Abstract
Cryptochromes are blue/ultraviolet-A light sensing photoreceptors involved in regulating various growth and developmental responses in plants. Investigations on the structure and functions of cryptochromes in plants have been largely confined to Arabidopsis (Arabidopsis thaliana), tomato (Lycopersicon esculentum), and pea (Pisum sativum). We report here the characterization of the cryptochrome 1 gene from Brassica napus (BnCRY1), an oilseed crop, and its functional validation in transgenics. The predicted BnCRY1 protein sequence shows a high degree of sequence identity (94%) to Arabidopsis CRY1. A semiquantitative reverse transcription-polymerase chain reaction and the western-blot analysis revealed that blue light up-regulates its transcript and protein levels in young seedlings. The BnCRY1 promoter harbors conventional light-responsive cis-acting elements, which presumably impart light activation to the GUS (β-glucuronidase) reporter gene expressed in Arabidopsis. Although the BnCRY1 transcript could be detected in all the tissues examined, its protein was virtually undetectable in mature leaves and the root, indicating a tissue-specific translational control or protein turnover. The antisense-BnCRY1 Brassica transgenic seedlings accumulated negligible levels of CRY1 protein and displayed an elongated hypocotyl when grown under continuous white or blue light (but not under red or far-red light); the accumulation of anthocyanins was also reduced significantly. The adult transformants were also found to be tall when grown under natural light environment in a containment facility without any artificial illumination. These data provide functional evidence for a role of blue light up-regulated cry1 in controlling photomorphogenesis in Brassica species.
Plants have evolved sophisticated sensory photoreceptors, which coordinately judge the quality, quantity, direction, and duration of light, to regulate diverse photomorphogenic responses throughout their life cycle (Gyula et al., 2003; Sullivan and Deng, 2003; Franklin and Whitelam, 2004). These sensory photoreceptors have been classified broadly into three groups based on the wavelength of light they perceive. Phytochromes, which are best characterized and extensively studied, comprise a small family of red/far-red (600–750 nm) sensing photoreceptors (Khurana et al., 1998, 2004; Quail, 2002; Chen et al., 2004). Cryptochromes and phototropins perceive the blue/UV-A (320–500 nm) part of the solar spectrum (Briggs and Olney, 2001; Khurana, 2001; Cashmore, 2003; Lin and Shalitin, 2003; Chen et al., 2004; Banerjee and Batschauer, 2005). The photoreceptors responsible for perceiving UV-B radiation (280–320 nm), however, remain elusive (Bharti and Khurana, 1997; Frohnmeyer and Staiger, 2003).
The first cryptochrome gene was cloned through the molecular analysis of T-DNA insertion mutant allele of hy4 (Ahmad and Cashmore, 1993). The HY4 gene encodes a protein of 681 amino acid residues, with a high degree of sequence match to photolyase, a DNA repair enzyme activated by blue light. Later, HY4 was designated as cryptochrome 1, cry1 (Lin et al., 1995). The second member of the cryptochrome gene family, At-PHH1 or CRY2, was isolated subsequently by screening an Arabidopsis (Arabidopsis thaliana) cDNA library using CRY1 as a probe (Hoffman et al., 1996; Lin et al., 1996b). The AtCRY1 and AtCRY2 proteins show approximately 58% identity within the N-terminal region, whereas the C-terminal extension shows only approximately 14% identity (Hoffman et al., 1996; Lin et al., 1998). Cryptochromes have now been identified from diverse species, including Chlamydomonas reinhardtii (Small et al., 1995), Physcomitrella patens (Imaizumi et al., 1999, 2002), Adiantum capillus-veneris (Kanegae and Wada, 1998; Imaizumi et al., 2000), rice (Oryza sativa; Kumar, 2000; Matsumoto et al., 2003), tomato (Lycopersicon esculentum; Ninu et al., 1999; Perrotta et al., 2000), pea (Pisum sativum; Platten et al., 2005a, 2005b), and a nonphotosynthetic holoparasitic plant, Orobanche minor (Okazawa et al., 2005). Using a random PCR approach, various cryptochrome members from angiosperms like melon (Cucumis melo), banana (Musa spp.), and barley (Hordeum vulgare) were isolated (Perrotta et al., 2001); however, their function remains unknown. Cryptochromes have also been identified and functionally characterized from Drosophila, zebrafish, mouse, and human (van der Spek et al., 1996; Emery et al., 1998; Kobayashi et al., 1998, 2000). Animal cryptochromes, in most cases, play a role in entrainment of circadian clock and act as components of the central oscillator (Cashmore, 2003; Sancar, 2004).
In plants, cryptochromes (cry1 and cry2) participate in many aspects of photomorphogenesis, such as inhibition of hypocotyl elongation (Ahmad and Cashmore, 1993; Lin et al., 1998; Lin, 2002), accumulation of anthocyanins (Ahmad et al., 1995), and cotyledon expansion (Botto et al., 2003). In addition, cryptochromes also regulate flowering time (Guo et al., 1998; Mockler et al., 1999; Giliberto et al., 2005) and circadian clock (Devlin and Kay, 1999, 2000; Millar, 2003). The processes like deetiolation, flowering, and circadian entrainment are in fact coordinately regulated by the combined action of phytochromes and cryptochromes (Casal, 2000; Sullivan and Deng, 2003).
In dark, cry1 is localized in the nucleus and detected primarily in the cytoplasm on exposure to light, whereas cry2 is confined to the nucleus in both dark and light (Guo et al., 1999; Yang et al., 2000). Light induced activity of cry1 and cry2 is mediated through its C-terminal (CCT) domain (Yang et al., 2000). The activation of CCT1 (of cry1) most likely is mediated through the blue light-dependent alteration in the dimerized N terminal of cry1 (Sang et al., 2005). The C-terminal domain was also shown to interact with the master regulator COP1 to control photomorphogenesis (Wang et al., 2001; Yang et al., 2001). Besides COP1, only a few more signaling components (e.g. SUB1, PP7, HFR1, OBP3, HRB1, and AtMYC2) involved in cryptochrome-mediated blue light signaling have been identified (Guo et al., 2001; Duek and Fankhauser, 2003; Moller et al., 2003; Kang et al., 2005; Ward et al., 2005; Yadav et al., 2005). Only recently, an insight into the primary photochemistry underlying the photoactivation of cry1 has been gained. It involves intraprotein electron transfer from conserved residues (Trp and Tyr) to the excited flavin adenine dinucleotide (FAD), which stimulates the autophosphorylation of cry1 and is responsible for its biological activity (Giovani et al., 2003; Zeugner et al., 2005).
Among higher plants, cryptochromes have been well studied and characterized only in Arabidopsis, tomato, and pea. To learn more about cryptochromes, we have initiated the characterization and functional analysis of the cryptochrome gene family from an agronomical important crop plant, Brassica napus, a close relative of Arabidopsis. The CRY1 gene was isolated from a variety ISN-706, which is cultivated in northern and cooler regions of India and is valued for oilseed. The BnCRY1 gene is represented as a single copy in the genome of B. napus, an allotetraploid, and its expression is up-regulated by light, both in terms of transcript abundance and the translational product. The analysis of anti-BnCRY1 transgenics has substantiated the role of CRY1 in regulating plant height and anthocyanin accumulation.
RESULTS
Gene Encoding CRY1 Protein in B. napus
The full-length BnCRY1 gene from B. napus was isolated by screening a genomic library using AtCRY1 gene as a probe. Two of the strongly hybridizing clones were verified by sequence analysis and found to be identical. The larger clone was processed for sequencing by primer walking. The genomic sequence thus obtained was used to design primers and the corresponding cDNA clone amplified by reverse transcription (RT)-PCR and completed by 5′ RACE and 3′ RACE. The sequence of the genomic and cDNA clones of BnCRY1 is available in the EMBL Nucleotide Sequence Database (accession nos. AJ344565 [gene] and AJ704628 [cDNA]).
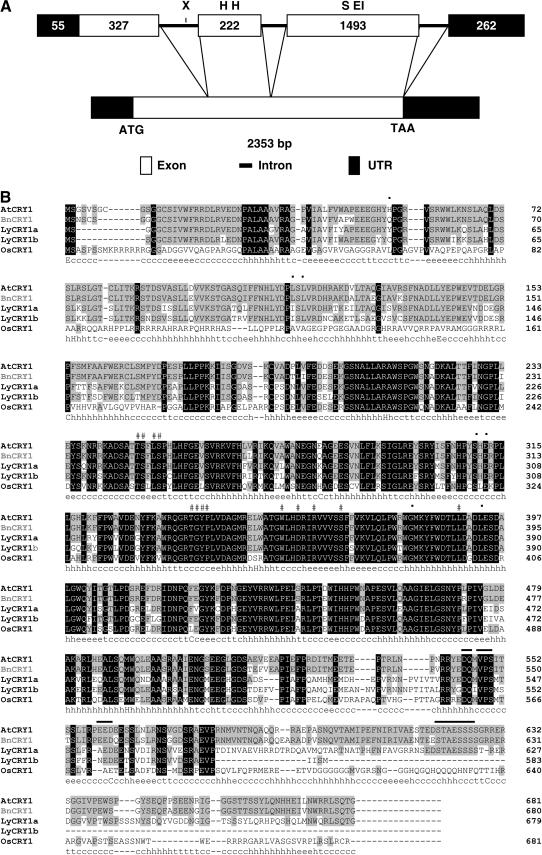
A comparative analysis of cDNA and genomic sequences revealed that BnCRY1 contains three introns and four exons (Fig. 1A). The third intron spans 188 bp and is followed immediately by a 261 bp 3′ untranslated region (UTR), which makes up the fourth exon. The stop codon (TAA) is generated by splicing of the third and fourth exons. The BnCRY1 cDNA contains a 5′ noncoding region of 55 nucleotides and a coding region of 2,040 nucleotides (680 amino acids, 76.7 kD). It harbors a polyadenylation signal (AATAAA) at position 2,269 to 2,274 bp just before the polyA tail. The Kyte-Doolittle hydropathy plot analysis does not show any hydrophobic region (data not shown), suggesting that BnCRY1 is a soluble protein consistent with the earlier reports on AtCRY1 (Ahmad and Cashmore, 1993) and other cryptochromes.
Figure 1.
A, Schematic diagram representing the alignment of BnCRY1 cDNA with the corresponding gene. Exon borders are indicated with a line connecting the cDNA and the exons. Numbers depict the size of UTR and exons. Restriction sites for EcoRI (EI), HindIII (H), SalI (S), and XbaI (X), which have been used for Southern analysis, are indicated on the horizontal bar representing gene structure. B, Amino acid sequence alignment of five representative plant cryptochromes using ClustalW. Black-boxed and gray-boxed letters represent residues that are identical in all or most cryptochromes, respectively. # and • symbols indicate the residues interacting with FAD and MTHF, respectively. Lines above the sequences mark the DAS domain present in the C-terminal region. The predicted secondary structure of BnCRY1 as determined using SOPM is shown below the alignment data and consists of α-helices (h), extended β-sheets (e), and coil (c) regions.
Using the ClustalW algorithm (Thompson et al., 1994), the deduced amino acid sequence of BnCRY1 was aligned with AtCRY1, LeCRY1, and OsCRY1 (Ahmad and Cashmore, 1993; Perrotta et al., 2000; Matsumoto et al., 2003). When compared to other cryptochromes, including dicot and monocot representatives, a high percentage of sequence identity was observed in the N-terminal PHR (photolyase-related) domain of BnCRY1 (Fig. 1B). In the C-terminal region, although overall similarity is low, all three hallmark motifs are conserved. Collectively, these three motifs are known as the DAS domain and comprise DQXVP (function unknown), an acidic (short stretch represented by E and D), and STAESSSS (implicated in interaction with phytochrome A [phyA]) motifs (Ahmad et al., 1998b; Kanegae and Wada, 1998). However, the traditional STAESSSS motif present in dicots is not conserved in OsCRY1 (Matsumoto et al., 2003).
Like Type I photolyases, AtCRY1 associates with two cofactors, the light-harvesting cofactor (MTHF) and a catalytic cofactor (FAD; Lin et al., 1995; Malhotra et al., 1995). All 13 amino acids, predicted to interact with FAD in AtCRY1, were found to be conserved in BnCRY1. The TGYP motif was also observed at the 337- to 340-amino acid position, which is conserved in all the Type I photolyases and forms a part of the FAD-binding domain (Malhotra et al., 1992). Six out of seven identical amino acid residues (His at position 52 is replaced by Gln), known to interact with the light-harvesting cofactor (MTHF), are also conserved in BnCRY1 (Fig. 1B).
The secondary structure of BnCRY1 was solved by the self-optimized prediction method (SOPM; Geourjon and Deleage, 1994). The SOPM results indicate that BnCRY1 consists of the α-helix (37.10%), β-strand (15.10%), and random coil (39.74%; Fig. 1B). The software did not provide the percentage of 310 helix, which plays a major role in the structural configuration of both photolyases and AtCRY1 (Brautigam et al., 2004). The α-helices and β-strands were randomly distributed throughout the BnCRY1 polypeptide and not organized into any specific domain.
Relationship with Other Cryptochromes
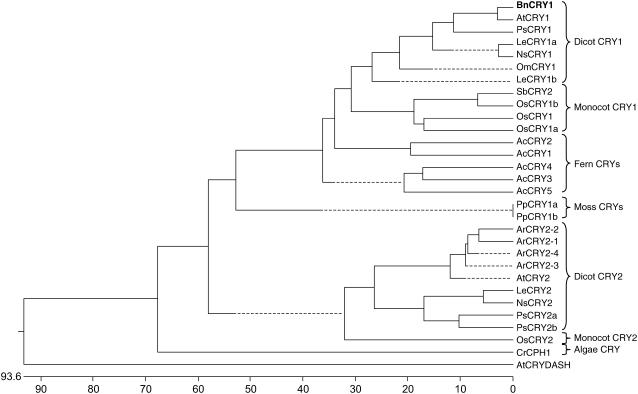
The phylogenetic analysis of 30 plant and near-plant cryptochromes representing 12 diverse species was carried out using the Dnastar MegAlign program by the Clustal method (Fig. 2). The BnCRY1 grouped under dicot CRY1 clade and showed maximum similarity with AtCRY1, which again reflects a close evolutionary relationship between Arabidopsis and Brassica. A distinct coevolution of cryptochromes along with the hierarchy of plant taxons from algae to angiosperms was also apparent. Duplication of the CRY gene into CRY1 and CRY2 predates the dicot-monocot divergence as these genes were found in both dicots and monocots. Interestingly, the presence of only CRY1-like genes in Adiantum and Physcomitrella suggests that the gene duplication that gave rise to CRY1 and CRY2 major lineages occurred after the divergence of lower plants and seed plants (Spermatophyta). Seed plants are believed to have evolved in the late Paleozoic era about 360 million years ago (Mya), whereas monocots and dicots diverged around 170 Mya (Sanderson et al., 2004). Thus, based on this analysis, we hypothesize that the split between the CRY1-like and CRY2-like lineages occurred between 170 Mya and 360 Mya.
Figure 2.
Phylogram of plant cryptochromes. The amino acid sequences of 30 plant and near-plant genes included in the analysis were obtained from the National Center for Biotechnology Information (NCBI) database. The alignment was conducted by Dnastar MegAlign program using Clustal method under default options. The abbreviations used are as follows: At, Arabidopsis; Ac, A. capillus-veneris; Ar, Armoracia rusticana; Bn, B. napus; Le, tomato; Om, O. minor; Os, rice; Pp, P. patens; Ps, pea; Sb, Sorgham bicolor; Cr, C. reinhardtii; and Ns, Nicotiana sylvestris.
BnCRY1 Is Represented as a Single Copy Gene on the Genome of an Allotetraploid
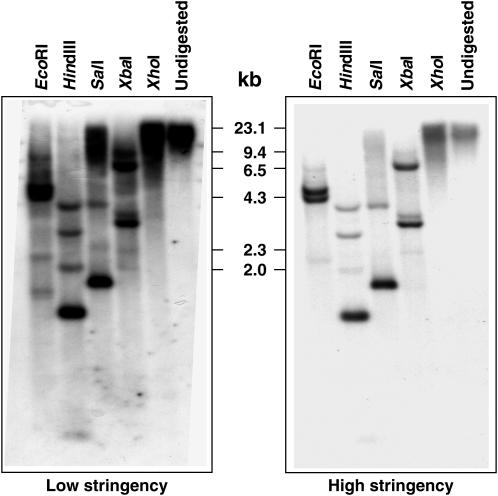
B. napus is a natural allotetraploid (2n = 38, AACC) derived from interspecific hybridization of the two diploid (A and C) genomes of Brassica rapa and Brassica oleracea, respectively, followed by spontaneous chromosome doubling (Song et al., 1988; Lakshmikumaran et al., 2003). Thus, two copies of BnCRY1 were expected. Southern-blot hybridization carried out with a gene probe harboring the entire coding region of BnCRY1, particularly under high (42°C) stringency conditions (Fig. 3), however, showed that BnCRY1 is represented as a single copy in the B. napus genome. The restriction map of BnCRY1 shows one site each for EcoRI, SalI, XbaI, and two sites for the HindIII restriction enzyme (Fig. 1A). The XhoI enzyme does not restriction digest BnCRY1. The in silico restriction profile matches well with the Southern profile obtained under high stringency conditions (Fig. 3). A few additional (but mostly faint) bands observed under both high and low stringency conditions may represent nonspecific hybridization with one or more copies of as yet uncharacterized CRY2 gene(s). It will be interesting to establish the genomic (A or C genome) localization of the BnCRY1 characterized in this study.
Figure 3.
Southern analysis of BnCRY1 under low and high stringency conditions. Numbers marked in the middle of the two sections represent the size of HindIII-digested λ DNA. Restriction enzymes used for digesting genomic DNA have been indicated on the top of the sections.
Light-Dependent and Spatial/Temporal Expression Profile
Gene Expression Analysis
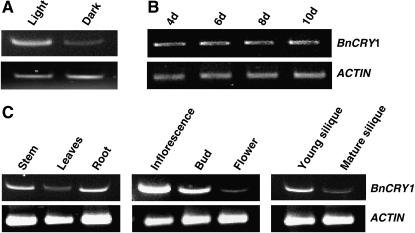
To examine whether transcript levels of BnCRY1 are regulated by light and developmental cues and display tissue specificity, its expression was examined by semiquantitative RT-PCR, using gene-specific primers. The BnCRY1 transcript levels were found to be higher in seedlings grown in white light (70 μmol m−2 s−1) in comparison to the dark-grown seedlings (Fig. 4A). However, the transcript levels were more or less similar in seedlings grown in light for various durations (Fig. 4B). The BnCRY1 transcript was present ubiquitously, to a detectable level, in all the organs examined, including stem, leaf, root, inflorescence, floral bud, flower, and silique (Fig. 4C); it was relatively more abundant in the stem, inflorescence (consisting of inflorescence meristem and emerging floral buds), young silique, and, surprisingly, in the root.
Figure 4.
Comparative RT-PCR analysis to show light and developmental regulation, and tissue-specific expression of BnCRY1. A shows the transcript levels in 6-d-old dark- and light-grown seedlings. B and C show the BnCRY1 transcript levels at different developmental stages and in various tissues/organs, respectively. The inflorescence tissue consisted of inflorescence meristem and young emerging floral buds. ACTIN transcript was used as internal control.
Immunoblot Analysis for Protein Profile
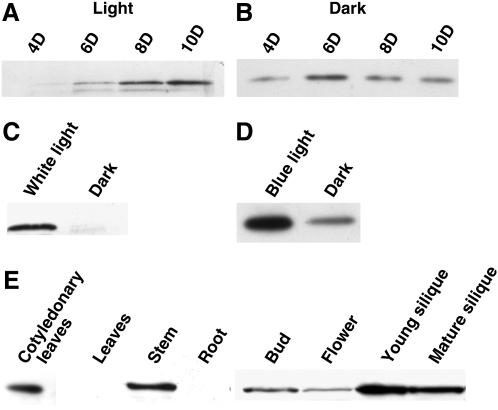
The BnCRY1 protein of approximately 76 kD could be detected (by immunoblot assay) in the extracts of whole seedlings grown in dark or white light (70 μmol m−2 s−1) for various durations (Fig. 5, A and B). Apart from the predominant 76-kD polypeptide, a fast-migrating polypeptide was always detected in the extracts of the light-grown tissue. This additional polypeptide may represent an altered phosphorylation status of CRY1 (Shalitin et al., 2002). The level of BnCRY1 was found to increase significantly in seedlings grown in light from 4 to 10 d. In comparison, however, the level of protein remained nearly constant in dark-grown seedlings, except some increase on day 6. The level of BnCRY1 was quite low in the dark-grown seedlings, in comparison to those grown in white light (70 μmol m−2 s−1) continuously for 6 d (Fig. 5C); in fact, a long exposure had to be given to obtain signals for the dark-grown samples (Fig. 5B). To study the effect of blue light on the accumulation of the BnCRY1 protein, 6-d-old etiolated Brassica seedlings were exposed to blue light (10 μmol m−2 s−1) for 36 h (Fig. 5D). On irradiation of seedlings with blue light, the BnCRY1 levels increased severalfold (the gel blot in Fig. 5D was exposed little longer than in Fig. 5C).
Figure 5.
Comparative analysis of BnCRY1 protein levels in Brassica seedlings grown in light (A) and dark (B) for various durations. Note that the blot in B was exposed for a longer duration to amplify signals. C shows BnCRY1 expression in 6-d-old light- and dark-grown Brassica seedlings. The effect of blue light on BnCRY1 levels in 6-d-old dark-grown seedlings irradiated with 36 h blue light is displayed in D. The dark-grown seedlings of the same developmental stage were taken as control; the blot in D was exposed for longer duration than the one shown in C. E shows the tissue-specific expression of BnCRY1. For western analysis, anti-6×His∷CT-BnCRY1 primary antibody was used.
The western-blot analysis revealed the presence of BnCRY1 in cotyledons, stems, buds, flowers, and siliques (Fig. 5E). The expression was particularly higher in cotyledons, stems, and siliques. However, despite repeated attempts, the BnCRY1 protein could not be detected in roots as well as mature leaves under the given conditions; note that the BnCRY1 transcript could be detected in both leaves and roots (Fig. 4C). The distribution pattern of BnCRY1 appears to be largely consistent with the role cry1 plays in regulating various growth and developmental processes in plants.
The BnCRY1 Promoter Imparts Light Regulation to β-Glucuronidase in Transgenic Arabidopsis
The core regulatory elements like TATA box and CAAT box were identified at positions −29 (AATATA) and −122 (TCCAAA), respectively. To demonstrate that BnCRY1 promoter is indeed light regulated, the 1,124-bp region upstream of BnCRY1 translational start site was analyzed using PLACE (plant cis-acting regulatory elements; http//www.dna.affrc.go.jp/htdocs/place; Higo et al., 1999). This search revealed the presence of light regulatory elements, like GT1 and GATA boxes (Terzaghi and Cashmore, 1995; Guilfoyle, 1997; Tyagi and Gaur, 2003), along with some circadian clock-regulated elements, like CIACADIANLELHC and CCA1ATLHCB1 (Wang et al., 1997; Piechulla et al., 1998; Table I), in the BnCRY1 promoter upstream region. To determine if the transcription of BnCRY1 is light inducible, the functional analysis of its promoter was carried out in stably transformed Arabidopsis plants. Two constructs, one of 1.1 kb (CRY1P1∷GUS [β-glucuronidase]) and the other harboring 348 bp (CRY1P2∷GUS) upstream region from the translational start site, were designed. The smaller fragment bears most of the well-known light regulatory elements. Both of these constructs were mobilized into Arabidopsis via Agrobacterium-mediated root explant transformation.
Table I.
Light and clock regulatory elements of the BnCRY1 promoter
| cis-Regulatory Elements | Nucleotide Position |
|---|---|
| CIACADIANLELHC | −982 |
| CCA1ATLHCB1 | −144 |
| GATA BOX | −68, −73, −80, −546, −877, −884 |
| GT1 BOX | −149, −817, −822 |
| I BOX | −65, −71 |
Light Activation of GUS Reporter by the BnCRY1 Promoter
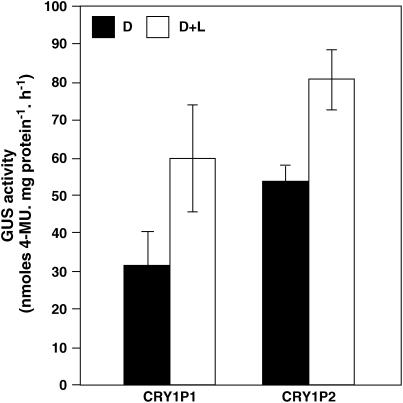
The T2 progeny seedlings of five independent transgenic events (for each construct) were grown in dark for 8 d and another set of 7-d-old dark-grown seedlings exposed to white light for 24 h. The analysis of both CRY1P1∷GUS and CRY1P2∷GUS harboring seedlings revealed that the GUS activity was higher in dark-grown seedlings irradiated with white light for 24 h, as compared to the dark control (Fig. 6). The increased GUS activity in seedlings exposed to light (for only 24 h) indicates that BnCRY1 promoter may be regulated by light. This study further provides evidence that the smaller deletion construct harboring several light regulatory elements may be sufficient for driving GUS expression in a light-dependent manner.
Figure 6.
Light induction of BnCRY1 promoter fused to GUS reporter gene. For light induction assay, the T2 transgenic Arabidopsis seedlings representing five independent lines for each construct (CRY1P1 and CRY1P2) were grown for 8 d in dark (D) or 7 d in dark followed by white light irradiation for 24 h (D + L). The data presented represent mean ± sd of GUS activity measured in seedlings of five independent lines for each construct.
Spatial Expression of the BnCRY1 Promoter-Driven GUS
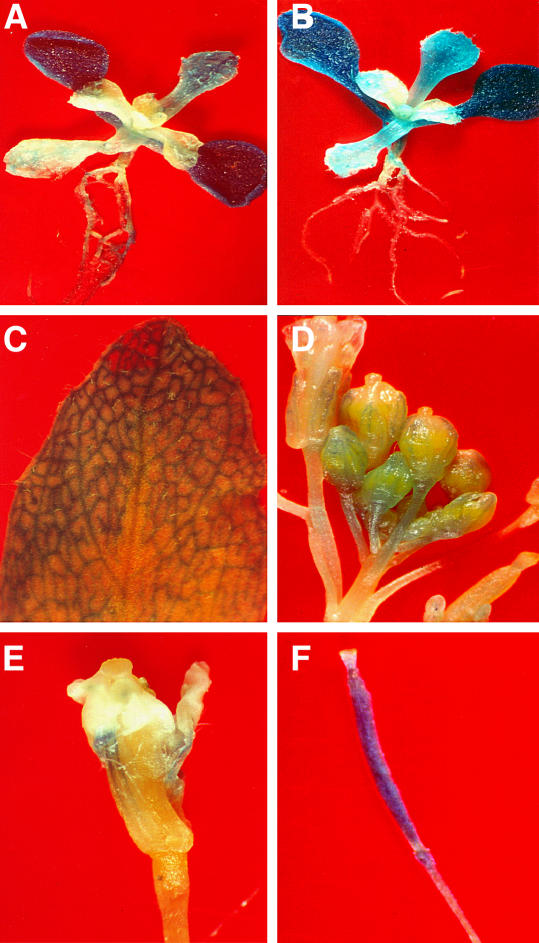
The GUS activity was determined histochemically to analyze the pattern of GUS expression and infer the promoter activity of the endogenous gene. The CRY1P1∷GUS construct, harboring 1.1-kb promoter, transcribed in all the organs like root, stem, leaf, floral bud, flower, and silique (Fig. 7, A–F). The GUS activity was distinctly high in the cotyledons of the 15-d-old transgenic plants in comparison to the first pair of leaves. A low level of GUS expression with nonuniform pattern was also observed in the roots (Fig. 7, A and B). Thus, the GUS reporter construct exhibits a regulation essentially similar to that of the endogenous gene as observed by RT-PCR (Fig. 4).
Figure 7.
Tissue-specific expression analysis of BnCRY1 gene promoter fused to the GUS reporter gene. A and B show the localization of CRY1P1:GUS in 15-d-old transgenic Arabidopsis seedlings. C to F show the GUS expression in leaf, inflorescence buds, flower, and silique of CRY1P1 (T2) transgenic Arabidopsis plants.
Stem Elongation and Decreased Anthocyanin Accumulation in Brassica Transgenics with Reduced BnCRY1 Levels
To study the in vivo function of Brassica cry1, the antisense transgenic approach was adopted. However, instead of B. napus, Brassica juncea was selected because of its amenability in tissue cultures and higher transformation efficiency. The C-terminal region of BnCRY1 was amplified and cloned in the reverse orientation between the 35S promoter and nopaline synthase (NOS) polyadenylation site as terminator in a modified pCAMBIA 2310 vector (Fig. 8A) and introduced into B. juncea via Agrobacterium-mediated transformation of hypocotyl segments. The transgenic plants were allowed to grow and the T1 seeds harvested for at least 10 independent plants.
Figure 8.
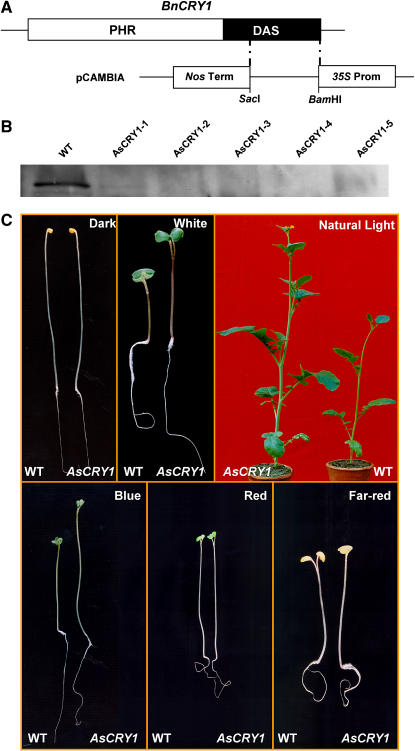
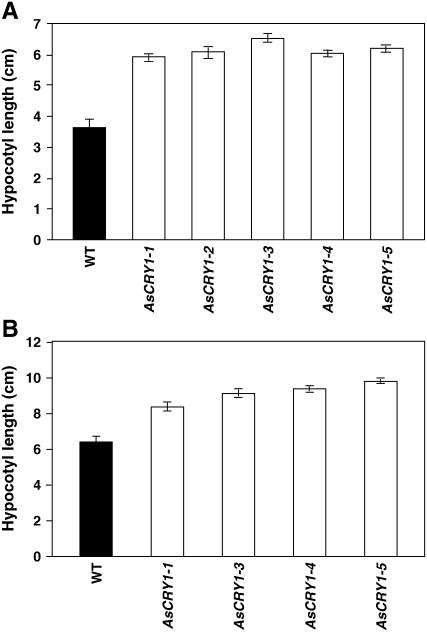
A, Diagrammatic representation depicting cloning strategy of the C terminus of BnCRY1 in modified pCAMBIA2301 vector for antisense construct. B, Western-blot analysis for quantitation of CRY1 in wild type and five different antisense transgenic lines (T2) of B. juncea (AsCRY1-1 to AsCRY1-5). C, Comparison of hypocotyl length between 8-d-old AsCRY1 and wild-type seedlings grown in dark or irradiated with white, blue, red, or far-red light. The phenotype of 45-d-old AsCRY1 and wild-type adult plants grown under field conditions during winter season in a containment facility is shown in the top right. AsCRY1, Antisense-CRY1 seedlings/plants. Please note that scale in different sections in C may not be same, although within the section the seedlings/plants are of same magnification.
To check for the phenotype (hypocotyl growth) of the antisense-BnCRY1 (AsCRY1) transgenics, the hypocotyl length of 15-d-old T1 seedlings was measured. Under continuous white light (70 μmol m−2 s−1), all the seedlings examined showed elongated hypocotyl and petioles when compared to the wild type (Figs. 8C and 9A). On illumination with continuous blue light (10 μmol m−2 s−1), all the transgenic lines showed decreased inhibition of hypocotyl elongation (Figs. 8C and 9B); because of shortage of seeds, the AsCRY1-2 lines could not be tested for hypocotyl growth inhibition assay under blue light. In comparison to seedlings grown under white light, the hypocotyl elongation growth was greater under blue light. This may be due to the inhibitory effect of far-red and red light present in the white light, which act in a combinatorial manner with blue light for complete realization of the hypocotyl/stem growth inhibition response (Folta and Spalding, 2001). However, the hypocotyl growth of the AsCRY1 transgenic seedlings was not affected differentially (vis-à-vis wild type) by either red or far-red light (Fig. 8C), indicating that impairment in Brassica cry1 function does not affect red or far-red response. The T1 plants were grown during the winter under a short photoperiod (November to April) in a containment facility. At the adult stage too, the AsCRY1 plants were distinctly taller (Fig. 8C). In addition, leaves too were relatively large and the stem diameter greater in the AsCRY1 plants. Whether cry1 in Brassica plays a direct role in regulating these traits or adversely affects the function of some other sensory photoreceptor will be our endeavor to examine.
Figure 9.
Comparison of the hypocotyl growth response of the wild-type and AsCRY1 Brassica seedlings grown under white light (70 μmol m−2 s−1; A) and blue light (10 μmol m−2 s−1; B). Histograms represent the hypocotyl growth of 10-d-old wild-type and antisense (T1) seedlings developed from independent transformation events. The data presented represent mean ± sd of 10 seedlings for each transgenic line.
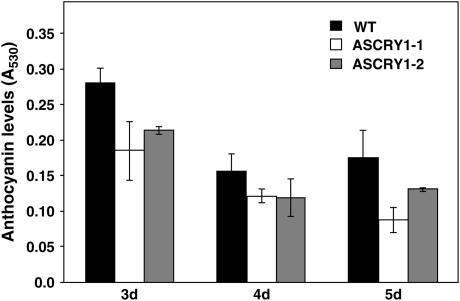
In addition to controlling plant height, cry1 also regulates anthocyanin accumulation. Earlier studies with Arabidopsis have shown that anthocyanin levels have an overriding effect of developmental cues, and are optimal in 3- to 4-d-old light-grown seedlings and decline thereafter (Feinbaum and Ausubel, 1988; Bharti and Khurana, 2003). The anthocyanin content was thus checked in two of the transgenic lines (AsCRY1-1 and AsCRY1-2) grown in blue light (10 μmol m−2 s−1) for various durations. In B. juncea too, anthocyanin levels were high in the 3-d-old seedlings and declined subsequently both in the wild type and the transgenic lines. However, the anthocyanin content was relatively lower in the transgenic lines, on any given day, with the effect being more pronounced in the line AsCRY1-1, particularly on days 3 and 5 (Fig. 10).
Figure 10.
Comparison of the anthocyanin content in the wild-type and AsCRY1 Brassica seedlings at different stages of development. For experimental details, see “Materials and Methods.”
To substantiate whether the long-hypocotyl phenotype and reduced anthocyanin accumulation in AsCRY1 transgenic seedlings was indeed due to reduced CRY1 levels, immunoblot analysis was performed with wild-type and transgenic seedlings. The CRY1 protein could not be detected or was considerably reduced in all five AsCRY1 transgenic lines examined (Fig. 8B). The copy number of AsCRY1 insert(s) was checked by Southern analysis, and one to three insertions in independent transgenic lines were detected (data not shown). A strict correlation between plant height and anthocyanin content, and gene dosage effect, will be possible only when a more detailed analysis of the homozygous lines of antisense-BnCRY1 transgenics becomes available.
DISCUSSION
As expected, owing to the genomic relatedness among Arabidopsis and Brassica, the BnCRY1 gene showed similar structural organization as AtCRY1. Sequence analysis of BnCRY1 revealed 94% similarity with the gene encoding HY4 flavin-type blue light photoreceptor (AF361588). The BLASTP analysis confirmed the sequence match of BnCRY1 with the other known cryptochromes, such as AtCRY2, LeCRY1, LeCRY2, OsCRY1, OsCRY2, CPH1, PpCRY1, AcCRY1, and SaPHR. A low percentage identity (31%–43%) was also observed with DNA photolyases (CPD photolyase [AE005817] and 6-4 photolyase [AB042254]). Although the length of introns varied, the intron and exon boundaries were conserved between BnCRY1 and AtCRY1 genes. The secondary structure of BnCRY1 consists mainly of α-helices and β-strands, which are randomly distributed throughout the primary amino acid sequence and thus do not cluster into groups like αβ domain and helical domain that are present in the secondary structure of photolyases (Brudler et al., 2003).
Based on the evolutionary history and ancient duplication events, angiosperm cryptochromes have been grouped into two classes, CRY1 and CRY2 (Perrotta et al., 2000), with a recent addition of a novel class, CRY-DASH (Kleine et al., 2003). BnCRY1 showed a close relationship with AtCRY1. The CRY1 species belonging to the dicot family are more closely related to the monocot CRY1 rather than to the dicot CRY2. The fern (A. capillus-veneris) cryptochromes can be classified into three groups, indicating three duplication events; AcCRY1/AcCRY2 and AcCRY3/AcCRY4 were grouped in pairs, demonstrating recent duplication events. Similar phylogenetic analyses of cryptochromes were also performed by various groups (Imaizumi et al., 2000; Perrotta et al., 2000, 2001). The apparent presence of only CRY1-like genes in lower plants suggests that the CRY1 and CRY2 duplication is specific to the spermatophytes, which were estimated to have evolved approximately 360 Mya. To date, we do not have sequence information of cryptochromes from gymnosperms; therefore, the sequence of more cryptochromes from diverse groups including gymnosperms would further refine this picture. In a database search, various homologs of OsCRY1 and OsCRY2 were observed with a minor percentage of mismatches at the nucleotide level. In fact, OsCRY1 (1b) sequence has been characterized as OsCRY2 by Matsumoto et al. (2003), despite the fact that overall similarity between OsCRY1 and the renamed OsCRY2 is 78.8%; such high similarity is usually not observed between the two classes of cryptochromes. The phylogenetic analysis also grouped OsCRY2 under the monocot CRY1 class. Moreover, we have identified and sequenced an OsCRY2 gene (accession no. AJ298877) from rice with 38% and 39% amino acid identity with the two OsCRY species identified earlier (D. Kumar, P. Sharma, A.K. Tyagi, and J.P. Khurana, unpublished data).
Along with ploidy, the chromosomal rearrangements like duplications and deletions play a major role in evolution. The copy number of CRY1 varies from species to species; for example, AtCRY1 is represented as a single copy in the Arabidopsis genome, whereas both tomato and barley harbor two copies of CRY1. However, despite the fact that B. napus is an amphidiploid (AACC), this study shows that the BnCRY1 gene in B. napus genome is most probably represented as a single copy. The genetic analysis of B. napus indicates that the genome of this amphidiploid is in a state of flux, and a large scale rearrangement due to duplication, deletion, and inversions or translocations of genetic segments has occurred (Sharpe et al., 1995). The loss of the other copy of CRY1 is an example of a secondary loss and supports the theory of major deletions in the C genome of B. napus, although it remains to be validated experimentally.
The BnCRY1 transcript abundance, as well as protein levels, is regulated by light and developmental cues. Severalfold induction in the level of BnCRY1 protein was observed on illumination with white or blue light. The expression profile, along with in vivo promoter∷GUS fusion analysis in transgenic Arabidopsis, indicates the abundance of CRY1 in young and meristematic tissues like cotyledonary leaves, emerging inflorescence buds with inflorescence meristem, and young siliques with developing embryos. Although the CRY1 transcript could be detected in the root tissue by RT-PCR analysis, no protein could be detected. Thus, the tissue-specific expression of BnCRY1 may also be regulated at the translational level and/or protein degradation. It is interesting to note here that the BnCRY1 protein was undetectable in leaves under our experimental conditions, whereas in Arabidopsis a substantial amount of CRY1 accumulates (Lin et al., 1996a). The transcript abundance of BnCRY1 is associated with its role in cotyledon expansion, inhibition of stem growth, initiation of flowering, and early stages of silique formation. The promoter∷GUS fusion analysis in transgenic Arabidopsis demonstrated the contribution of BnCRY1 putative light-responsive elements in light-regulated expression, and indicates that the promoter fragment −348 bp upstream from the transcription start site may be sufficient to confer up-regulation by light. The activation of BnCRY1 promoter by light is consistent with the observation by Toth et al. (2001), who demonstrated that the AtCRY1 promoter-driven luciferase gene expression is up-regulated by light. However, earlier reports claim that the CRY1 protein levels do not change on exposure of Arabidopsis seedlings to light (Lin et al., 1996a; Shalitin et al., 2003), although it undergoes blue light-induced protein phosphorylation (Shalitin et al., 2003). In contrast to the light-independent expression of AtCRY1, light caused down-regulation of CRY1 levels in tomato and tobacco in a manner essentially similar to AtCRY2 (Ahmad et al., 1998a). On the other hand, this study provides evidence that BnCRY1 levels are rather up-regulated by white and blue light under the given experimental conditions. There is a possibility that regulation occurs both at the level of protein accumulation/stability and at the transcript level, such that one compensates for the other. The analysis of CRY1 from diverse species may divulge more on the molecular mechanism of light regulation for CRY1 itself.
The decrease in growth inhibition of the AsCRY1 transgenics of Brassica under field conditions and also at the seedling stage provides evidence for the in vivo function of cry1 in regulating stem growth. The effect of cry1 in regulating plant height appears to be more pronounced in Brassica than reported for tomato and pea (Ninu et al., 1999; Platten et al., 2005a). This may either be a species-specific response or due to interplay with other sensory receptors involved in regulating plant height, as observed in case of pea in particular (Platten et al., 2005a). In the moss P. patens too, the analysis of the disruptants of two cryptochromes, PpCRY1a and PpCRY1b, revealed that they act redundantly to induce side branching in protonema and leaf growth in gametophores but cause inhibition of stem growth of gametophores specifically in response to blue light (Imaizumi et al., 2002), thus drawing a parallel between higher plants and the moss system with respect to cry1 responses. It was further shown that cryptochromes regulate moss development by repressing auxin signals usually involved in cell elongation and/or cell division. In higher plants like Arabidopsis, tomato, and Brassica too, the loss of inhibition of hypocotyl growth by light in the antisense plants may be due to altered expression of genes regulating cell division and cell wall expansion. In addition, cryptochrome modulates stem growth by repressing GA3 and auxin levels (Folta et al., 2003). The microarray analysis of mRNA isolated from blue light-treated wild-type and cry1 mutant seedlings revealed that CRY1 activates genes in the GA3 biosynthetic pathway. On illumination with blue light, GA20 oxidase and gibberellin β-hydroxylase are activated in cry1, which increases the active GA4. Thus, it is conceivable that all these factors may regulate hypocotyl and stem growth in antisense Brassica plants as well. The AsCRY1 transgenics also display reduced accumulation of anthocyanins and increased internode length and early flowering in at least some transgenic lines (data not shown), implicating the role of BnCRY1 in regulating these blue light-mediated responses. Although cry2 plays a more predominant role in controlling flowering time (Guo et al., 1998), in some studies cry1 too has been shown to influence flowering in Arabidopsis and pea, mostly through its interaction with other sensory receptors, including cry2, phyA, and phyB (Yang et al., 2000; Platten et al., 2005a). In fact, cry1 has a small inhibitory effect on flowering, especially in the absence of functional phyA (Platten et al., 2005a), and our preliminary finding with AsCRY1 lines of Brassica may represent a similar scenario. Besides analyzing the antisense BnCRY1 transgenics in more detail in the above context, it will be our endeavor to also raise the CRY1 overexpression lines and analyze the performance of both types of transgenics in the field (in a containment facility) to examine if their yield is not compromised due to altered photosensitivity to blue light.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Brassica napus var. ISN-706 and Brassica juncea var. RLM-198 were obtained from the Indian Agricultural Research Institute, New Delhi. Seeds were washed thoroughly and soaked overnight in running tap water. The imbibed seeds were spread on cotton saturated with reverse osmosis water. Plants were grown either in dark or light (16-h photoperiod) for desired duration in a culture room/growth chamber maintained at 24°C ± 1°C.
Library Construction, Isolation, and Sequencing of the Genomic BnCRY1 Clone
Total plant DNA was extracted from 8-d-old dark-grown Brassica seedlings following the procedure of Dellaporta et al. (1983). A genomic library of the high Mr DNA partially digested with MboI restriction enzyme was constructed in a Lambda Dash II replacement vector (Stratagene) according to manufacturer's instructions. A total of 4 × 105 recombinant plaques were screened under low stringency conditions (at 55°C) with an [α-32P]dATP-labeled (Megaprime DNA Labeling system) 2.3-kb AtCRY1 gene as a probe; it was amplified by PCR using a primer pair 5′-ATGTCTGGTTCTGTATCTGGTTGTG-3′ and 5′-TTACCCGGTTTGTGAAAGCCGTC-3′. For details of hybridization and washings, see Kulshreshtha et al. (2005). After three successive rounds of screening, phage DNA was isolated from the putative clones following the protocol of Santos (1991) and subjected to Southern analysis after digestion with desired restriction enzymes. One of the positive clones was sequenced using the Thermosequenase Dye Terminator Cycle sequencing kit (Amersham International) and a DNA sequencer (ABI Prism 377).
Amplification of the BnCRY1 cDNA
The cDNA was amplified by RT-PCR using primer pair (5′-CCATCGATATGTCTAATTCATGTTCAGGTG-3′ and 5′-GTCTCGAGGTGACAGCCGTCTCCA-3′) designed based upon BnCRY1 gene sequence obtained. Using 1 μg total RNA isolated from 4-d-old light-grown seedlings (Nagy et al., 1988), RT-PCR was carried out with Titan One Tube RT-PCR system (Roche). The PCR conditions were: 30 min at 50°C; 2 min at 94°C; 10 cycles (30 s at 94°C; 30 s at 55°C; 1 min at 68°C); 15 cycles (30 s at 94°C; 30 s at 55°C; 1 min at 68°C); and 5 s extension every cycle. The amplified cDNA was cloned in pBluescript SK+ and sequenced by primer walking. The 5′ UTR and 3′ UTR of BnCRY1 mRNA were completed using the Smart RACE cDNA amplification kit (BD Biosciences), according to manufacturer's protocol using gene-specific primers: 5′-GTGGAGAAAGGAACGAGGTTGTGGCACTG-3′ and 5′-CATGAGGCACTCTCGCAGATGTGGCAAC-3′. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced.
Southern Blot and RT-PCR Analysis
An aliquot of 15 μg of the plant DNA was digested independently with EcoRI, HindIII, SalI, XbaI, and XhoI restriction enzymes (Roche Molecular Biolabs) and Southern analysis performed as described earlier (Thakur et al., 2003). The full-length BnCRY1 gene labeled with [α-32P]dATP was used as a probe. Hybridization was carried out at 37°C or 42°C in hybridization buffer containing 50% formamide, 5× SSC, 5× Denhard's solution, 50 mm sodium phosphate, pH 6.5, and 250 μg/mL of denatured herring sperm DNA. The filters were washed at ambient temperature (25°C ± 1°C) with the following buffers in sequence: 5× SSC and 0.1% SDS for 10 min, 2× SSC and 0.1% SDS for 15 min, and 1× SSC and 0.1% SDS for 5 min. The filters were exposed to Kodak X-OMAT film with an intensifying screen at −80°C for the desired duration depending upon the counts retained on the filter.
For expression analysis, total RNA was isolated from various tissues (frozen in liquid nitrogen) using a LiCl method (Nagy et al., 1988). To avoid DNA contamination, all samples were treated with DNase I (Roche Applied Science). RT-PCR was performed using primer pair 5′-GGCACCAGAGGAAGAAGGGCACT-3′, 5′-CATGGTGGTTCTGCAAGTAGC-3′, and PCR conditions were: 30 min at 50°C for RT; 2 min at 94°C; 10 cycles (30 s at 94°C; 30 s at 55°C; 1 min at 68°C); 15 cycles (30 s at 94°C; 30 s at 55°C; 1 min at 68°C); and 5 s extension every cycle. ACTIN served as the internal control.
Antibody Production
The BnCRY1 gene was restriction digested with BamHI and SalI enzymes, which have the internal sites present in the third exon, and the 350-bp fragment thus generated cloned into pQE-30 vector in the same reading frame as 6×His affinity tag. The fusion protein was expressed in Escherichia coli strain M15 and purified using a Ni-NTA affinity column (Qiagen). Immunizations were done by subcutaneous injection of 20 μg of emulsified protein per mouse followed by two booster doses with 15 μg of protein after every 2 weeks. Serum collected was stored at −80°C for later use.
Immunoblot Analysis
The total protein from plant tissue was extracted following the procedure of Zivy et al. (1983). Aliquots of the samples were denatured in the presence of SDS-PAGE sample buffer (15.5 mm Tris-Cl, pH 6.8, 720 mm 2-mercaptoethanol, 10% glycerol, 3% SDS). Equal amount (100 μg) of protein samples was resolved using 12.5% SDS-PAGE and subjected to western blotting (Towbin et al., 1979). The blots were probed with anti-6×His∷CT-BnCRY1 (1:1,000) antibody. The rabbit anti-mouse IgG (1:10,000) conjugated to horseradish peroxidase (Sigma) was used as secondary antibody. Proteins were detected using the ECL Plus Chemiluminescence kit (Amersham) according to manufacturer's instructions.
Promoter Deletion Constructs and Transformation of Arabidopsis
The 1.1-kb and 348-bp genomic fragments upstream of translation start site were PCR amplified using primers 5′-GCTCTAGACATGAGTTGGAATCAGTT-3′, 5′-GCTCTAGAATACATGTGCGGAGGTACG-3′, and 5′-CCTCTAGACTCAATCTTAAAGCTCTTAC-3′. The amplified promoter fragments were cloned in pBI101 vector (Jefferson et al., 1987) and mobilized to Agrobacterium tumefaciens strain GV3101 by chemical transformation (An et al., 1988). These deletion constructs were then transferred to Arabidopsis (Arabidopsis thaliana) using Agrobacterium-mediated root transformation protocol (Valvekens et al., 1992). The primary transformants were denoted as T0 and seeds (T1) obtained from various independent lines were analyzed for single copy insertion. The kanamycin-resistant lines segregating in 3:1 ratio were selected and allowed to self-fertilize. The T2/T3 seedlings were utilized for promoter analysis.
Histochemical and Quantitative Analysis of GUS Activity
Arabidopsis seedlings harboring the transgene promoter∷GUS fusions were stained overnight at 37°C in GUS assay buffer (1 m NaHPO4 buffer, pH 7.0, 50 mm EDTA, pH 8.0, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 0.1% Triton X-100, 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid). After overnight staining, destaining was done with 70% ethanol and chlorophyll removed by washing at least two to three times. The samples were photographed employing an epifluorescence microscope (Nikon EFD-3).
For quantitative analysis, GUS activity was measured as described by Jefferson et al. (1987), using the substrate 4-methylumbelliferyl glucuronide. For each promoter∷GUS construct, several independent insertion lines were analyzed.
Brassica Transformation for Raising Antisense-BnCRY1 Transgenics
The BnCRY1 cDNA was amplified by PCR: 5 min at 94°C for 30 cycles (94°C for 30 s, 65°C for 30 s, and 70°C for 45 s), followed by incubation at 70°C for 7 min, using the primer pair 5′CRYSacI GGGAGCTCGAAGAAGGACTTGGCGAT and 3′CRYBamHI CGCGGATCCAACTATTTCATGGTGGTTC. The antisense fragment (corresponding to the C terminus of BnCRY1) thus obtained was cloned in the modified pCAMBIA 2310 vector using BamHI and SacI restriction sites and introduced in B. juncea var. RLM-198 hypocotyl sections via Agrobacterium. For regeneration and shoot formation from putative transgenics, the hypocotyl sections cocultivated with Agrobacterium were placed on agar-gelled Murashige and Skoog medium containing 1 mg/L naphthylacetic acid, 1 mg/L benzylaminopurine, 3.4 mg/L AgNO3, 250 mg/L cefotaxime, and 50 mg/L kanamycin. The callus or regenerating plantlets were subcultured on fresh medium after every 15 d, for two to three times, until the shootlets appeared. The healthy shoots were transferred to the rooting medium containing 0.1 mg/L naphthylacetic acid and 50 mg/L kanamycin. As soon as a small root mass was observed, the plantlets were transferred to earthen pots containing garden soil. The T0 plants raised in a growth room were allowed to set seed at 24°C ± 1°C, under continuous light (100 μmol m−2 s−1). The adult (T1) transgenic plants were grown under field conditions in a containment facility.
Hypocotyl Elongation Assay and Anthocyanin Estimation
For hypocotyl elongation growth assay, seeds were germinated in clay pots containing garden soil and irradiated with white light, blue light, red light, or far-red light in the cabinets kept in a growth room. After 10 d of growth, the hypocotyl length of 10 seedlings each of wild type and antisense-BnCRY1 line was measured and averaged. The experiments were repeated at least once with essentially similar results, and, thus, the data of only a representative experiment are presented.
For anthocyanin estimation, the wild-type and antisense seedlings were grown on Murashige and Skoog medium supplemented with 2% Suc and 0.8% agar under continuous blue light (10 μmol m−2 s−1) for 3 to 5 d. The anthocyanins from three seedlings of each line were extracted independently overnight in 3 mL of acidic (1% HCl) methanol in a dark chamber. To the acidic methanol extract, 2 mL of water and 3 mL of chloroform were added and mixed thoroughly. The absorbance of aqueous phase was determined at 530 nm as a measure of anthocyanin levels on a per-seedling basis.
Light Source and Energy Measurements
For irradiation of Brassica seedlings with monochromatic lights, blue, red, and far-red light sources were custom designed. The blue and red light sources consist of 31 × 12 array of light-emitting diodes (LED) selected for their spectral quality. Each LED (λmax 465 nm for blue and λmax 652 nm for red light) was powered by a variable voltage source capable of 10 mA forward current. For far-red irradiation, four epoxy lens type infrared illuminators (LED735-66-60; Roithner Lasertechnik), comprising 60 high efficiency diode chips each (λmax 735 nm), were mounted to give uniform illumination. Each LED was powered by 9-V, 1,000-mA regulator with heat sink mounted series pass transistor. The forward current and the height of the source from the plant material were adjusted to yield uniform irradiation of blue (10 μmol m−2 s−1), red (8 μmol m−2 s−1), and far-red light (2.5 μmol m−2 s−1), respectively, as measured by the LI-189 radiometer (LI-COR) over an area measuring 40 cm × 30 cm. White light (70 μmol m−2 s−1) was provided from a bank of Cool Daylight fluorescent lamps (Philips, TL 5800 K).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtCRY, Q43125; LeCRY1, AAD44161; PsCRY1, AAO23970; OmCRY1, AAR08429; LeCRY1B, AAL02092; OsCRY1b, BAB70688; SbCRY2, AAN37909; OsCRY1a, BAB70686; AcCRY2, BAA32808; AcCRY1, BAA32807; PpCRY1a, BAA83338; PpCRY1b, BAB70665; AcCRY3, BAA32809; AcCRY4, BAA88423; OsCRY2, BAC78798; LeCRY2, AAF72556; AcRY5, BAA88424; AtCRY2, AAL16379; ArCRY2-3, BAC67178; ArCRY2-4, BAC67179; ArCRY2-1, BAC67176; PsCRY2b AAO23972; OsCRY1, BAA82885; ArCRY2-2, BAC67177; PsCRY2a, AAO23971; OsCRY2 (indica var.), CAC82538; CrCPH1, AAC37438; AtCRYDASH, NP_568461; and SaPHR, X72019.
Acknowledgments
We sincerely thank Drs. Margaret Ahmad and Akhilesh K. Tyagi for useful suggestions, Dibyendu Kumar for assistance in phylogenetic analysis, and Dr. Anil K. Tyagi for providing facilities and assistance in raising anti-BnCRY1 antibodies. We are grateful to Arvind Dixit for design and assembly of the light sources.
This work was supported by the Department of Biotechnology, Government of India, and through infrastructural support from the University Grants Commission, New Delhi, by the Department of Science and Technology, Government of India, and by the Council of Scientific and Industrial Research, New Delhi (research fellowship to M.C. and P.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jitendra P. Khurana (khuranaj@genomeindia.org).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076323.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Cashmore AR (1998. a) Chimeric proteins between CRY1 and CRY2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998. b) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1: 939–948 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Lin C, Cashmore AR (1995) Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8: 653–658 [DOI] [PubMed] [Google Scholar]
- An G, Ebert P, Mitra A, Ha S (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3 1–19
- Banerjee R, Batschauer A (2005) Plant blue-light receptors. Planta 220: 498–502 [DOI] [PubMed] [Google Scholar]
- Bharti AK, Khurana JP (1997) Mutants of Arabidopsis as tools to understand the regulation of phenylpropanoid pathway and UV-B protection mechanisms. Photochem Photobiol 65: 765–776 [DOI] [PubMed] [Google Scholar]
- Bharti AK, Khurana JP (2003) Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana (ecotype Estland) impaired in flavonoid biosynthesis pathway. Plant Sci 165: 1321–1332 [Google Scholar]
- Botto JF, Alonso-Blanco C, Garzaron I, Sanchez RA, Casal JJ (2003) The Cape Verde Islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiol 133: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA 33: 12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Olney MA (2001) Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol 125: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, et al (2003) Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell 11: 59–67 [DOI] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol 71: 1–11 [DOI] [PubMed] [Google Scholar]
- Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version 2. Plant Mol Biol Rep 1: 19–22 [Google Scholar]
- Devlin PF, Kay SA (1999) Cryptochromes: bringing the blues to circadian rhythms. Trends Cell Biol 9: 295–298 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679 [DOI] [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM (1988) Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol Cell Biol 8: 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36: 203–214 [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP (2001) Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J 28: 333–340 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2004) Light signals, phytochromes and cross-talk with other environmental cues. J Exp Bot 55: 271–276 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D (2003) Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geourjon C, Deleage G (1994) SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng 7: 157–164 [DOI] [PubMed] [Google Scholar]
- Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit anti-oxidant content. Plant Physiol 137: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovani B, Byrdin M, Ahmad M, Brettel K (2003) Light-induced electron transfer in a cryptochrome blue-light photoreceptor. Nat Struct Biol 10: 489–490 [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ (1997) The structure of plant gene promoters. In JK Setlow, ed, Genetic Engineering. Plenum Press, New York, pp 15–47
- Guo H, Duong H, Ma N, Lin C (1999) The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light dependent post-translational mechanism. Plant J 19: 279–287 [DOI] [PubMed] [Google Scholar]
- Guo H, Mockler T, Duong H, Lin C (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291: 487–490 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Gyula P, Schafer E, Nagy F (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6: 446–452 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PD, Batschauer A, Hays JB (1996) PHH1, a novel gene from Arabidopsis thaliana that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol Gen Genet 253: 259–265 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kanegae T, Wada M (2000) Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12: 81–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kiyosue T, Kanegae T, Wada M (1999) Cloning of the cDNA encoding the blue light photoreceptor cryptochrome from the moss Physcomitrella patens (accession no. AB027528). Plant Physiol 120: 120510490396 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae T, Wada M (1998) Isolation and characterization of homologues of plant blue-light photoreceptor (cryptochrome) genes from the fern Adiantum capillus-veneris. Mol Gen Genet 259: 345–353 [DOI] [PubMed] [Google Scholar]
- Kang X, Chong J, Ni M (2005) HYPERSENSITIVE TO RED AND BLUE 1, a ZZ-type zinc finger protein, regulates phytochrome B-mediated red and cryptochrome-mediated blue light responses. Plant Cell 17: 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana JP (2001) Cryptic blues: mechanism in sight! Curr Sci 80: 189–198 [Google Scholar]
- Khurana JP, Dasgupta U, Laxmi A, Kumar D, Paul LK (2004) Light control of plant development by phytochromes: a perspective. Proc Indian Natl Sci Acad Part B Biol Sci 70: 379–411 [Google Scholar]
- Khurana JP, Kochhar A, Tyagi AK (1998) Photosensory perception and signal transduction in higher plants: molecular genetic analysis. CRC Crit Rev Plant Sci 17: 465–539 [Google Scholar]
- Kleine T, Lockhart P, Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35: 93–103 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kanno S, Smit B, Van der Horst GT, Takao M, Yasui A (1998) Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res 26: 5086–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Ishikawa T, Hirayama J, Daiyasu H, Kanai S, Toh H, Fukuda I, Tsujimura T, Terada N, Kamei Y, et al (2000) Molecular analysis of zebrafish photolyase/cryptochrome family: two types of cryptochromes present in zebrafish. Genes Cells 5: 725–738 [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Kumar N, Balyan HS, Gupta PK, Khurana P, Tyagi AK, Khurana JP (2005) Structural characterization, expression analysis and evolution of the red/far-red sensing photoreceptor gene, PHYTOCHROME C (PHYC), localized on the ‘B’ genome of hexaploid wheat (Triticum aestivum L.). Planta 221: 675–689 [DOI] [PubMed] [Google Scholar]
- Kumar D (2000) Isolation and characterization of OsCRY2 cDNA from rice (Oryza sativa L.) encoding a sensory blue light receptor, cryptochrome 2. M.Phil. thesis. University of Delhi, Delhi, India
- Lakshmikumaran M, Das S, Srivastava PS (2003) Application of molecular markers in Brassica coenospecies: comparative mapping and tagging. In T Nagata, S Tabata, eds, Brassicas and Legumes: From Genome Structure to Breeding. Springer-Verlag, Berlin, pp 37–68
- Lin C (2002) Blue light receptors and signal transduction. Plant Cell (Suppl) 14: S207–S225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Cashmore AR (1996. a) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10: 893–902 [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Chan J, Cashmore AR (1996. b) CRY2: a second member of the Arabidopsis cryptochrome gene family (accession no. U43397). Plant Physiol 110: 1047
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970 [DOI] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra K, Baer M, Li YF, Sancar GB, Sancar A (1992) Identification of chromophore binding domains of yeast DNA photolyase. J Biol Chem 267: 2909–2914 [PubMed] [Google Scholar]
- Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A (1995) Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34: 6892–6899 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Hirano T, Iwasaki T, Yamamoto N (2003) Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol 133: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ (2003) A suite of photoreceptors entrains the plant circadian clock. J Biol Rhythms 18: 217–226 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Moller SG, Kim YS, Kunkel T, Chua NH (2003) PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15: 1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Kay SA, Chua N-H (1988) Analysis of gene expression in transgenic plants. In SB Gelvin, RA Schilperoort, DPS Verma, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp B4 1–29
- Ninu L, Ahmad M, Miarelli C, Cashmore AR, Giuliano G (1999) Cryptochrome 1 controls tomato development in response to blue light. Plant J 18: 551–556 [DOI] [PubMed] [Google Scholar]
- Okazawa A, Trakulnaleamsai C, Hiramatsu H, Fukusaki E, Yoneyama K, Takeuchi Y, Kobayashi A (2005) Cloning of a cryptochrome homologue from the holoparasitic plant Orobanche minor Sm. Plant Physiol Biochem 43: 499–502 [DOI] [PubMed] [Google Scholar]
- Perrotta G, Ninu L, Flamma F, Weller JL, Kendrick RE, Nebuloso E, Giuliano G (2000) Tomato contains homologues of Arabidopsis cryptochromes 1 and 2. Plant Mol Biol 42: 765–773 [DOI] [PubMed] [Google Scholar]
- Perrotta G, Yahoubyan G, Nebuloso E, Renzi L, Giuliano G (2001) Tomato and barley contain duplicated copies of cryptochrome 1. Plant Cell Environ 24: 991–997 [Google Scholar]
- Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38: 655–662 [DOI] [PubMed] [Google Scholar]
- Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, Weller JL (2005. a) Cryptochrome 1 contributes to blue light sensing in pea. Plant Physiol 139: 1472–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Foo E, Foucher F, Hecht V, Reid JB, Weller JL (2005. b) The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol 59: 683–696 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180–188 [DOI] [PubMed] [Google Scholar]
- Sancar A (2004) Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem 279: 34079–34082 [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Thorne JL, Wikstrom N, Bremer K (2004) Molecular evidence on plant divergence times. Am J Bot 91: 1656–1665 [DOI] [PubMed] [Google Scholar]
- Sang Y, Li Q-H, Rubio V, Zhang Y-C, Mao J, Deng X-W, Yang H-Q (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis cryptochrome 1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA (1991) An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res 19: 5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yu X, Maymon M, Mockler T, Lin C (2003) Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe AG, Parker IA, Keith DJ, Lydiate DJ (1995) Frequent nonreciprocal translocations in the amphidipliod genome of oilseed rape (Brassica napus). Genome 38: 1112–1121 [DOI] [PubMed] [Google Scholar]
- Small GD, Min B, Lefebvre PA (1995) Characterization of a Chlamydomonas reinhardtii gene encoding a protein of the DNA photolyase/blue light photoreceptor family. Plant Mol Biol 28: 443–454 [DOI] [PubMed] [Google Scholar]
- Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphism (RFLPs). 1. Genome evolution of diploid and amphidiploid species. Theor Appl Genet 75: 784–794 [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445–474 [Google Scholar]
- Thakur JK, Malik MR, Bhatt V, Reddy MK, Sopory SK, Tyagi AK, Khurana JP (2003) A POLYCOMB group gene of rice (Oryza sativa L. subspecies indica), OsiEZ1, codes for a nuclear localized protein expressed preferentially in young seedlings and during reproductive development. Gene 314: 1–13 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi AK, Gaur T (2003) Light regulation of nuclear photosynthetic genes in higher plants. CRC Crit Rev Plant Sci 22: 417–452 [Google Scholar]
- Valvekens D, Lijsebettens MV, Montagu MV (1992) Arabidopsis regeneration and transformation (root explant system). In K Lindsay, ed, Plant Tissue Culture Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A8 1–17
- van der Spek PJ, Kobayashi K, Bootsma D, Takao M, Eker AP, Yasui A (1996) Cloning, tissue expression, and mapping of a human photolyase homolog with similarity to plant blue-light receptors. Genomics 37: 177–182 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control of development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallapa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu Y, Tang RH, Liu D, Liu Y, Cashmore AR (2000) The C-termini of Arabidopsis cryptochrome mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Zeugner A, Byrdin M, Bouly J-P, Bakrim N, Giovani B, Brettel K, Ahmad M (2005) Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem 280: 19437–19440 [DOI] [PubMed] [Google Scholar]
- Zivy M, Thiellement H, de Vienne D, Hofmann JP (1983) Study on nuclear and cytoplasmic genome expression in wheat by two dimensional gel electrophoresis. Theor Appl Genet 66: 1–7 [DOI] [PubMed] [Google Scholar]