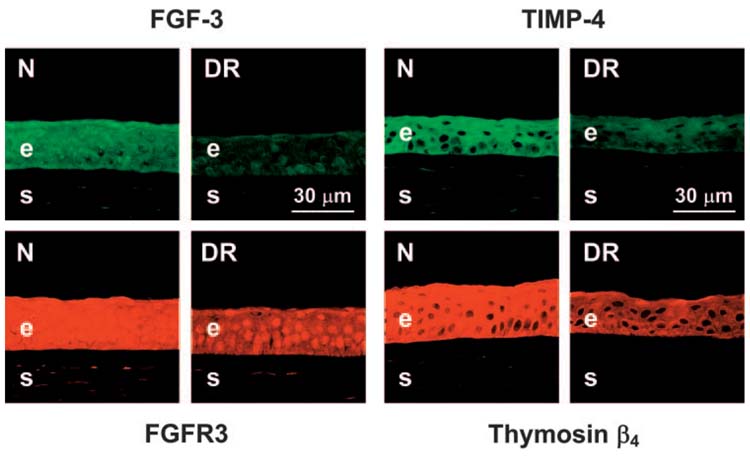
Distribution of FGF-3, its receptor FGFR3, TIMP-4, and thymosin β4 in ex vivo and organ-cultured normal (N) and DR corneas. FGF-3 (upper left panels, pAb AF1206) and FGFR3 (lower left panels, pAb C-15) are diminished in DR corneal epithelium. The same is true for TIMP-4 (upper right panels, mAb 153934) and thymosin β4 (lower right panels, pAb 609270). Data for ex vivo and organ-cultured corneas are similar; only ex vivo corneas are presented. Indirect immunofluorescence. e, epithelium; s, stroma.
TIMP-4 was found in corneal epithelium in accordance with previous data37 and in some keratocytes. Staining was decreased in DR corneal epithelium (Fig. 5; Table 4), confirming gene microarray and QPCR data. Most MMPs that were decreased in diabetic corneas (Fig. 1; Table 3) could not be revealed by immunostaining even in normal corneas (Ref. 21 and Ljubimov AV, unpublished data, 2004).
Because DR corneas have delayed wound healing,25 we were interested in the expression of cell migration modulators such as HGF (see above). Another migration stimulator, thymosin β4 (Tβ4), was found to be significantly less expressed in DR than in normal corneas by gene microarray and QPCR (Figs. 1, 2). Consistent with this result, epithelial staining for Tβ4 was decreased significantly in ex vivo and organ-cultured DR corneas compared with normal corneas (Fig. 5; Table 4). Two different antibodies to Tβ4 (Table 2) gave identical staining.
We have previously documented laminin-10 (α5β1γ1) changes in the epithelial BM of DR corneas.20,25 Gene microarray analysis additionally identified laminin α4 chain (not described in the cornea previously) as decreased in DR corneas. Although QPCR did not confirm gene microarray data on α4 (not shown here), this chain was further analyzed by immunhistochemistry. The antibody to the α4 chain IIIa domain34 gave a uniform staining of the normal corneal epithelial BM (not shown). Weak staining was seen at the endothelial side of Descemet membrane (not shown), where laminin-10 is also found. Given that in these locations laminin β1 chain is present but β2 chain is absent,51 the data are consistent with the presence of laminin-8 (α4β1γ1).52 In ex vivo and organ-cultured DR corneas, the epithelial BM staining was weaker than normal and locally discontinuous in most cases (Table 4), similar to changes observed previously for laminin-10 α5 chain.20,25
Treatment of Organ-cultured Corneas with Cystatin C or MMP-10
To gain further insight into the functional significance of elevated proteinase expression in DR corneas, wounded organ cultures were used. Treatment of DR corneas with a natural cathepsin inhibitor, cystatin C, resulted in increased and more continuous immunostaining for laminin-10 α5 chain in the epithelial BM and for integrin α3β1 in epithelial cells (Fig. 6, left). After treatment, their patterns became similar to those seen in normal corneas. Epithelial wound healing rates did not change significantly after cystatin C treatment, possibly because not enough corneas were analyzed. Treatment of normal organ cultures with cathepsin F was not attempted because the purified protein was not commercially available.
FIGURE 6.
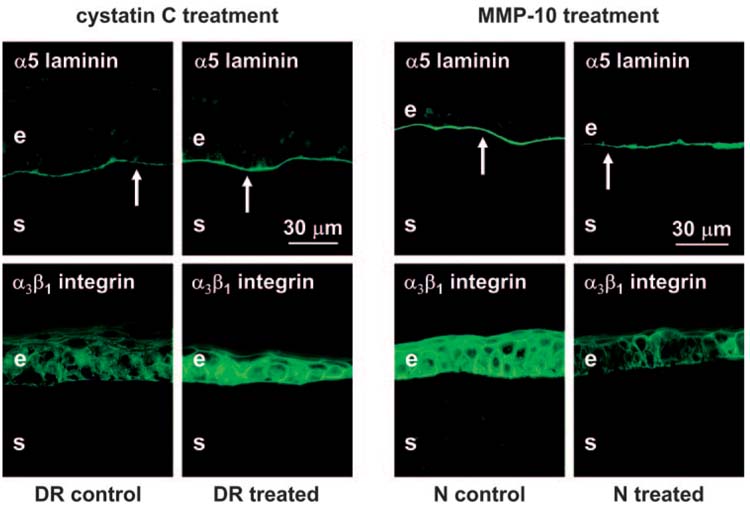
Left panels: Treatment of wounded DR organ-cultured corneas with cathepsin inhibitor cystatin C. Healed corneas are presented. Laminin-10 α5 chain staining (arrows) becomes continuous and similar to normal after cystatin C treatment (upper panels). Integrin α3β1 staining is weak and irregular in control corneas but becomes significantly stronger and more regular in cystatin C-treated corneas (lower panels). Right panels: Treatment of wounded normal organ-cultured corneas with MMP-10. Healed corneas are presented. Note continuous staining of the epithelial BM (arrows) for laminin-10 α5 chain in BSA-treated (control) corneas and discontinuous staining in MMP-10-treated corneas (upper panels). Strong staining for integrin α3β1 is seen in control corneas; it is weak and irregular in treated corneas (lower panels). Indirect immunofluorescence. e, epithelium; s, stroma.
Because our previous data showed elevated expression of MMP-10 in DR corneas,21 we were interested in examining whether in normal corneas this proteinase would change diabetic markers and wound healing rates toward a more diabetic phenotype. To this end, normal organ-cultured wounded corneas were treated with MMP-10, which resulted in slower epithelial wound healing (5.5 ± 1.9 days for MMP-10-treated vs 3.4 ± 0.8 days for BSA-treated). Although statistical significance was not reached, possibly because of individual variability, there was a tendency toward slower healing in MMP-10-treated organ cultures. Immunofluorescence analysis showed that the laminin-10 α5 chain in MMP-10-treated corneas was locally discontinuous in the epithelial BM, whereas BSA-treated corneas typically had strong and continuous staining (Fig. 6, upper right). After MMP-10 treatment, epithelial staining for integrin α3β1 became weaker than normal and less regular (Fig. 6, lower right). These changes were similar to those previously observed in DR corneas.23,28
DISCUSSION
Gene microarray analysis is a powerful tool for identifying gene expression profiles of healthy and diseased tissue. In the cornea, this technology has been successfully used to identify gene expression in specific cell types48 and its changes after growth factor, cytokine, drug, or hypoxia treatment,53-55 during wound healing,56,57 and in keratoconus and bullous keratopathy.47,58
Although gene microarray results are generally accurate and reproducible,57 many authors agree that these data must be verified by subsequent mRNA and protein analysis.52,57 The present microarray results were validated by QPCR and immunostaining. However, we did not detect significant changes in the expression of MMP-3 and MMP-10 that were previously shown to be increased in DR corneas compared with normal corneas using various methods.21,25 The expression of these genes was only increased in a minority of diabetic and DR samples. Previously noted discrepancies between gene array and QPCR or protein data were ascribed to errors in conventional array analysis software, errors in cRNA amplification, lower array sensitivity compared with QPCR, or individual subject variability by age and sex.59-61 For these reasons, validation of microarray results by QPCR and immunohistochemistry or Western blotting remains a necessary step in identifying gene expression changes in disease.
Here we have attempted to identify gene and protein expression abnormalities in diabetic and DR human corneas compared with normal ones using Agilent 22,000-gene microarrays. Because diabetes affects all corneal cells, whole corneal gene expression was analyzed. We hypothesized21,28 that molecular alterations in diabetic corneas could have been caused by increased proteolysis, possibly triggered by altered growth factor/cytokine expression. Consequently, we focused on gene microarray analysis of proteinases and growth factors/cytokines comparing the expression of each gene in individual diabetic and DR corneas with the expression of the same gene in a pool of normal corneas. This approach21 allowed us to minimize individual variations between normal corneas, compare individual diseased corneas to reveal most common changes, and reduce the cost of analysis. Results were verified by QPCR (mRNA expression) and immunohistochemistry (protein expression) for select differentially expressed genes. None of these genes were altered in the same way in keratoconus or bullous keratopathy (Refs. 47, 58 and Spirin KS, Brown DJ, Ljubimov AV, Kenney MC, unpublished data, 2000) as they were in diabetic corneas, suggesting that their changes were specific for diabetes.
Previously, cathepsins A, B, D, G, K, L, and V/L2 were found in corneal cells.62-66 We provide here the first account of cathepsins F and H in human cornea. They were mostly localized in the epithelium, and cathepsin F expression was significantly increased in DR corneas (Fig. 3; Table 3). Two cathepsin inhibitors, cystatins F and S, were significantly decreased in diabetic and DR corneas (Fig. 1), which could lead to a net increase in cathepsin activity. Cathepsin F was described in macrophages and in atherosclerotic lesions, where it participates in major histocompatibility complex class 2 processing and can modify low-density lipoprotein particles by cleaving apolipoprotein B-100.67-69 Cathepsins are a family of proteolytic enzymes, many of which, including cathepsin F, are cysteine proteinases; some cathepsins are serine or aspartic proteinases.38 Cathepsins are mostly lysosomal proteinases that are increased in tumors and may degrade extracellular substrata such as laminin. The half-life of a cathepsin at neutral pH increases when it uses extracellular matrix proteins as substrata.70,71 Therefore, an increase in cathepsin F could play a role in the degradation of diabetic corneal epithelial BM and integrins.20 In fact, our data show that treating organ-cultured DR corneas with a potent cathepsin F inhibitor, cystatin C, restored normal laminin-10 and integrin α3β1 staining patterns (Fig. 6, left). In experimental nephrosis, the activation of cathepsin B is accompanied by a decrease of α3 integrin72;a similar situation may exist in diabetic corneas.
We previously observed an increased expression and activity of stromelysins MMP-3 and, especially, MMP-10 in diabetic and DR corneas.21 Therefore, it seemed important to examine a direct effect of MMP-10 (contrary to MMP-3, it was primarily upregulated in the epithelium in DR corneas) on diabetic markers and epithelial wound healing by treating normal organ-cultured corneas with MMP-10. The treatment delayed epithelial wound healing and led to diabetic-like alterations of laminin-10 α5 chain and integrin α3β1 (Fig. 6, right). Therefore, the upregulation of BM-degrading MMP-10 in DR corneas may play a functional role in diabetic corneal epithelial abnormalities. Overall, the data on cathepsin inhibitor and MMP-10 treatment of organ-cultured corneas provide additional support to our hypothesis of the importance of increased proteolysis in corneal diabetic alterations.
Stromelysin activity may be increased in diabetes by the downregulation of TIMPs. Previously, no increase of TIMP-1, TIMP-2, or TIMP-3 was found in diabetic corneas.21 However, the present analysis revealed the downregulation of TIMP-4 in such corneas (Figs. 1, 2, 5), suggesting an increase in net proteolytic activity.72 TIMP-4 can inhibit MMP-2 and MMP-9. To date, its interactions with MMP-3 and MMP-10 have not been studied and may be of interest for future investigations. TIMP-4 is able to induce cell apoptosis.73 Therefore, its decrease in DR corneas may also be related to a possible attempt of the corneal tissue to reduce cell death in high glucose conditions.
The HGF/c-met system is important for wound healing, cell migration, epithelial branching morphogenesis, and angiogenesis.41,42,75 In DR corneas, epithelial HGF increased, whereas c-met expression decreased (Tables 3, 4). The downregulation of c-met might impair HGF signaling, which could lead to reduced migration of DR corneal epithelium and delayed wound healing. A somewhat similar situation exists in fulminant hepatic failure, with abundant circulating HGF but lack of c-met in the liver, which results in massive cell death and liver failure.42 It would be interesting to identify the effects of increasing c-met in the cornea by gene therapy on wound healing in DR corneas.
Another indirect indication for decreased HGF activity in DR corneas may be the downregulation of Tβ4. This 5-kDa polypeptide is an actin-sequestering protein and a mediator of cell proliferation, differentiation, angiogenesis, and metastasis.45 It also promotes cell migration and wound healing.43-45 In DR corneas, Tβ4 expression is decreased in the epithelium (Figs. 2, 5; Tables 3, 4). Because HGF can enhance Tβ4 expression,76 its reduction in DR corneas may result from possible downregulation of HGF signaling because of decreased c-met. In turn, reduced expression of laminin expression-stimulating77 and migration-promoting43 Tβ4 may contribute to BM degradation and delayed wound healing in DR corneas.
FGF-3 plays a role in corneal development and epithelial differentiation.46 In DR corneas, it is decreased coordinately with one of its receptors, FGFR3 (Table 4). FGF-3 is identical to the int-2 proto-oncogene upregulated in different tumors.78 Certain data suggest that the FGF-3 locus on chromosome 11q may be implicated in genetic susceptibility to IDDM.79 FGFR3 is upregulated in skin wounds and in migrating neural crest cells during development.80,81 Therefore, FGF-3 and FGFR3 might participate in wound healing and cell migration. Their distribution in corneal epithelium may imply that a concomitant decrease of FGF-3 and FGFR3 in DR corneas may contribute to impaired corneal cell motility and delayed wound healing.
The laminin α4 chain is identified here for the first time in cornea. In corneal epithelial BM, it is probably part of laminin-8 (α4β1γ1).52 Laminin-8 is expressed in endothelial BM but is also found in skin epithelial BM.82 This laminin isoform is increased in human glioblastomas, participates in tumor invasion, and mediates cell migration.52,83,84 Therefore, it may mediate cell migration and corneal wound healing; its disruption in DR corneas (Table 4) may thus impair these processes. Given that there was no reduction in mRNA for the laminin α4 chain by QPCR (not shown), one may suggest that its decrease in DR corneas was caused by degradation by proteinases, such as MMP-1022,23 or cathepsin F.
In summary, the analysis of gene microarrays validated by QPCR and immunostaining of tissue sections allowed us to identify distinct abnormalities in the expression of several proteinases and growth factors in DR corneas compared with normal ones. Induction of a more normal phenotype in organ-cultured DR corneas by cathepsin F inhibition and of a more diabetic phenotype in normal corneas by MMP-10 treatment were also demonstrated, suggesting functional significance of elevated proteolysis in diabetic corneal alterations. It would be important to show that changes in specific growth factors (see also Refs. 26-28) would influence proteinase expression and to identify an exact role of select proteinases in epithelial BM degradation and impaired epithelial wound healing in DR corneas. Most of the growth factor, laminin, and proteinase changes described here primarily concern corneal epithelium, would lead to alterations of cell migration and wound healing, and may thus be directly related to clinically observed diabetic corneal abnormalities. The presented evidence may help elucidate the roles of distinct components in diabetic corneal alterations using antibodies, inhibitors, and purified proteins. It can also pave the way for an efficient future gene therapy85 for corneal diabetes. Select genes (MMP-10, cathepsin F) may be silenced using siRNA or antisense technology, whereas others (c-met, FGF-3/FGFR3, Tβ4, TIMP-4) may be upregulated using virally mediated gene transfer. In this respect, the corneal organ culture system would provide an ideal testing ground for these emerging therapies.
Supplementary Material
Acknowledgments
The authors thank Gregory Schultz (University of Florida, Gainesville, FL) for valuable advice on cluster analysis.
Footnotes
Presented in part at the annual meeting of the Association for Research in Vision and Ophthalmology, Fort Lauderdale, Florida, April 2004.
Supported by National Eye Institute Grants R01 EY13431 (AVL) and K08 13412 (GS), General Clincial Research Center Grant M01 RR00425 to Cedars-Sinai Medical Center (N-NC, CW, AVL), and the Skirball Program in Molecular Ophthalmology (MS, AAK, AMA, AVL). GS is a recipient of the Career Development Award from Research to Prevent Blindness.
Disclosure: M. Saghizadeh, None; A.A. Kramerov, None; J. Tajbakhsh, None; A.M. Aoki, None; C. Wang, None; N.-N. Chai, None; J.Y. Ljubimova, None; T. Sasaki, None; G. Sosne, None; M.R.J. Carlson, None; S.F. Nelson, None; A.V. Ljubimov, None
References
- 1.Aiello LP, Gardner TW, King GL, et al. Diabetic retinopathy. Diabetes Care. 1998;21:143–156. doi: 10.2337/diacare.21.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Frank RN. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 3.Didenko TN, Smolyakova GP, Sorokin YL, Egorov VV. Clinical and pathogenetic features of neurotrophic disorders in the cornea of diabetics [in Russian] Vestn Oftalmol. 1999;115:7–11. [PubMed] [Google Scholar]
- 4.Sanchez-Thorin JC. The cornea in diabetes mellitus. Int Ophthalmol Clin. 1998;38:19–36. [PubMed] [Google Scholar]
- 5.Saito J, Enoki M, Hara M, Morishige N, Chikama T, Nishida T. Correlation of corneal sensation, but not of basal or reflex tear secretion, with the stage of diabetic retinopathy. Cornea. 2003;22:15–18. doi: 10.1097/00003226-200301000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Gekka M, Miyata K, Nagai Y, et al. Corneal epithelial barrier function in diabetic patients. Cornea. 2004;23:35–37. doi: 10.1097/00003226-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Dogru M, Katakami C, Inoue M. Tear function and ocular surface changes in noninsulin-dependent diabetes mellitus. Ophthalmology. 2001;108:586–592. doi: 10.1016/s0161-6420(00)00599-6. [DOI] [PubMed] [Google Scholar]
- 8.Friberg TR, Ohji M, Scherer JJ, Tano Y. Frequency of epithelial debridement during diabetic vitrectomy. Am J Ophthalmol. 2003;135:553–554. doi: 10.1016/s0002-9394(02)02014-7. [DOI] [PubMed] [Google Scholar]
- 9.Kallinikos P, Berhanu M, O’Donnell C, Boulton AJ, Efron N, Malik RA. Corneal nerve tortuosity in diabetic patients with neuropathy. Invest Ophthalmol Vis Sci. 2004;45:418–422. doi: 10.1167/iovs.03-0637. [DOI] [PubMed] [Google Scholar]
- 10.Morikubo S, Takamura Y, Kubo E, Tsuzuki S, Akagi Y. Corneal changes after small-incision cataract surgery in patients with diabetes mellitus. Arch Ophthalmol. 2004;122:966–969. doi: 10.1001/archopht.122.7.966. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto A, Sasaki H, Kitagawa K. Successful treatment of diabetic keratopathy with punctal occlusion. Acta Ophthalmol Scand. 2004;82:115–117. doi: 10.1111/j.1395-3907.2004.0189h.x. [DOI] [PubMed] [Google Scholar]
- 12.Azar DT, Spurr-Michaud SJ, Tisdale AS, Gipson IK. Altered epithelial-basement membrane interactions in diabetic corneas. Arch Ophthalmol. 1992;110:537–540. doi: 10.1001/archopht.1992.01080160115045. [DOI] [PubMed] [Google Scholar]
- 13.Kaji Y, Usui T, Oshika T, et al. Advanced glycation end products in diabetic corneas. Invest Ophthalmol Vis Sci. 2000;41:362–368. [PubMed] [Google Scholar]
- 14.Sato E, Mori F, Igarashi S, et al. Corneal advanced glycation end products increase in patients with proliferative diabetic retinopathy. Diabetes Care. 2001;24:479–482. doi: 10.2337/diacare.24.3.479. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe H, Katakami C, Miyata S, Negi A. Corneal disorders in KKAy mouse: a type 2 diabetes model. Jpn J Ophthalmol. 2002;46:130–139. doi: 10.1016/s0021-5155(01)00487-7. [DOI] [PubMed] [Google Scholar]
- 16.Akimoto Y, Kawakami H, Yamamoto K, Munetomo E, Hida T, Hirano H. Elevated expression of O-GlcNAc-modified proteins and O-GlcNAc transferase in corneas of diabetic Goto-Kakizaki rats. Invest Ophthalmol Vis Sci. 2003;44:3802–3809. doi: 10.1167/iovs.03-0227. [DOI] [PubMed] [Google Scholar]
- 17.Fujita H, Morita I, Takase H, Ohno-Matsui K, Mochizuki M. Prolonged exposure to high glucose impaired cellular behavior of normal human corneal epithelial cells. Curr Eye Res. 2003;27:197–203. doi: 10.1076/ceyr.27.4.197.16598. [DOI] [PubMed] [Google Scholar]
- 18.McDermott AM, Xiao TL, Kern TS, Murphy CJ. Non-enzymatic glycation in corneas from normal and diabetic donors and its effects on epithelial cell attachment in vitro. Optometry. 2003;74:443–452. [PubMed] [Google Scholar]
- 19.Ljubimov AV, Burgeson RE, Butkowski RJ, et al. Basement membrane abnormalities in human eyes with diabetic retinopathy. J Histochem Cytochem. 1996;44:1469–1479. doi: 10.1177/44.12.8985139. [DOI] [PubMed] [Google Scholar]
- 20.Ljubimov AV, Huang ZS, Huang GH, et al. Human corneal epithelial basement membrane and integrin alterations in diabetes and diabetic retinopathy. J Histochem Cytochem. 1998;46:1033–1041. doi: 10.1177/002215549804600907. [DOI] [PubMed] [Google Scholar]
- 21.Saghizadeh M, Brown DJ, Castellon R, et al. Overexpression of matrix metalloproteinase-10 and matrix metalloproteinase-3 in human diabetic corneas: a possible mechanism of basement membrane and integrin alterations. Am J Pathol. 2001;158:723–734. doi: 10.1016/S0002-9440(10)64015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krampert M, Bloch W, Sasaki T, et al. Activities of the matrix metalloproteinase stromelysin-2 (MMP-10) in matrix degradation and keratinocyte organization in wounded skin. Mol Biol Cell. 2004;15:5242–5254. doi: 10.1091/mbc.E04-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rechardt O, Elomaa O, Vaalamo M, et al. Stromelysin-2 is upregulated during normal wound repair and is induced by cytokines. J Invest Dermatol. 2000;115:778–787. doi: 10.1046/j.1523-1747.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- 24.Alexander CM, Howard EW, Bissell MJ, Werb Z. Rescue of mammary epithelial cell apoptosis and entactin degradation by a tissue inhibitor of metalloproteinases-1 transgene. J Cell Biol. 1996;135:1669–1677. doi: 10.1083/jcb.135.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabosova A, Kramerov AA, Aoki AM, Murphy G, Zieske JD, Ljubimov AV. Human diabetic corneas preserve wound healing, basement membrane, integrin and MMP-10 differences from normal corneas in organ culture. Exp Eye Res. 2003;77:211–217. doi: 10.1016/s0014-4835(03)00111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HS, Shang T, Chen Z, Pflugfelder SC, Li DQ. TGF-β1 stimulates the production of gelatinase (MMP-9), collagenases (MMP-1, -13) and stromelysins (MMP-3, -10, -11) by human corneal epithelial cells. Exp Eye Res. 2004;79:263–274. doi: 10.1016/j.exer.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Gerber A, Welte T, Ansorge S, Buhling F. Expression of cathepsins B and L in human lung epithelial cells is regulated by cytokines. Adv Exp Med Biol. 2000;477:287–292. doi: 10.1007/0-306-46826-3_31. [DOI] [PubMed] [Google Scholar]
- 28.Saghizadeh M, Chwa M, Aoki A, et al. Altered expression of growth factors and cytokines in keratoconus, bullous keratopathy and diabetic human corneas. Exp Eye Res. 2001;73:179–189. doi: 10.1006/exer.2001.1028. [DOI] [PubMed] [Google Scholar]
- 29.Fonovic M, Bromme D, Turk V, Turk B. Human cathepsin F: expression in baculovirus system, characterization and inhibition by protein inhibitors. Biol Chem. 2004;385:505–509. doi: 10.1515/BC.2004.059. [DOI] [PubMed] [Google Scholar]
- 30.Bannikov GA, Karelina TV, Collier IE, Marmer BL, Goldberg GI. Substrate binding of gelatinase B induces its enzymatic activity in the presence of intact propeptide. J Biol Chem. 2002;277:16022–16027. doi: 10.1074/jbc.M110931200. [DOI] [PubMed] [Google Scholar]
- 31.Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62:4364–4368. [PubMed] [Google Scholar]
- 32.Desai KV, Michalowska AM, Kondaiah P, Ward JM, Shih JH, Green JE. Gene expression profiling identifies a unique androgen-mediated inflammatory/immune signature and a PTEN (phosphatase and tensin homolog deleted on chromosome 10)-mediated apoptotic response specific to the rat ventral prostate. Mol Endocrinol. 2004;18:2895–2907. doi: 10.1210/me.2004-0033. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Wong WH. DNA-Chip Analyzer (dChip) In: Parmigiani G, Garrett ES, Irizarry R, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. Springer; New York: 2003. [Google Scholar]
- 34.Sasaki T, Mann K, Timpl R. Modification of the laminin α4 chain by chondroitin sulfate attachment to its N-terminal domain. FEBS Lett. 2001;505:173–178. doi: 10.1016/s0014-5793(01)02812-5. [DOI] [PubMed] [Google Scholar]
- 35.Talts JF, Sasaki T, Miosge N, et al. Structural and functional analysis of the recombinant G domain of the laminin α4 chain and its proteolytic processing in tissues. J Biol Chem. 2000;275:35192–35199. doi: 10.1074/jbc.M003261200. [DOI] [PubMed] [Google Scholar]
- 36.Hansel DE, Rahman A, Hidalgo M, et al. Identification of novel cellular targets in biliary tract cancers using global gene expression technology. Am J Pathol. 2003;163:217–229. doi: 10.1016/S0002-9440(10)63645-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma DH, Zhang F, Shi W, et al. Expression of tissue inhibitor of metalloproteinase-4 in normal human corneal cells and experimental corneal neovascularization. Ophthalmic Res. 2003;35:199–207. doi: 10.1159/000071171. [DOI] [PubMed] [Google Scholar]
- 38.Turk D, Guncar G. Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr D Biol Crystallogr. 2003;59(pt 2):203–213. doi: 10.1107/s0907444902021479. [DOI] [PubMed] [Google Scholar]
- 39.Wilson SE, Mohan RR, Mohan RR, Ambrosio R, Jr, Hong J, Lee J. The corneal wound healing response: cytokine-mediated interaction of the epithelium, stroma, and inflammatory cells. Prog Retin Eye Res. 2001;20:625–637. doi: 10.1016/s1350-9462(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 40.Daniels JT, Limb GA, Saarialho-Kere U, Murphy G, Khaw PT. Human corneal epithelial cells require MMP-1 for HGF-mediated migration on collagen I. Invest Ophthalmol Vis Sci. 2003;44:1048–1055. doi: 10.1167/iovs.02-0442. [DOI] [PubMed] [Google Scholar]
- 41.Ljubimova JY, Petrovic LM, Wilson SE, Geller SA, Demetriou AA. Expression of HGF, its receptor c-met, c-myc, and albumin in cirrhotic and neoplastic human liver tissue. J Histochem Cytochem. 1997;45:79–87. doi: 10.1177/002215549704500111. [DOI] [PubMed] [Google Scholar]
- 42.Ljubimova JY, Petrovic LM, Arkadopoulos N, Blanc P, Geller SA, Demetriou AA. Lack of hepatocyte growth factor receptor (c-met) gene expression in fulminant hepatic failure livers before transplantation. Dig Dis Sci. 1997;42:1675–1680. doi: 10.1023/a:1018805330280. [DOI] [PubMed] [Google Scholar]
- 43.Sosne G, Hafeez S, Greenberry AL, 2nd, Kurpakus-Wheater M. Thymosin β4 promotes human conjunctival epithelial cell migration. Curr Eye Res. 2002;24:268–273. doi: 10.1076/ceyr.24.4.268.8414. [DOI] [PubMed] [Google Scholar]
- 44.Sosne G, Szliter EA, Barrett R, Kernacki KA, Kleinman H, Hazlett LD. Thymosin beta 4 promotes corneal wound healing and decreases inflammation in vivo following alkali injury. Exp Eye Res. 2002;74:293–299. doi: 10.1006/exer.2001.1125. [DOI] [PubMed] [Google Scholar]
- 45.Philp D, Goldstein AL, Kleinman HK. Thymosin β4 promotes angiogenesis, wound healing, and hair follicle development. Mech Ageing Dev. 2004;125:113–115. doi: 10.1016/j.mad.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Robinson ML, Ohtaka-Maruyama C, Chan CC, et al. Dysregulation of ocular morphogenesis by lens-specific expression of FGF-3/int-2 in transgenic mice. Dev Biol. 1998;198:13–31. doi: 10.1006/dbio.1998.8879. [DOI] [PubMed] [Google Scholar]
- 47.Spirin KS, Ljubimov AV, Castellon R, et al. Analysis of gene expression in human bullous keratopathy corneas containing limiting amounts of RNA. Invest Ophthalmol Vis Sci. 1999;40:3108–3115. [PubMed] [Google Scholar]
- 48.Chakravarti S, Wu F, Vij N, Roberts L, Joyce S. Microarray studies reveal macrophage-like function of stromal keratocytes in the cornea. Invest Ophthalmol Vis Sci. 2004;45:3475–3484. doi: 10.1167/iovs.04-0343. [DOI] [PubMed] [Google Scholar]
- 49.Philipp WE, Speicher L, Goöattinger W. Histological and immunohistochemical findings after laser in situ keratomileusis in human corneas. J Cataract Refract Surg. 2003;29:808–820. doi: 10.1016/s0886-3350(02)01611-5. [DOI] [PubMed] [Google Scholar]
- 50.Hughes SE. Differential expression of the fibroblast growth factor receptor (FGFR) multigene family in normal human adult tissues. J Histochem Cytochem. 1997;45:1005–1019. doi: 10.1177/002215549704500710. [DOI] [PubMed] [Google Scholar]
- 51.Ljubimov AV, Burgeson RE, Butkowski RJ, Michael AF, Sun T-T, Kenney MC. Human corneal basement membrane heterogeneity: topographical differences in the expression of type IV collagen and laminin isoforms. Lab Invest. 1995;72:461–473. [PubMed] [Google Scholar]
- 52.Ljubimova J, Lakhter AJ, Loksh A, et al. Overexpression of α4 chain-containing laminins in human glial tumors identified by gene microarray analysis. Cancer Res. 2001;61:5601–5610. [PubMed] [Google Scholar]
- 53.Hong JW, Liu JJ, Lee JS, et al. Proinflammatory chemokine induction in keratocytes and inflammatory cell infiltration into the cornea. Invest Ophthalmol Vis Sci. 2001;42:2795–2803. [PubMed] [Google Scholar]
- 54.Lo WR, Rowlette LL, Caballero M, Yang P, Hernandez MR, Borraás T. Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steroid glaucoma. Invest Ophthalmol Vis Sci. 2003;44:473–485. doi: 10.1167/iovs.02-0444. [DOI] [PubMed] [Google Scholar]
- 55.Harvey SA, Anderson SC, SundarRaj N. Downstream effects of ROCK signaling in cultured human corneal stromal cells: microarray analysis of gene expression. Invest Ophthalmol Vis Sci. 2004;45:2168–2176. doi: 10.1167/iovs.03-1218. [DOI] [PubMed] [Google Scholar]
- 56.Varela JC, Goldstein MH, Baker HV, Schultz GS. Microarray analysis of gene expression patterns during healing of rat corneas after excimer laser photorefractive keratectomy. Invest Ophthalmol Vis Sci. 2002;43:1772–1782. [PubMed] [Google Scholar]
- 57.Cao Z, Wu HK, Bruce A, Wollenberg K, Panjwani N. Detection of differentially expressed genes in healing mouse corneas, using cDNA microarrays. Invest Ophthalmol Vis Sci. 2002;43:2897–2904. [PubMed] [Google Scholar]
- 58.Nielsen K, Birkenkamp-Demtroder K, Ehlers N, Orntoft TF. Identification of differentially expressed genes in keratoconus epithelium analyzed on microarrays. Invest Ophthalmol Vis Sci. 2003;44:2466–2476. doi: 10.1167/iovs.02-0671. [DOI] [PubMed] [Google Scholar]
- 59.Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19:1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- 60.Asyali MH, Shoukri MM, Demirkaya O, Khabar KS. Assessment of reliability of microarray data and estimation of signal thresholds using mixture modeling. Nucleic Acids Res. 2004;32:2323–2335. doi: 10.1093/nar/gkh544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowers I, Liu D, Farkas RH, et al. Gene expression variation in the adult human retina. Hum Mol Genet. 2003;12:2881–2893. doi: 10.1093/hmg/ddg326. [DOI] [PubMed] [Google Scholar]
- 62.Obuchowska I, Stankiewicz A, Mariak Z. Cathepsin A activity of normal bovine ocular tissues and pathological human intraocular fluids. Acta Biochim Pol. 1996;43:687–692. [PubMed] [Google Scholar]
- 63.Dong Z, Katar M, Linebaugh BE, Sloane BF, Berk RS. Expression of cathepsins B, D and L in mouse corneas infected with Pseudomonas aeruginosa. Eur J Biochem. 2001;268:6408–6416. doi: 10.1046/j.0014-2956.2001.02607.x. [DOI] [PubMed] [Google Scholar]
- 64.Haeckel C, Krueger S, Buehling F, et al. Expression of cathepsin K in the human embryo and fetus. Dev Dyn. 1999;216:89–95. doi: 10.1002/(SICI)1097-0177(199910)216:2<89::AID-DVDY1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 65.Whitelock RB, Fukuchi T, Zhou L, et al. Cathepsin G, acid phosphatase, and α1-proteinase inhibitor messenger RNA levels in keratoconus corneas. Invest Ophthalmol Vis Sci. 1997;38:529–534. [PubMed] [Google Scholar]
- 66.Adachi W, Kawamoto S, Ohno I, et al. Isolation and characterization of human cathepsin V: a major proteinase in corneal epithelium. Invest Ophthalmol Vis Sci. 1998;39:1789–1796. [PubMed] [Google Scholar]
- 67.Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Brömme D. Human cathepsin F: molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem. 1998;273:32000–32008. doi: 10.1074/jbc.273.48.32000. [DOI] [PubMed] [Google Scholar]
- 68.Lindstedt L, Lee M, Oorni K, Bromme D, Kovanen PT. Cathepsins F and S block HDL3-induced cholesterol efflux from macrophage foam cells. Biochem Biophys Res Commun. 2003;312:1019–1024. doi: 10.1016/j.bbrc.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 69.öörni K, Sneck M, Brömme D, et al. Cysteine protease cathepsin F is expressed in human atherosclerotic lesions, is secreted by cultured macrophages, and modifies low density lipoprotein particles in vitro. J Biol Chem. 2004;279:34776–34784. doi: 10.1074/jbc.M310814200. [DOI] [PubMed] [Google Scholar]
- 70.Heck LW, Blackburn WD, Irwin MH, Abrahamson DR. Degradation of basement membrane laminin by human neutrophil elastase and cathepsin G. Am J Pathol. 1990;136:1267–1274. [PMC free article] [PubMed] [Google Scholar]
- 71.Buck MR, Karustis DG, Day NA, Honn KV, Sloane BF. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem J. 1992;282(pt 1):273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reiser J, Oh J, Shirato I, et al. Podocyte migration during nephrotic syndrome requires a coordinated interplay between cathepsin L and α3 integrin. J Biol Chem. 2004;279:34827–34832. doi: 10.1074/jbc.M401973200. [DOI] [PubMed] [Google Scholar]
- 73.Schulze CJ, Wang W, Suarez-Pinzon WL, Sawicka J, Sawicki G, Schulz R. Imbalance between tissue inhibitor of metalloproteinase-4 and matrix metalloproteinases during acute myocardial ischemia-reperfusion injury. Circulation. 2003;107:2487–2492. doi: 10.1161/01.CIR.0000065603.09430.58. [DOI] [PubMed] [Google Scholar]
- 74.Mannello F, Gazzanelli G. Tissue inhibitors of metalloproteinases and programmed cell death: conundrums, controversies and potential implications. Apoptosis. 2001;6:479–482. doi: 10.1023/a:1012493808790. [DOI] [PubMed] [Google Scholar]
- 75.Ma PC, Maulik G, Christensen J, Salgia R. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003;22:309–325. doi: 10.1023/a:1023768811842. [DOI] [PubMed] [Google Scholar]
- 76.Oh IS, So SS, Jahng KY, Kim HG. Hepatocyte growth factor upregulates thymosin β4 in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2002;296:401–405. doi: 10.1016/s0006-291x(02)00888-4. [DOI] [PubMed] [Google Scholar]
- 77.Sosne G, Xu L, Prach L, et al. Thymosin beta 4 stimulates laminin-5 production independent of TGF-beta. Exp Cell Res. 2004;293:175–183. doi: 10.1016/j.yexcr.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 78.Li Y, Hively WP, Varmus HE. Use of MMTV-Wnt-1 transgenic mice for studying the genetic basis of breast cancer. Oncogene. 2000;19:1002–1009. doi: 10.1038/sj.onc.1203273. [DOI] [PubMed] [Google Scholar]
- 79.Hashimoto L, Habita C, Beressi JP, et al. Genetic mapping of a susceptibility locus for insulin-dependent diabetes mellitus on chromosome 11q. Nature. 1994;371:161–164. doi: 10.1038/371161a0. [DOI] [PubMed] [Google Scholar]
- 80.Kubota Y, Ito K. Chemotactic migration of mesencephalic neural crest cells in the mouse. Dev Dyn. 2000;217:170–179. doi: 10.1002/(SICI)1097-0177(200002)217:2<170::AID-DVDY4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 81.Takenaka H, Yasuno H, Kishimoto S. Immunolocalization of fibroblast growth factor receptors in normal and wounded human skin. Arch Dermatol Res. 2002;294:331–338. doi: 10.1007/s00403-002-0333-z. [DOI] [PubMed] [Google Scholar]
- 82.Petäjäniemi N, Korhonen M, Kortesmaa J, et al. Localization of laminin α4-chain in developing and adult human tissues. J Histochem Cytochem. 2002;50:1113–1130. doi: 10.1177/002215540205000813. [DOI] [PubMed] [Google Scholar]
- 83.Fujiwara H, Gu J, Sekiguchi K. Rac regulates integrin-mediated endothelial cell adhesion and migration on laminin-8. Exp Cell Res. 2004;292:67–77. doi: 10.1016/j.yexcr.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 84.Khazenzon NM, Ljubimov AV, Lakhter AJ, et al. Antisense inhibition of laminin-8 expression reduces invasion of human gliomas in vitro. Mol Cancer Ther. 2003;2:985–994. [PubMed] [Google Scholar]
- 85.Rosenblatt MI, Azar DT. Gene therapy of the corneal epithelium. Int Ophthalmol Clin. 2004;44:81–90. doi: 10.1097/00004397-200404430-00010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


