The International Rice Functional Genomics Consortium (IRFGC) has initiated a project to provide the rice research community with access to extensive information on genetic variation present within and between diverse rice cultivars and landraces, as well as the genetic resources to exploit that information. Among crop plants, rice is uniquely positioned to achieve this goal due to the release of a high-quality, whole-genome sequence; advances in the use of high-density arrays to compare complex genomes; and the availability of large collections of genetic materials rich in trait variation. In this project, the international rice research community will collaborate with Perlegen Sciences to identify a large fraction of the single nucleotide polymorphisms (SNPs) present in cultivated rice through whole-genome comparisons of 21 rice genomes, including cultivars, germplasm lines, and landraces. The SNP data will be entirely public (www.oryzasnp.org) and can be used to identify a collection of SNPs for undertaking whole-genome scans. Initial funding for this effort has been provided by the International Rice Research Institute (IRRI), the Generation Challenge Program, and the U.S. Department of Agriculture's Cooperative State Research, Education and Extension Service. In this communication, we wish to inform the research community about this project, to mobilize the research community to participate in detailed phenotyping of these lines, and to provide the opportunity to nominate additional candidate lines for a potential extension of this study.
THE IMPORTANCE OF SNPs
DNA sequence variation accounts for a large fraction of observed differences between plant individuals or varieties, including plant development, yield, stress tolerance, and nutritional quality. The bulk of natural genetic variation in organisms is represented by SNPs or small insertions or deletions. The potentially large number of SNPs in the genomes of individuals within a population or species (Schafer and Hawkins, 1998; Cargill et al., 1999; Kwok, 2001; Syvanen, 2001) provides the foundation for novel approaches to genomic mapping of quantitative trait loci. In humans, SNP variation is approximately 0.5% per nucleotide site (Cargill et al., 1999), while in maize (Zea mays) the variation is closer to 1% to 2% per site (Tenaillon et al., 2001). In the rice genome, SNP occurrences are estimated at approximately three to four SNPs per 1,000 bases, depending on the examined chromosomal regions (Feng et al., 2002; Yu et al., 2005). Recent estimates for SNP variation in rice, based on the indica and japonica genome sequence data, are less than 0.4% (Feltus et al., 2004). On an applied level, the very high densities of SNPs in a genome have made them a preferred molecular marker for fine-mapping studies (Rafalski, 2002). More fundamentally, SNPs are the basic units of genomic diversity, and understanding the evolutionary dynamics of plant genomes involves, in part, assessing the levels, patterning, and distribution of these SNPs (Aquadro, 1992). Studies of SNPs provide a framework for examining how population history, breeding system, and selection affect variation at genetic loci, and delineate the mechanisms that lead to evolutionary diversification of genomes (Nordborg and Innan, 2002; Palaisa et al., 2004). For example, molecular population genomics uses SNP data to probe the levels and patterning of nucleotide polymorphisms within and between loci to test whether specific genes are evolving under selection or in a neutral manner (Bergelson et al., 1998; Purugganan and Suddith, 1999; Bustamante et al., 2002; Palaisa et al., 2004). Of particular utility for whole-genome scans will be the identification of tag SNPs, SNPs that define haplotype regions (Johnson et al., 2001) and can be used to track these regions across populations for testing associations (Gonzalez-Neira et al., 2006).
From a practical perspective, SNP discovery is valuable for rice improvement in two fundamental ways. First, it reveals DNA variation among varieties, thus providing the tools for selection in breeding programs (Rafalski, 2002). Second, it provides the “ultimate anchor” to relate all forms of polymorphisms, including biochemical, metabolic, physiological, and phenotypic performance. Due to the availability of extensive SNP datasets, the theory and practice of using SNP data to identify causal genetic factors of phenotypes have largely come from human and mouse research (Botstein and Risch, 2003; Frazer et al., 2004; Hirschhorn and Daly, 2005; Wang et al., 2005). These systems provide excellent examples where relationships between SNPs and phenotypes in case versus control studies or in well-characterized inbred mice lines (Pletcher et al., 2004; Guenet, 2005) have been established. Aranzana et al. (2005) have shown in Arabidopsis (Arabidopsis thaliana) that association mapping in a selfing species is possible through identification of previously known flowering-time and pathogen-resistance alleles; however, in their study, a high level of false positives were found for all traits, indicating the need for appropriate genomic controls for association studies. Eliminating spurious genotype-phenotype associations could be possible by adopting the novel approach of Yu et al. (2006) that includes tests of both the population structure and kinship relationships. Considering the wide experimental options possible with plants using natural populations, historical breeding pedigrees, and specially designed segregating populations (Rafalski and Morgante, 2004), we expect great utility of genome-wide association analysis for gene discovery in rice. With the exception of Arabidopsis (Bevan and Walsh, 2005; Nordborg et al., 2005), as yet no other plant species has an extensive, genome-wide SNP dataset.
A HIGH-THROUGHPUT TECHNOLOGY TO IDENTIFY RELEVANT SNPs IN COMPLEX GENOMES
The IRFGC will collaborate with Perlegen Sciences to obtain a rich resource of rice SNPs through the “resequencing” of the nonrepetitive portions of the genomes in multiple rice lines using a high-density microarray technology pioneered at Perlegen Sciences (http://www.perlegen.com; Patil et al., 2001). Using the high-quality whole rice genome sequence as a template, Perlegen will design SNP-discovery arrays that contain oligomers designed to include all possible SNP variations with multiple levels of redundancy. When used in hybridizations with “challenge” genomes, sequence differences between the regions in common to the two genomes will be revealed, and through comparison a sequence for these regions of the new “challenge” genome can be deduced. By comparing the information from 24 human genomes, Perlegen discovered and developed assays for approximately 1.5 million SNPs distributed throughout the human genome (Patil et al., 2001; Hinds et al., 2005). Projects are currently in progress to identify SNPs in the mouse and Arabidopsis genomes (K. Frazer, Perlegen Sciences, personal communication).
RICE IS AN EXCELLENT PLANT MODEL FOR CONNECTING WHOLE-GENOME VARIATION WITH PHENOTYPE
Rice is ideally positioned to exploit the Perlegen technology because of the availability of a high-quality, whole-genome sequence in combination with a large store of genetic materials exhibiting extensive trait variation. A high-quality, finished sequence of the japonica subspecies (var Nipponbare) was recently published by the International Rice Genome Sequencing Project (2005), and a draft sequence (approximately 6× sequence coverage) of the indica subspecies (var 93-11; Yu et al., 2005) generated by the Beijing Genomics Institute also is available (Sasaki and Burr, 2000; Barry, 2001; Goff et al., 2002; Yu et al., 2005). In addition, high-quality, uniform annotation of the rice genome is ongoing at the structural and functional levels (Yuan et al., 2005). Furthermore, rice functional genomic resources for assessing gene function on a genome-wide scale are well established or ongoing in rice. At the transcript level, serial analysis of gene expression projects are under way for rice (Matsumura et al., 1999; Gowda et al., 2004). Serial analysis of gene expression data, coupled with the approximately 32,000 full-length cDNAs (Kikuchi et al., 2003) and approximately 400,000 expressed sequence tags in GenBank, provide a rich resource for transcript structure and expression patterns in rice. In addition, a large, public massively parallel signature sequencing project has commenced in rice (http://mpss.udel.edu/rice/) to more deeply sample the transcriptome. A collection of microarray platforms is available for genome-wide expression studies, including a publicly available long oligonucleotide array (www.ricearray.org), an Affymetrix expression array (http://www.affymetrix.com/products/arrays/specific/rice.affx), and an Agilent expression array (http://www.chem.agilent.com/Scripts/PDS.asp?lPage=12133). These are complemented by a project to develop an atlas of expression in a panel of rice tissues throughout development (http://plantgenomics.biology.yale.edu/riceatlas). Collections of tagged lines are available in rice using Tos17, Ac/Ds, and T-DNA (Hirochika et al., 2004; http://orygenesdb.cirad.fr/) and provide induced variation.
Of critical importance for future application of the SNP data to plant breeding is the availability of rice germplasm that contains a wealth of trait diversity (Rafalski and Morgante, 2004). The International Rice GenBank Collection (IRGC) at IRRI comprises the largest collection of rice germplasm held in trust for the world community, with more than 102,547 accessions from the Asian cultivated rice (Oryza sativa), 1,651 accessions from the African cultivated rice (Oryza glaberrima), and 4,508 accessions from 22 related wild relatives. IRRI maintains records of breeding pedigrees of all modern rice varieties derived from mating of traditional varieties (International Rice Information System; http://www.iris.irri.org/). The power of the genetic resources in rice is that they allow a detailed characterization of important traits, such as tolerance to biotic and abiotic stresses, yield, nutrition, and grain quality. The deep collection also enables analysis of traits undergoing selection in the course of domestication. These existing diverse germplasm collections are “gold mines” for analysis of allelic diversity of all rice genes.
Furthermore, application of the SNP data will also depend on the extent of linkage disequilibrium (LD) present in rice. Garris et al. (2003) showed that at the xa5 locus, LD was found to be significant across 100 kb. Another study indicates that for other loci, LD extends up to 200 kb or more (M. Purugganan, personal communication). If the lower estimate is used, haplotype blocks of 100 kb imply that about 4,000 tag SNPs—SNPs representative of the haplotype within a region—would be adequate for whole-genome scans in rice. While SNP data from 21 varieties may not be sufficient for robust inferences of association depending on the magnitude of phenotypic differences for the trait, tag SNPs identified from the data would be the entry point to genotype sufficient additional varieties with contrasting phenotypes for improving the power of association tests.
Rice, like other plants, offers an advantage for genetic analysis that is not possible (as in humans) or straightforward in animal species (e.g. in mouse). Genetic crosses can be readily performed in plants to produce segregating populations. Efforts are under way to develop genetic stocks (i.e. mapping populations, such as recombinant inbred lines) using the rice lines nominated for resequencing to facilitate application of the SNP and phenotyping data to exploit genetic diversity for crop improvement.
CANDIDATE RICE LINES FOR SNP DISCOVERY
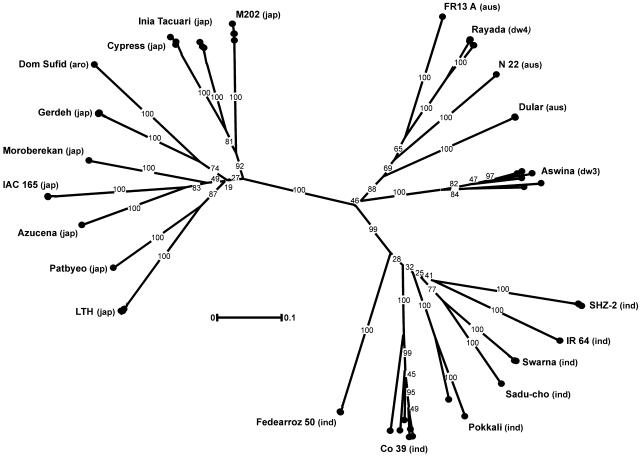
After consultation with the international rice research community, a list of varieties was developed that will be used for SNP discovery (Table I). The varieties were selected in terms of their value in breeding and genetic studies and relative diversity to each other. Figure 1 shows a dendrogram derived by simple sequence repeat fingerprinting. It depicts the genetic relationship of these lines and also illustrates some degree of genetic heterogeneity within some lines. The population structure in the dendrogram is similar to that observed by Garris et al. (2005) in their study of a larger collection of germplasm varieties, some of which are in common. Together with the two already sequenced varieties (Nipponbare and 93-11), the selected lines span the genetic diversity from major rice varietal groups and include important breeding lines and varieties with a wealth of derived pedigrees, mapping populations, mutants, and introgression lines. All varieties to be resequenced will be subjected to single-seed descent purification.
Table I.
Potential candidate varieties for the SNP discovery projecta
| Variety | Origin | Type (VG)b | Sourcec | Agronomic Attributes |
|---|---|---|---|---|
| Co 39 | India | Indica (VG I) | IRGC 51231 | Parent in a mapping population, good yield in aerobic drought screening trials; blast susceptible, very short duration. |
| Fedearroz 50 | Colombia | Indica (VG I) | CIAT-FLAR BCF 1530 | Popular variety in several countries, stay green, quality traits, disease tolerance, progenitor of many breeding lines. |
| IR64 | The Philippines | Indica (VG I) | IRGC 66970 | Quality traits, multiple stress tolerance (diseases and insects), progenitor of many breeding and mapping populations, 40,000 chemical- and irradiation-induced mutants available, widely grown for many years. |
| Pokkali | India | Indica (VG I) | IRGC 108921 | Salt tolerance, parent of multiple mapping populations, microarray gene expression data available. |
| Sadu-cho | Korea | Indica (VG I) | IRGC 2243 | Korean landrace with long grain and indica-type endosperm. |
| Shan-Huang Zhan-2 (SHZ-2) | China | Indica (VG I) | IRTP 19808 | Disease-resistant, high-yielding variety. Demonstrated source of at least five superior defense gene alleles for durable resistance to blast. In the pedigrees of many varieties in south China. |
| Swarna | India | Indica (VG I) | IRTP 12715 | High yield potential, wide adaptability. Widely planted variety in India. |
| Dular | India | Aus/boro (VG II) | IRGC 32561 | Possible β-carotene donor. Red pericarp. |
| FR13 A | India | Aus/boro (VG II) | IRGC 6144 | Submergence tolerance, carrying the effective Sub1 allele. Red pericarp. |
| N 22 | India | Aus/boro (VG II) | IRGC 4819 | Iron, red pericarp. Heat tolerant. Considered drought tolerant. Good yield in managed drought trials. Drought-response expressed sequence tag collection. |
| Aswina | Bangladesh | Deep-water III (VG III) | IRGC 26289 | Deep-water rice. |
| Rayada | Bangladesh | Deep-water IV (VG IV) | IRGC 77210 | Deep-water rice. |
| Dom Sufid | Iran | Aromatic (VG V) | IRGC 12880 | Basmati plant type, aromatic rice. |
| Azucena | The Philippines | Trop japonica (VG VI) | IRGC 328 | Grain-iron quantitative trait loci. Quality traits. Parent of two mapping populations. Near-isogenic lines with high yield under water stress. Deep root distribution. |
| Cypress | United States | Trop japonica (VG VI) | IRTP 19532 | Good grain quality; cold tolerant. |
| IAC 165 | South America | Trop japonica (VG VI) | IRGC 82275 | Parent in mapping population, deep roots. |
| Inia Tacuari | Uruguay | Trop japonica (VG VI) | CIAT-FLAR BCF 828 | Popular early variety, good adaptation and stability, excellent grain quality. |
| Moroberekan | Guinea | Trop japonica (VG VI) | IRGC 12048 | Source of blast quantitative resistance traits and drought-tolerance factors. Multiple advanced breeding populations. Functional evidence for drought tolerance observed in mapping populations and backcross progeny. |
| Gerdeh | Iran | Temp japonica (VG VI) | IRGC 32301 | Near East origin. |
| Li-Jiang-Xin-Tuan-Hei-Gu (LTH) | China | Temp japonica (VG VI) | IRGC 59323 | High disease susceptibility, cold tolerant. |
| M 202 | United States | Temp japonica (VG VI) | U.S. Department of Agriculture; IRGC 77142 | Popular variety. |
| Patbyeo | Korea | Temp japonica (VG VI) | IRGC 55607 | Traditional Korean variety. |
Initial selection of 22 rice lines based on geographic representation, diversity, traits, and usage. Final list for resequencing may differ pending on suitable nominations by the community.
Type refers to the variety group (VG) as determined by isozyme analysis. For the japonica group, lines are designated as either tropical (trop) or temperate (temp).
Source designations are given as accession numbers from the IRGC, the Latin Fund for Irrigated Rice genebank at the International Center for Tropical Agriculture (CIAT-FLAR BCF), or the International Network for Genetic Evaluation of Rice (previously the International Rice Testing Program [IRTP]).
Figure 1.
Dendrogram of potential candidate varieties showing between- and within-variety diversity. For each variety, four to seven individual plants were genotyped using 49 simple sequence repeat markers distributed across the genome. Pairwise distances were calculated using the Dice similarity coefficient for 500 bootstrap replicates and subjected to clustering by unweighted pair group method with arithmetic means. Numbers on branches indicate the percentage of times that group occurred in the strict majority-rule consensus tree. Variety types are annotated as ind (indica), jap (japonica), aro (aromatic), aus (aus/boro), dp3 (deep-water III), and dp4 (deep-water IV).
DETAILED PHENOTYPING OF THE RICE LINES
Application of the SNP data generated by this effort for association genetics will require detailed and comprehensive phenotyping of the rice lines for multiple traits, such as tolerance to abiotic and biotic stresses, grain quality, and nutrition. Currently, through collaborations among the IRFGC, phenotyping of some of these traits is planned. Yet, additional phenotyping in a range of environments and conditions will be necessary before the SNP data can be fully utilized. Hence, we are seeking experts in physiology and biochemistry willing to collaborate on the phenotyping of these rice lines for traits of interest.
We encourage you to visit our project Web site at http://www.oryzasnp.org. At this site, you can nominate lines for inclusion into a potential second phase to extend the coverage of the SNP data across additional varieties and indicate your interest to participate in phenotyping of the nominated rice lines. Comments are also welcome through e-mail with the corresponding author.
Acknowledgments
We thank the International Rice Genome Sequencing Project for access to build 4 of their pseudomolecule assemblies for sequence analysis, the Beijing Genome Institute (Gane Wong) for access to the indica (93-11) genome assembly and suggestions on experimental design, Thomas Bureau and Douglas Moen of McGill University for contributing repeat masking protocols, and Shaohuang Zhang for conducting simple sequence repeat analysis of the candidate varieties.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kenneth L. McNally (k.mcnally@cgiar.org).
References
- Aquadro CF (1992) Why is the genome variable? Insights from Drosophila. Trends Genet 8: 355–362 [DOI] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horto M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, et al (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G (2001) The use of the Monsanto draft rice genome sequence in research. Plant Physiol 125: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Stahl E, Dudek S, Kreitman M (1998) Genetic variation within and among populations of Arabidopsis thaliana. Genetics 148: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M, Walsh S (2005) The Arabidopsis genome: a foundation for plant research. Genome Res 15: 1632–1642 [DOI] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet (Suppl) 33: 228–237 [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL (2002) The cost of inbreeding in Arabidopsis. Nature 416: 531–534 [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22: 231–238 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14: 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Wade CM, Hinds DA, Patil N, Cox DR, Daly MJ (2004) Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 mb of mouse genome. Genome Res 14: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, McCouch SR, Kresovich S (2003) Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genomes (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Neira A, Ke X, Lao O, Calafell F, Navarro A, Comas D, Cann H, Bumpstead S, Ghori J, Hunt S, et al (2006) The portability of tagSNPs across populations: a worldwide survey. Genome Res 16: 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Jantasuriyarat C, Dean RA, Wang GL (2004) Robust-LongSAGE (RL-SAGE): a substantially improved LongSAGE method for gene discovery and transcriptome analysis. Plant Physiol 134: 890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenet JL (2005) The mouse genome. Genome Res 15: 1729–1740 [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079 [DOI] [PubMed] [Google Scholar]
- Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, et al (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54: 325–334 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95–108 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, et al (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29: 233–237 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Dishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Kwok PY (2001) Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet 2: 235–258 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Terauchi R (1999) Technical advance: transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J 20: 719–726 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196 [DOI] [PMC free article] [PubMed]
- Nordborg M, Innan H (2002) Molecular population genetics. Curr Opin Plant Biol 5: 69–73 [DOI] [PubMed] [Google Scholar]
- Palaisa K, Morgante M, Tingey S, Rafalski A (2004) Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci USA 101: 9885–9890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, et al (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719–1723 [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batlov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al (2004) Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol 2: e393 [DOI] [PMC free article] [PubMed]
- Purugganan MD, Suddith JI (1999) Molecular population genetics of floral homeotic loci. Departures from the equilibrium-neutral model at the APETALA3 and PISTILLATA genes of Arabidopsis thaliana. Genetics 151: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski A, Morgante M (2004) Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet 20: 103–111 [DOI] [PubMed] [Google Scholar]
- Rafalski JA (2002) Novel genetic mapping tools in plants: SNPs and LD-based approaches. Plant Sci 162: 329–333 [Google Scholar]
- Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3: 138–141 [DOI] [PubMed] [Google Scholar]
- Schafer AJ, Hawkins JR (1998) DNA variation and the future of human genetics. Nat Biotechnol 16: 33–39 [DOI] [PubMed] [Google Scholar]
- Syvanen AC (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev Genet 2: 930–942 [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98: 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Barratt BJ, Clayton DG, Todd JA (2005) Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet 6: 109–118 [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Wang A, Zhu W, Maiti R, Lin H, Hamilton J, Haas B, Sultana R, Cheung F, et al (2005) The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol 138: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]