Abstract
Plant steroid hormones, brassinosteroids (BRs), are essential for normal photomorphogenesis. However, the mechanism by which light controls physiological functions via BRs is not well understood. Using transgenic plants carrying promoter-luciferase reporter gene fusions, we show that in Arabidopsis (Arabidopsis thaliana) the BR-biosynthetic CPD and CYP85A2 genes are under diurnal regulation. The complex diurnal expression profile of CPD is determined by dual, light-dependent, and circadian control. The severely decreased expression level of CPD in phytochrome-deficient background and the red light-specific induction in wild-type plants suggest that light regulation of CPD is primarily mediated by phytochrome signaling. The diurnal rhythmicity of CPD expression is maintained in brassinosteroid insensitive 1 transgenic seedlings, indicating that its transcriptional control is independent of hormonal feedback regulation. Diurnal changes in the expression of CPD and CYP85A2 are accompanied by changes of the endogenous BR content during the day, leading to brassinolide accumulation at the middle of the light phase. We also show that CPD expression is repressed in extended darkness in a BR feedback-dependent manner. In the dark the level of the bioactive hormone did not increase; therefore, our data strongly suggest that light also influences the sensitivity of plants to BRs.
Plant development is determined by complex interaction between endogenous programs and environmental signals, with light being one of the most important environmental cues. Light controls essential biological processes, such as germination, shade avoidance, de-etiolation, phototropism, chloroplast movement, stomatal opening, circadian entrainment, and flowering. The plasticity of plant responses to light is ensured by multiple photoreceptors having different wavelength specificities and sensitivities, and activating target genes through either distinct or integrated signaling mechanisms. In Arabidopsis (Arabidopsis thaliana), red/far-red signals are perceived by five phytochromes, PHYA to PHYE, whereas two types of cryptochromes and phototropins function as blue and UVA light receptors (for review, see Sullivan and Deng, 2003; Chen et al., 2004). Light regulation is crucial for coordinating the metabolic and physiological functions of plants with the alternating light and dark periods during the day. In addition to direct, light-dependent diurnal regulation of gene activities, light signals are crucial for entrainment of the endogenous circadian clock that ensures cyclic expression of circadian-controlled genes with a roughly 24-h period length, thereby contributing to the precise timing/phasing of numerous biochemical and physiological processes during the day. On the other hand, a hallmark of circadian rhythms is that they persist for several days under constant environmental conditions (McClung, 2001). The importance of these processes can be gauged by the fact that approximately 11% of the genes in Arabidopsis show diurnal patterns of expression and that about 25% of these are also under circadian regulation (Schaeffer et al., 2001).
Whereas the perception of light signals is an intracellular process, responses at the tissue and organ levels are mediated and coordinated through the action of phytohormones. Several lines of evidence suggest that almost all hormone groups participate in the control of photomorphogenic processes and that this involves complex cross talk between the signaling routes of the different hormones. Light can control the effect of phytohormones by influencing their biosynthesis, metabolism, transport, and/or perception (Nemhauser and Chory, 2002).
One hormone group that has been implicated in photobiological responses is brassinosteroids (BRs). In addition to the role of these steroidal phytohormones in promoting growth, fertility, and stress resistance, they also regulate photomorphogenesis, and seedlings of some of the first characterized BR-deficient mutants were shown to have short hypocotyls, open cotyledons, and elevated expression of light-regulated nuclear genes when grown in the dark (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996). Physiological effects of BRs depend on local hormone concentration and responsiveness at the sites of action. The actual levels of bioactive BRs depend on the balance between their tightly controlled biosynthesis and inactivation (Fujioka and Yokota, 2003).
BRs are synthesized from phytosterols through multiple, mostly oxidative reactions leading to brassinolide (BL), the biologically most active BR. In vivo conversion studies revealed two alternative reaction routes, the early and late C-6 oxidation pathways that utilize 6-oxo or 6-deoxo intermediates, respectively (Choi et al., 1997). In Arabidopsis, several of the biosynthetic enzymes have been identified through the characterization of BR-deficient mutants, and those catalyzing oxidative conversions were found to be cytochrome P450 monooxygenases of the CYP85 or CYP90 families (for recent review, see Fujioka and Yokota, 2003). Relaxed substrate specificities of these P450 enzymes allow conversions along parallel routes, and recent analyses suggest that BRs are synthesized via a complex network of reactions (Fujioka et al., 2002; Shimada et al., 2003).
The severe BR-deficient phenotype caused by brassinazole, a specific inhibitor of BR biosynthesis, suggests that de novo synthesis is a key factor in determining the level of the bioactive hormone (Asami et al., 2000). Analysis of the intermediate pools in Arabidopsis, tomato (Lycopersicon esculentum), and rice (Oryza sativa) revealed conserved regulation of the BR pathway, in which C-6 oxidation, as well as steroid side chain hydroxylation at C-22 and C-23, were identified as potential rate-limiting reactions (Nomura et al., 2001). In Arabidopsis, C-6 oxidation is catalyzed by two enzymes, CYP85A1 and CYP85A2, of partially redundant functions. Under in vitro conditions, yeast-expressed CYP85A1 was shown to generate teasterone, typhasterol, and bioactive castasterone from their respective 6-deoxo precursors, whereas CYP85A2, in addition to these reactions, also catalyzed the Baeyer-Villiger oxidation of castasterone to BL (Shimada et al., 2001, 2003; Kim et al., 2005; Nomura et al., 2005). The C-22 and C-23 hydroxylation reactions were found to be mediated by DWF4/CYP90B1 and CPD/CYP90A1, respectively (Szekeres et al., 1996; Choe et al., 1998). Recent studies indicated that in Arabidopsis the expression of all BR-biosynthetic P450 genes is under both developmental and organ-specific regulation that take place primarily at the level of transcription (Bancos et al., 2002; Shimada et al., 2003; Kim et al., 2006). This can influence the efficiency of biosynthesis because activities of certain genes encoding rate-limiting enzymes, such as CPD in Arabidopsis or DWARF/CYP85A1 and CYP85A3 in tomato, show good correlation with the accumulation of active BRs (Bancos et al., 2002; Montoya et al., 2005; Nomura et al., 2005).
Homeostatic regulation, ensuring the adjustment of optimal BR concentrations under various physiological conditions, also requires temporal or permanent inactivation of excess hormone. In Arabidopsis, mutants overexpressing the BAS1/CYP734A1 and CHI2/SHK1/SOB7/CYP72C1 genes were shown to reduce the level of active BRs (Neff et al., 1999; Nakamura et al., 2005; Takahashi et al., 2005). In vivo conversion assays revealed that CYP734A1 (formerly CYP72B1) converts castasterone and BL to biologically inactive C-26-hydroxylated products, while in the case of CYP72C1 inactivation of the same substrates is achieved by a different, yet unknown mechanism (Turk et al., 2003, 2005). Biosynthesis and inactivation are coordinated via BR-dependent transcriptional control of the genes involved in both processes. All BR-biosynthetic P450 genes of Arabidopsis are under negative feedback regulation by active BRs (Bancos et al., 2002; Tanaka et al., 2005), whereas of those encoding inactivating enzymes only BAS1 was found BR inducible (Choe et al., 2001; Takahashi et al., 2005). Feedback regulation of CPD is abolished in the BR-insensitive bin2 and brassinosteroid insensitive 1-2 (bri1-2)/cbb2 mutants (Li et al., 2001; Bancos et al., 2002), indicating a key role of BR signaling in the hormonal self-regulation of BR biosynthesis and perhaps also inactivation. Recently BRASSINAZOLE RESISTANT 1 (BZR1), a nuclear component of this signaling cascade, has been identified as the transcriptional repressor that can down-regulate BR-biosynthetic genes by direct binding to their promoters (He et al., 2005).
Expression studies of the Arabidopsis CYP85 and CYP90 genes by reverse transcription (RT)-PCR provided valuable clues regarding the timing and spatial distribution of BR synthesis (Bancos et al., 2002; Shimada et al., 2003), but low transcript levels limited the resolution and reliability of these analyses. More precise localization of transcriptional activity could be achieved in transgenic plants carrying reporter genes fused to the CPD, CYP85A1, or CYP85A2 promoters (Mathur et al., 1998; Castle et al., 2005), proving the efficiency of reporter-based expression analysis. Because CPD is more actively transcribed than other BR-biosynthetic P450 genes (Shimada et al., 2003), we generated transgenic Arabidopsis lines expressing firefly (Photynus pyralis) luciferase (LUC) under control of the CPD promoter in order to allow in vivo monitoring of temporal changes in promoter activity by luminescence imaging. Here, we report that expression of the CPD:LUC fusion follows a complex diurnal pattern that is determined by both light and circadian regulation. Very similar diurnal cycles were observed with LUC driven by the promoter of CYP85A2, the gene required for BL synthesis. We also show that light induction of CPD is mediated primarily by phytochrome signaling, and that both light and circadian control of promoter activity are independent of the feedback regulation by BRs. Our analyses of the endogenous hormone content revealed BL accumulation in the light, suggesting that the level of active BRs is influenced by the diurnal expression of rate-limiting biosynthetic enzymes.
RESULTS
Diurnal Changes in the Expression of BR-Biosynthetic Genes
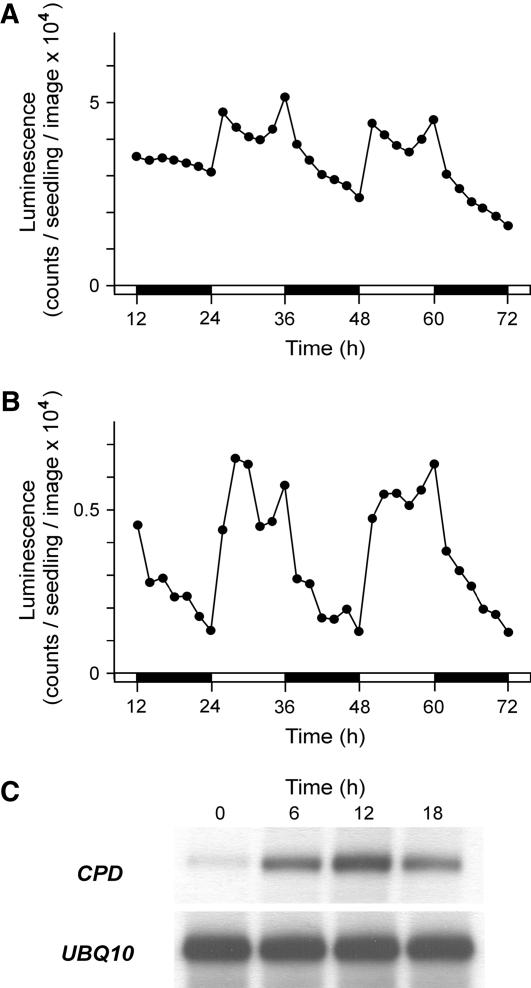
Fusion constructs with firefly LUC reporter offer a flexible, noninvasive system for studying the in vivo activity of BR-biosynthetic gene promoters. Initial characterization of Arabidopsis lines carrying CPD promoter-driven LUC revealed that the luminescence levels detected by a CCD camera system vary depending on the time of the day. In time-course experiments, CPD:LUC transgenic seedlings that were raised under alternating 12-h-light/12-h-dark photoperiods (LD) showed a characteristic diurnal pattern of LUC activity (Fig. 1A). Following lights-on, a sudden increase of luminescence intensity resulted in a maximum, then values gradually decreased, reaching a minimum around midday, then increased to a second maximum at lights-off. Thereafter, the detected luminescence decreased again throughout the night until a second minimum at the end of the dark phase.
Figure 1.
Diurnal expression of the BR-biosynthetic genes CPD and CYP85A2. A and B, LUC activity measured from the eighth day of development in transgenic seedlings harboring the CPD:LUC (A) or CYP85A2:LUC (B) transgenes. White and black bars at the time scale indicate light and dark periods, respectively. Zero time is the beginning of the eighth light period. Each section shows the result of a representative measurement. C, Autoradiogram of the RT-PCR products generated from the endogenous CPD and constitutive control UBQ10 transcripts during a daily LD cycle in wild-type seedlings. Zero time is the beginning of the ninth light period.
To see if other BR-biosynthetic genes can show similar diurnal changes of activity, we also generated LUC fusion with the promoter of CYP85A2. This gene encodes a BR C-6 oxidase catalyzing the last conversion steps of BL synthesis in Arabidopsis (Kim et al., 2005; Nomura et al., 2005), and its organ-specific expression pattern has been shown to be similar to that of CPD (Bancos et al., 2002; Castle et al., 2005). Although the luminescence emitted by CYP85A2:LUC transgenic seedlings was about 10-fold lower than that of the CPD:LUC-carrying ones, it also exhibited a diurnal profile with maxima and minima at similar time points as seen in the case of CPD (Fig. 1B). But, compared to CPD, CYP85A2 promoter activity showed a broader morning peak, and its daily changes had higher amplitude.
To ascertain that the expression data obtained with the CPD:LUC construct faithfully reflect the activity of the CPD promoter, in 1-week-old seedlings we determined the steady-state levels of the CPD mRNA during the LD cycle using semiquantitative RT-PCR assay (Fig. 1C). The amount of the transcript was found lowest before lights-on and highest before lights-off, whereas intermediate values were observed at midday and in the middle of the night. These data are in good agreement with the luminescence levels of the CPD:LUC transgenic line (Fig. 1B).
Circadian and Light Regulation of CPD Expression
Under LD conditions, the sudden changes of CPD expression at lights-on and lights-off suggested a regulatory role of light in control of transcriptional activity. To determine the temporal profile of CPD activity in the absence of light signals, we measured the luminescence levels of CPD:LUC seedlings in both continuous white light (LL) and continuous dark (DD), following a 7-d LD entrainment period. In LL, the LUC activity showed a free-running circadian oscillation that was maintained for several days. The expression cycles had roughly 24-h period lengths and approximately 2-fold rhythmic amplitude, with minimum points at subjective midday and maxima in the subjective night (Fig. 2A). Circadian activity of the CPD promoter could also be observed in DD, but under these conditions the cycles became attenuated after 3 d, at which point the expression level decreased to about 10% of the initial value (Fig. 2B).
Figure 2.
Circadian regulation of CPD expression under constant light conditions. A and B, LUC activity measured following 7 d of LD entrainment in transgenic seedlings harboring the CPD:LUC transgene during growth in LL (A) or DD (B). Time scale shows zeitgeber time elapsed from the onset of the last (eighth) light period; white and black bars indicate light and dark periods, respectively. Each section shows the result of a representative measurement.
Compared to the LL activity of CPD, the diurnal expression curve shows a strong increase at lights-on and a sudden decrease following lights-off, suggesting a positive role of light in the regulation of CPD (Fig. 1A). An activity minimum in the middle of the light phase shows temporal coincidence with the one observed at the subjective light period of the free-running LL cycle (Fig. 2A), and a shoulder in the second half of the dark phase seems to correspond to the postmaximum part of the LL curve measured at the same time of the subjective dark period (Fig. 2A). These similarities between the LD and LL profiles indicate that CPD expression is under dual control, with a light induction superimposed upon the circadian oscillation of the transcriptional activity.
Light Induction of CPD Is Dependent on Phytochrome Signaling
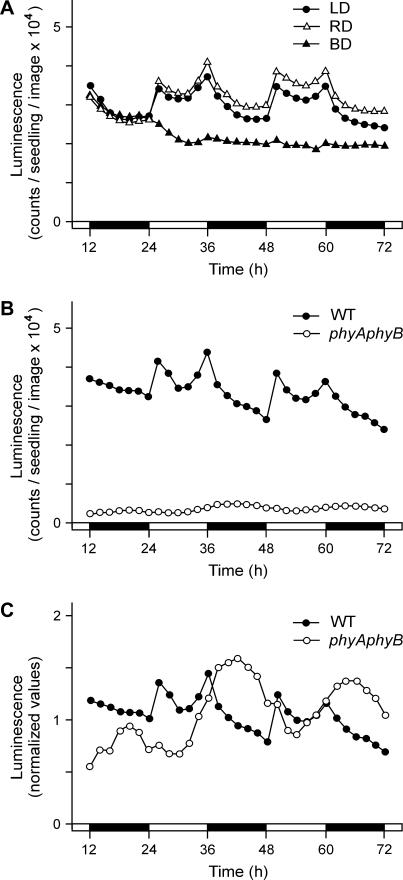
To clarify which light signaling pathway(s) is involved in the induction of CPD expression, we measured the activity of the CPD:LUC transgene in LD using light sources of different spectral characteristics. Three samples of seedlings were grown to 1 week old under LD photoperiods, and then their LUC activity was measured in LD cycles with the same light source, or by replacing white illumination with monochromatic red or blue light periods (RD and BD conditions). As shown in Figure 3A, diurnal cycling of CPD activity was maintained in RD, and the expression values were very similar to those seen in LD. By contrast, BD regimes caused a decrease in the expression level and severe dampening of the diurnal oscillation, resulting in an activity profile resembling the one detected in DD (Fig. 2B). Because red light was sufficient for enabling diurnal cycles, whereas blue light of the same intensity was not, these data suggest a major role for phytochrome photoreceptors in the light induction of CPD.
Figure 3.
The effects of wavelength and phytochrome deficiency on light induction of CPD. A, LUC activity of CPD:LUC transgenic plants under photoperiods of different wavelengths. Seedlings were entrained in LD (white light/dark cycles) for 7 d from germination, then grown further under photoperiods of the same length using white, red, or blue light sources (LD, RD, or BD, respectively). Zero time is the beginning of the last (eighth) common white illumination. B, Diurnal CPD:LUC transgene activity in wild-type and phyA-201phyB-5 backgrounds. C, To aid the comparison of the rhythmic component of expression, data from B were normalized for the average luminescence of the individual lines, as detected during the measurements. Zero time is the start of the eighth light period. White and black bars at the time scale indicate white light and dark periods, respectively. Each section shows the result of a representative measurement.
To verify the importance of phytochrome signaling in the light response, we measured the diurnal expression of CPD:LUC in transgenic seedlings lacking functional PHYA and PHYB, the two most abundant phytochromes. In these seedlings, the luminescence intensity in LD was only about 10% of that measured in wild-type background, and activity changes at lights-on and lights-off were barely recognizable (Fig. 3B). Instead, the LD expression detected in phyAphyB plants indicates the dominance of circadian regulation (Fig. 3C). The experiments showing diminished transgene induction by blue light or in phytochrome-deficient background reveal the primary role of phytochrome signaling in the diurnal regulation of CPD activity, and also in determining the level of expression.
Diurnal Fluctuation of CPD Expression Is Not Determined by Feedback Regulation
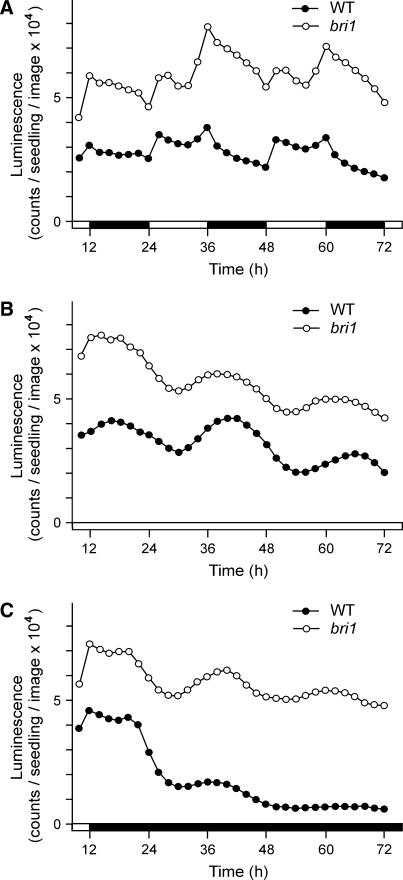
The transcription of CPD is stringently controlled by the level of biologically active BRs, and in turn the expression of CPD can influence BR accumulation. Therefore, it was important to clarify whether the hormonal feedback regulation is involved in the light and/or circadian control of CPD expression. To address this question, we measured the expression of the CPD:LUC transgene in the BR-insensitive bri1 mutant that had been shown to express CPD at an elevated level (Bancos et al., 2002). We found that, although in LD the activity CPD:LUC was about 2-fold higher in bri1 seedlings than in the wild-type control, the diurnal profile of expression was similar in both transgenic lines (Fig. 4A). Compared to the plants with wild-type background, in the CPD:LUC-carrying bri1 mutant the lights-on response was weaker and slower, whereas the decrease of activity following lights-off was more gradual. These data reveal that diurnal changes in CPD transcript levels do not depend on BR signaling and that the feedback regulation has only a minor modulating effect on the daily expression profile.
Figure 4.
The influence of BR insensitivity on the diurnal and circadian activity of CPD. Changes of CPD:LUC expression in wild-type or bri1 backgrounds are shown. Transgenic seedlings were grown in LD for 8 d from germination, and then LUC activity was measured under different light regimes. A, LD; B, LL; C, DD. Zero time is the beginning of the eighth light cycle. White and black bars at the time scale indicate light and dark periods, respectively. Each section shows the result of a representative measurement.
We also measured the LL and DD expression of CPD:LUC in bri1 seedlings to find out whether the circadian expression is dependent on BR signaling. In LL a similar oscillation pattern was observed as in the wild-type background, although the amplitude was lower in both absolute and relative terms (Fig. 4B). By contrast, the activities measured in DD were quite different from those of the wild-type control. Although a gradual dampening of the cycle amplitudes was also observed in bri1 plants, this was not accompanied by a rapid decrease of the expression level, which remained about as high as under LL conditions (Fig. 4C). These results show that the circadian regulation of CPD activity is maintained in the absence of BR signaling, whereas its repression in DD is caused primarily by hormonal feedback regulation.
Diurnal Changes of the Endogenous BR Content
Diurnal oscillation in the expression of BR-biosynthetic genes suggests that the rate of BR synthesis varies during the day/night cycle. To determine whether daily changes of CPD and CYP85A2 expression correlate with the steroid hormone content, we performed quantitative gas chromatography (GC)-mass spectrometry (MS) analysis of the endogenous BRs in 1-week-old wild-type seedlings during the LD cycle at 6-h intervals (Table I). The early C-6 oxidation intermediates cathasterone, teasterone, and 3-dehydro-6-deoxoteasterone were not detectable in the samples taken in the morning just before lights-on, at midday, in the evening before lights-off, and at midnight (0, 6, 12, and 18 h from lights-on, respectively). Levels of the late C-6 oxidation BRs and castasterone showed only minor differences, without any apparent diurnal variation. But, in contrast to the steadiness of the intermediate pools, at midday the seedlings accumulated large amounts of BL, the highly active BR that remained below the detection limit in samples collected at the other three time points of the day. These data show a remarkable increase of the active hormone content at the middle of the light period and also that the synthesis of BL does not appreciably affect the pools of BR intermediates.
Table I.
Endogenous BR levels during a LD cycle
BR data for each time point show the results of three independent experiments, yielding samples of 95 g, 83 g, 80 g, and 98 g (first lane); 37 g, 32 g, 40 g, and 31 g (second lane); and 40 g, 36 g, 40 g, and 37 g (third lane), all fresh weight.
| BR | BR Content | |||
|---|---|---|---|---|
| Hours from Lights-On
| ||||
| 0 | 6 | 12 | 18 | |
| ng kg−1 fresh weight | ||||
| BL | NDa | 151 | ND | ND |
| ND | 87 | ND | ND | |
| ND | 136 | ND | ND | |
| Castasterone | 47 | 45 | 38 | 46 |
| 72 | 83 | 75 | 64 | |
| 73 | 84 | 83 | 64 | |
| 6-Deoxocastasterone | 1,237 | 1,379 | 1,470 | 1,422 |
| 1,114 | 1,152 | 1,044 | 1,242 | |
| 1,196 | 1,156 | 1,178 | 1,239 | |
| 6-Deoxotyphasterol | 445 | 491 | 438 | 420 |
| 496 | 495 | 527 | 548 | |
| 740 | 412 | 584 | 653 | |
| 3-Dehydro-6-deoxoteasterone | 192 | 232 | 205 | 223 |
| 287 | 275 | 270 | 324 | |
| 276 | 318 | 269 | 434 | |
| 6-Deoxoteasterone | 68 | 71 | 87 | 66 |
| 182 | 71 | 98 | 77 | |
| 80 | 70 | 111 | 79 | |
| 6-Deoxocathasterone | 1,394 | 1,689 | 1,638 | 1,346 |
| 2,124 | 1,819 | 1,661 | 1,756 | |
| 2,779 | 1,856 | 1,554 | 1,590 | |
ND, Not detectable.
Because in DD the expression level of CPD decreased rapidly in the wild type but not in the bri1 background, we also wanted to find out the effect of prolonged dark growth on the endogenous BR content. Wild-type seedlings were grown to 1 week old in LD, then BRs were analyzed in samples that were either placed in darkness for 48 h, starting from the end of the eighth light period, or grown further for the same time in LD. As shown in Table II, the amounts of BR intermediates were only slightly influenced by extended dark treatment, and the level of the bioactive castasterone remained essentially unchanged. These measurements indicate that the repression of CPD activity in DD does not the result from an increase of the endogenous BR content.
Table II.
Endogenous BR levels after 48 h in DD
BR data for each time point show the results of three independent experiments, yielding samples of 125 g and 132 g (first lane); 87 g and 109 g (second lane); and 101 g and 100 g (third lane), all fresh weight.
| BR | BR Content
|
|
|---|---|---|
| 48 h DD | LD Control | |
| ng kg−1 fresh weight | ||
| BL | NDa | ND |
| ND | ND | |
| ND | ND | |
| Castasterone | 79 | 85 |
| 63 | 54 | |
| 55 | 65 | |
| 6-Deoxocastasterone | 531 | 977 |
| 802 | 905 | |
| 891 | 1,032 | |
| 6-Deoxotyphasterol | 387 | 521 |
| ND | 79 | |
| ND | 155 | |
| 3-Dehydro-6-deoxoteasterone | 192 | 201 |
| 188 | 182 | |
| 220 | 182 | |
| 6-Deoxoteasterone | 52 | 49 |
| 143 | 99 | |
| 120 | 114 | |
| 6-Deoxocathasterone | 1,405 | 1,608 |
| 887 | 1,356 | |
| 1,336 | 2,198 | |
ND, Not detectable.
Feedback Repression of CPD Reveals Enhanced BR Responsiveness in the Dark
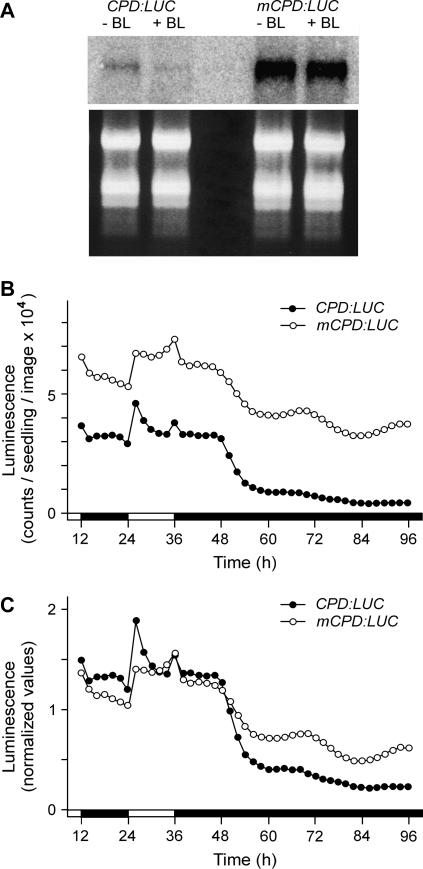
Our finding that BR-dependent down-regulation of CPD expression in DD was not accompanied by an increase of BR levels suggested that light conditions might influence the efficiency of feedback regulation. To find out whether dark repression of CPD is controlled by the BZR1 transcription factor, we measured how DD affected the activity of a modified CPD promoter (mCPD) that contains a point mutation within the BZR1-binding sequence. Seedlings harboring the mCPD:LUC fusion accumulated about 10-fold higher amount of the LUC mRNA than those with the CPD:LUC control, and the transcript level decreased only to 92% of the initial value, opposed to the 8% of the control, upon treatment with 100 nm BL (Fig. 5A). CCD camera measurements revealed that the luminescence profiles of mCPD:LUC seedlings in LD and subsequent DD are very similar to those of CPD:LUC in the bri1 background (Fig. 5B). This result shows that depletion of the CPD transcript in DD is primarily the consequence of BZR1-mediated feedback regulation. Because in seedlings GC-MS analyses did not detect an increase of the BR content in DD, our data strongly suggest that the feedback response is a result of enhanced BR susceptibility in the dark.
Figure 5.
Impaired feedback regulation alleviates dark repression of CPD. A, Feedback regulation of the CPD:LUC and mCPD:LUC transcripts by exogenously applied BL. Northern hybridization and ethidium bromide-stained images of the RNA samples (top and bottom sections, respectively) are shown. B and C, LUC activity of CPD:LUC and mCPD:LUC transgenic plants. Seedlings were germinated and grown in LD for 8 d, and then LUC activity was measured during LD to DD transition from the end of the eighth light period (zero time). Data from the same experiment are plotted both as absolute (B) and normalized (C) values. White and black bars at the time scale indicate light and dark periods, respectively. B and C show a representative measurement.
DISCUSSION
De novo synthesis plays an important role in setting the endogenous levels of bioactive BRs. In transgenic Arabidopsis, BR biosynthesis could be increased by overexpression of DWF4/CYP90B1, one of the rate-limiting enzymes (Choe et al., 2001), and parallel changes in the amounts of CPD:β-glucuronidase and native CPD transcripts revealed that the mRNA level of CPD is controlled primarily at the level of transcription (Mathur et al., 1998). Accordingly, reporter fusions with promoters of BR-biosynthetic genes, such as those of CPD, CYP85A1, and CYP85A2, could be used successfully to determine developmental and temporal expression patterns, as well as active sites of BR synthesis (Mathur et al., 1998; Castle et al., 2005). Whereas the crucial role and relatively high transcription level of CPD make its promoter ideal for in vivo expression assays using LUC reporter, such studies require faithful correspondence between the activities of endogenous CPD and the LUC reporter. During the diurnal cycle, we found good agreement between the levels of the CPD transcript and CPD:LUC-derived luminescence intensity, showing that this reporter construct is a reliable indicator of CPD expression.
Our data on the daily changes of CPD transcript levels correspond to those detected by a microarray analysis designed to identify diurnally expressed Arabidopsis genes (supplemental data to Schaeffer et al., 2001: genome-www.stanford.edu/microarray). These measurements show transcript accumulation of CPD (and also DET2, another BR-biosynthetic gene) at the end of the light period. Finer resolution of the diurnal expression by LUC imaging revealed that dual, light, and circadian regulation of CPD leads to two distinct expression maxima during the day: after lights-on and at lights-off. We found that CYP85A2:LUC activity follows a similar pattern, suggesting coordinated diurnal control of CPD and CYP85A2 transcription. Although the diurnal amplitude of CYP85A2 promoter activity was about 2 times higher than that of CPD, the 5- to 10-fold lower expression level of CYP85A2 (as indicated by cDNA microarray data available at the Genevestigator Web site: www.genevestigator.ethz.ch; Zimmermann et al., 2004) made its reporter-based detection more erratic.
In CPD:LUC-carrying seedlings, LUC activity increased rapidly following lights-on. Whereas the observed light induction is consistent with the concomitant accumulation of CPD mRNA, the initial increase can be slightly exaggerated by a weak, nonspecific, and transient enhancement of LUC activity, perhaps resulting form ATP pool expansion, in response to light (Millar et al., 1992). Circadian expression of CPD, which peaks in the middle of the subjective night, could be maintained for several days in LL, but decreased and became attenuated in DD in about 48 h. The amplitude of cycling is comparable to that of several circadian-regulated biosynthetic genes in Arabidopsis (Harmer et al., 2000), and also of the diurnal changes of CPD activity. Both the light induction and circadian control of the CPD promoter are functional in the BR-insensitive bri1 mutant; therefore, these processes are independent of the hormonal feedback regulation. It is tempting to speculate that the diurnal control of transcription may involve the AAAATCT motif present in both the CPD and CYP85A2 promoters. This sequence is a potential binding site of CIRCADIAN CLOCK ASSOCIATED 1, a transcription factor that has been shown to mediate circadian control via both negative and positive transcriptional complexes (Harmer and Kay, 2005), and also phytochrome-regulated gene expression (Green and Tobin, 1999).
Red light-specific induction of CPD:LUC, as well as its severely reduced activity and light response in the phyAphyB mutant, suggest a primary role for phytochrome signaling in the light regulation of CPD. Although in wild-type background we could also observe a slight increase of LUC activity following blue light exposure, which was used within the photobiologically active fluence range (Devlin and Kay, 2000), this may be attributed to PHYA effect or/and the aforementioned nonspecific light-induced increase of LUC activity, rather than cryptochrome-mediated signals. In green seedlings PHYB is the most abundant phytochrome species, whereas PHYA is the least (Sharrock and Clack, 2002). Therefore, the dramatic decrease of light inducibility in plants deficient for both PHYA and PHYB seems to indicate a predominant role for PHYB in controlling CPD expression. Light regulation of GAs has been studied in detail. Remarkably, most light-responsive genes of GA synthesis were also found to be under wavelength- or photoreceptor-specific control. In Arabidopsis, lettuce (Lactuca sativa), and pea (Pisum sativum), light was shown to regulate the expression of GA 3β-hydroxylase and GA 20-oxidase isoforms via either phytochrome-dependent or -independent mechanisms (Toyomasu et al., 1998; Yamaguchi et al., 1998; Ait-Ali et al., 1999; Jackson et al., 2000).
An increasing amount of evidence indicates the involvement of phytohormones in various light-dependent physiological processes, and in some cases the role of de novo hormone synthesis has been clearly demonstrated. For instance, light was shown to promote the germination of Arabidopsis and lettuce seeds by increasing their GA content via the induction GA 3β-hydroxylases (Toyomasu et al., 1998; Yamaguchi et al., 1998). Alabadí et al. (2004) reported that both mutational and chemical disruption of GA synthesis can interfere with normal skotomorphogenesis and cause aberrant expression of light-regulated nuclear genes. Furthermore, ethylene and GA were found to act coordinately in the phytochrome-mediated shade avoidance response, but their effect could be abolished by paclobutrazol, an inhibitor of GA synthesis (Pierik et al., 2004).
So far only a few studies have addressed the role of light in influencing the biosynthesis and removal of BRs. In Arabidopsis, higher BR levels were detected in light-grown plants than in dark-grown ones, though the plants used in these analyses were not of the same age (Choe et al., 2001). Light-induced accumulation of active BRs was more convincingly demonstrated in light-grown pea seedlings that, compared to the etiolated control, contained 17-fold and 4-fold excess of BL and castasterone, respectively (Symons et al., 2002). Nevertheless, dark-to-light shift of pea seedlings (Symons and Reid, 2003) or light-to-dark shift of Arabidopsis seedlings in our experiment did not appreciably change the levels of castasterone and upstream BR intermediates.
Recently, it has been reported that the expression of Arabidopsis CYP734A1/CYP72B1 and CYP72C1, both encoding BR-catabolizing P450s, are light repressed (Turk et al., 2003; Takahashi et al., 2005), indicating the possibility of coordinated regulation of these genes and light-inducible CPD and CYP85A2 that encode biosynthetic enzymes. Light-activated CYP85A2 and light-repressed CYP734A1/CYP72B1 and CYP72C1 expression seem consistent with the observed appearance of BL in young seedlings at midday. However, it remains to be clarified whether diurnal changes of BL synthesis correlate with similar changes in the amount of the CYP85A2 enzyme. Intriguingly, this transient accumulation of BL does not noticeably affect the pools of BR intermediates, even that of its immediate precursor castasterone, which is synthesized by the same CYP85A2 enzyme (Kim et al., 2005; Nomura et al., 2005). But castasterone itself serves as substrate for BL synthesis, and in tomato fruit intense BL accumulation was found accompanied by an actual decrease of the castasterone level (Montoya et al., 2005). That light-regulated accumulation of active BRs can contribute to photobiological responses is strongly suggested by reports on reduced phytochrome or light effects in BR-deficient mutants (Luccioni et al., 2002), as well as in transgenic lines that overexpress CYP734A1/CYP72B1 and CYP72C1 enzymes (Turk et al., 2005).
The rate of hypocotyl elongation in Arabidopsis shows circadian cycling, with maxima at 4 to 6 h after subjective midday (Dowson-Day and Millar, 1999). Accordingly, an mRNA microarray analysis revealed circadian expression of several genes associated with cell elongation (Harmer et al., 2000). This type of regulation is essential for normal morphogenesis because mutations affecting the circadian clock can also result in abnormal hypocotyl elongation (Dowson-Day and Millar, 1999). The toc1-3 mutation was shown to elevate the activity of GA5 that encodes a GA 20-oxidase (Blázquez et al., 2002). In potato (Solanum tuberosum), the complex diurnal profiles of StGA20ox1 and StGA20ox2 expression (Carrera et al., 1999; Jackson et al., 2000) suggest the involvement of circadian regulation. In line with the observed activity changes of GA-biosynthetic genes, the accumulation of bioactive GAs was also found to follow diurnal rhythmicity (Foster and Morgan, 1995). In addition to the daily oscillation of GA levels, the accumulation of elongation-controlling indole-3-acetic acid and ethylene were shown to be determined by the circadian clock (Jouve et al., 1999; Thain et al., 2004). These results imply that diurnal adjustments of phytohormone synthesis are required for normal development, and circadian regulation can ensure proper hormone levels in anticipation of the regular daily changes of environmental conditions. Our data show that the expression of BR-biosynthetic genes and the endogenous BR content are also under circadian control.
Earlier studies revealed that all P450 genes of BR biosynthesis are feedback regulated by bioactive BRs (Bancos et al., 2002; Shimada et al., 2003). Whereas the diurnal expression of CPD is only weakly influenced by this control mechanism, we found that the repression of this gene in DD is feedback dependent because it requires BRI1 receptor function and proper binding of the BZR1 repressor to its target site (He et al., 2005) in the CPD promoter. That the feedback effect is enhanced in DD without an increase of the BR content strongly suggests that light can decrease BR sensitivity, thereby providing another means of light control over the biosynthesis of BRs. Our finding is in good agreement with earlier reports on different intensities of BR responses under light or dark conditions (Fujioka et al., 1997; Choe et al., 1998; Yang et al., 2005). These regulatory mechanisms of BR synthesis and responsiveness remarkably resemble those of the GAs that also rely on the combination of feedback control of GA 20-oxidases and 3β-hydroxylases and reduced hormone sensitivity in the light (Reed et al., 1996).
The physiological benefits of diurnally controlled BR synthesis are yet to be elucidated. This complex regulation may ensure optimal hormone levels in accordance with the daily changes in growth and metabolic functions. It may also be important in maintaining BR homeostasis by coordinating oppositely regulated biosynthesis and catabolism during the light and dark periods of the day. Elevated CPD and CYP85A2 expression, as well as BL accumulation, are in apparent contradiction with the reduced hypocotyl elongation during the light periods. This anomaly is likely caused by the observed light-dependent decrease in BR responsiveness. Our data and earlier reports indicate concerted regulation of BR levels and sensitivity, while flexibility of this dual control may be ensured by developmental and organ-specific differences between these two effects. Although the impact of de novo BR synthesis is not clear, the complex transcriptional regulation of the biosynthetic enzymes indicates its apparent physiological importance.
An interesting possibility is that, by controlling the accumulation of BL during the light phases, diurnal regulation of BR-biosynthetic genes may influence flowering time in Arabidopsis. Recently, it has been shown that intense synthesis of bioactive BRs is associated with reproductive development (Montoya et al., 2005; Nomura et al., 2005; Symons et al., 2006); therefore, it seems feasible that in Arabidopsis light induction of CYP85A2 can contribute to flower induction through enhancing BL accumulation under long-day conditions. In long-day plants, GAs were reported to promote flowering following long photoperiod up-regulation of light-inducible GA 20-oxidase genes (Blázquez et al., 2002; Lee and Zeevaart, 2002). In Arabidopsis, BRs may have a similar role in determining flower induction because BR-deficient mutants like det2 show characteristic late flowering phenotype (Chory et al., 1991).
MATERIALS AND METHODS
Plant Material
Seeds of transgenic and wild-type Arabidopsis (Arabidopsis thaliana; ecotype Columbia) were surface sterilized and sown on Murashige and Skoog medium (Duchefa) supplemented with 1% (w/v) Suc and 0.2% (w/v) Phytagel (Sigma). Seedlings were grown at 22°C in controlled-environment chambers (SANYO Electronic) under alternating regimes of 12 h white fluorescent light (50–60 μmol m−2 s−1) and 12 h dark (LD). LL and DD were provided using the same conditions as in the corresponding phases of LD. Blue and red light emitted by LED panels at wavelengths 470 ± 5 and 660 ± 5 nm, respectively, were used at a fluence rate of approximately 10 μmol m−2 s−1, approximately equal to their spectral proportions in the white light.
Generation of Transgenic Plants
The CPD:LUC+ reporter construct was generated by cloning the −968 to −5 segment (relative to the translation start) of the CPD promoter (Mathur et al., 1998) into the T-DNA-based binary vector pPCV812 (Koncz et al., 1994), in which the uidA (β-glucuronidase) part had been replaced by the firefly (Photynus pyralis) LUC-derived LUC+ (Promega). The CYP85A2 promoter (−971 to −5) was cloned in the same vector, as was described by Castle et al. (2005). The modified promoter used in the mCPD:LUC+ was isolated from among PCR-mutagenized derivatives of the CPD promoter and, except for a G > A transition at −80 (that strongly inactivates the BZR1-binding sequence CGTGTG by altering its first guanine; He et al., 2005), is identical with the wild-type −968 to −5 segment. The reporter gene-containing binary constructs were conjugated into Agrobacterium tumefaciens GV3101 (pMP90RK) as described by Koncz et al. (1994), and stable Arabidopsis transformants were generated in wild-type and phyA-201phyB-5 (Reed et al., 1994) backgrounds using the flower dip method of Clough and Bent (1998). At least 10 primary transformants were isolated, and from T2 plants showing 3:1 segregation of the transgene homozygous lines were obtained by self-pollination. Following assays of LUC activity, one representative line of each construct was selected for further analysis. (For each reporter construct, the expression profiles of eight primary transformants, including the selected line, are shown in Supplemental Fig. 1.) The CPD:LUC+ fusion was introduced in bri1-101 (Li and Chory, 1997) by crossing this mutant with the selected transgenic line of wild-type background.
Measurement of LUC Activity
Seven-day-old LD-grown transgenic seedlings harboring LUC reporter constructs were transferred to plastic plates with fresh medium. Patches of 100 seedlings were then sprayed twice with filter-sterilized 25 mm d-luciferin (Biosynth A.G.) and 0.01% (v/v) Triton X-100 solution in 5 mm Tris-phosphate buffer, pH 8.0. The last spraying was done 12 h before zero time (the onset of the eighth light period), 24 h before the start of the LUC activity measurements. Alternatively, following surface sterilization, seeds were sown on Murashige and Skoog medium supplemented with 0.5 mm luciferin, placed to 4°C for 12 h, then germinated and raised in DD. Bioluminescence imaging was done at 22°C with a liquid nitrogen-cooled backilluminated digital CCD camera (Princeton Instruments), taking 25-min exposures every second hour. Luminescence intensities were evaluated using Metamorph imaging software (Meta Series 4.5; Universal Imaging). All measurements were repeated at least four times. Since the data were highly reproducible (see Supplemental Fig. 2), in each case we show the result of a representative experiment.
GC-MS Analysis of BR Content
For determining endogenous BR levels during the diurnal cycle, wild-type seedlings were grown in LD, then, starting from the onset of the eighth light period, harvested at 6-h intervals. In three experiments, the plant samples collected at 0 h (subjective morning), 6 h (subjective noon), 12 h (subjective evening), and 18 h (subjective midnight) were immediately frozen in liquid nitrogen, and then subjected to lyophilization. For measuring the BR content in DD, LD-entrained wild-type seedlings were transferred to DD or kept further in LD (control) from the end of the eighth light period (0 h, subjective evening). In three experiments, dark-treated samples and their LD controls were harvested after 48 h (subjective evening), as described above. During and at the end of the dark periods, seedlings were harvested under green safe light.
BR extraction, purification, and analysis were carried out as described by Nomura et al. (2001). In brief, methanol extracts of the plant material were spiked with deuterated BRs prior to reduction to aqueous residues. After partitioning between ethyl acetate and 0.5 m K2HPO4, the former was evaporated to dryness and then partitioned between hexane and 80% methanol. The latter was evaporated and purified using a charcoal column and then a Sephadex LH-20 column. BR fractions were combined and further purified on a Senshu Pak ODS 3251-D column (250- × 8-mm i.d.). The resulting BR samples were analyzed by GC-MS on a Shimadzu-QP2010 GC-MS system using selected ion monitoring mode.
Transcript Analysis
Total RNA was isolated using TRI reagent (Sigma) as recommended by the manufacturer. For semiquantitative RT-PCR assay, RNA was obtained from the same plant batches that were used for BR analysis during the diurnal cycle. Each cDNA sample was prepared from 5 μg of RNA using Ready-To-Go T-primed first-strand kit (Pharmacia Biotech). Primers, PCR conditions, and detection of the amplified products derived from the CPD and UBQ10 (constitutive control) transcripts were as described by Bancos et al. (2002). For northern analysis, 20 μg of RNA samples were separated on formaldehyde-agarose gel (1.2%; w/v) and blotted on Nytran N transfer membrane (Schleicher & Schuell). The RNA blot was hybridized with 32P-labeled probe derived from the full LUC+ CDS, and following stringent washes the distribution of the label was displayed and quantified using a PhosphorImager 445 SI (Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank Katalin Jószai (Institute of Plant Biology, BRC HAS) for her technical support, Yumiko Yamada (Department of Biosciences, Teikyo University) for the purification of plant extracts, Jianming Li (Department of Cellular and Developmental Biology, University of Michigan) for providing the bri1-101 mutant, and Suguru Takatsuto (Department of Chemistry, Joetsu University of Education) for supplying deuterated BR standards.
This work was supported by the Hungarian Scientific Research Fund (grant nos. T 42639 to M.S. and F 47013 to L.K.-B.), an International Joint Project grant of the Royal Society (to G.J.B. and M.S.), research grants from the Biotechnology and Biological Sciences Research Council (to G.J.B.) and the Human Frontiers Science Program (RG00162–2000 to T.Y. and G.J.B), and a Grant-in-Aid for Scientific Research (B13460050 to T.Y.) from the Ministry of Education, Science, Sports and Culture of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Miklós Szekeres (szekeres@nucleus.szbk.u-szeged.hu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079145.
References
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y (1999) Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol 121: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gil J, Blázquez MA, García-Martínez JL (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid inhibitor. Plant Physiol 123: 93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancos S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M (2002) Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Trénor M, Weigel D (2002) Independent control of gibberellin biosynthesis and flowering time by the circadian clock in Arabidopsis. Plant Physiol 130: 1770–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Jackson SD, Prat S (1999) Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol 119: 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle J, Szekeres M, Jenkins G, Bishop GJ (2005) Unique and overlapping expression patterns of Arabidopsis CYP85 genes involved in brassinosteroid C-6 oxidation. Plant Mol Biol 57: 129–140 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 83–117 [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome-P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann K (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Choi YH, Fujioka S, Nomura T, Harada A, Yokota T, Takatsuto S, Sakurai A (1997) An alternative brassinolide biosynthetic pathway via late C-6 oxidation. Phytochemistry 44: 609–613 [Google Scholar]
- Chory J, Nagpal P, Peto C (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17: 63–71 [DOI] [PubMed] [Google Scholar]
- Foster KR, Morgan PW (1995) Genetic regulation of development in Sorghum bicolor. The ma3R allele disrupts diurnal control of gibberellin biosynthesis. Plant Physiol 108: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, Sakurai A (1997) The Arabidopsis de-etiolated 2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96: 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, James PE, Carrera E, Prat S, Thomas B (2000) Regulation of transcript levels of a potato gibberellin 20-oxidase gene by light and phytochrome B. Plant Physiol 124: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve L, Gaspar T, Kevers C, Greppin H, Degli Agosti R (1999) Involvement of indole-3-acetic acid in the circadian growth of the first internode of Arabidopsis. Planta 209: 136–142 [DOI] [PubMed] [Google Scholar]
- Kim HB, Kwon M, Ryu H, Fujioka S, Takatsuto S, Yoshida S, An CS, Lee I, Hwang I, Choe S (2006) The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis. Plant Physiol 140: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-W, Hwang J-Y, Kim Y-S, Joo S-H, Chang SC, Lee JS, Takatsuto S, Kim S-K (2005) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Martini N, Szabados L, Hrouda M, Brachmair A, Schell J (1994) Specialized vectors for gene tagging and expression studies. In SB Gelvin, AR Schilperoort, eds, Plant Molecular Biology Manual, Vol B2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–22
- Lee DJ, Zeevaart JAD (2002) Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiol 130: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J (2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccioni LG, Oliviero KA, Yanofsky MJ, Boccalandro HE, Casal JJ (2002) Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol 128: 173–181 [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- McClung CR (2001) Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol 52: 139–162 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Hiratsuka K, Chua N-H, Kay SA (1992) Firefly luciferase as a reporter of regulated gene expression in higher plants. Plant Mol Biol Rep 10: 324–337 [Google Scholar]
- Montoya T, Nomura T, Yokota T, Farrar K, Harrison K, Jones JGD, Kaneta T, Kamiya Y, Szekeres M, Bishop GJ (2005) Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J 42: 262–269 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Satoh T, Tanaka SI, Mochizuki N, Yokota T, Nagatani A (2005) Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J Exp Bot 56: 833–840 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, Chory J (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J, Chory J (2002) Photomorphogenesis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199, www.aspb.org/publications/arabidopsis
- Nomura T, Kushiro T, Yokota T, Kamiya Y, Bishop GJ, Yamaguchi S (2005) The last reaction producing brassinolide is catalyzed by cytochrome P450s, CYP85A3 in tomato and CYP85A2 in Arabidopsis. J Biol Chem 280: 17873–17879 [DOI] [PubMed] [Google Scholar]
- Nomura T, Sato T, Bishop GJ, Kamiya Y, Takatsuto S, Yokota T (2001) Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57: 171–178 [DOI] [PubMed] [Google Scholar]
- Pierik R, Cuppens MLC, Voesenek LACJ, Visser EJW (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136: 2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J (1996) Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol 112: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T (2002) Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol 130: 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Symons GM, Davies C, Shavrukov Y, Dry IB, Reid JB, Thomas MR (2006) Grapes on steroids. Brassinosteroids are involved in grape berry ripening. Plant Physiol 140: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB (2003) Hormone levels and response during de-etiolation in pea. Planta 216: 422–431 [DOI] [PubMed] [Google Scholar]
- Symons GM, Schultz L, Kerckhoffs LH, Davies NW, Gregory D, Reid JB (2002) Uncoupling brassinosteroid levels and de-etiolation in pea. Physiol Plant 115: 311–319 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei G, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nakazawa M, Shibata K, Yokota T, Ishikawa A, Suzuki K, Kawashima M, Ichikawa T, Shimada H, Matsui M (2005) shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J 42: 13–22 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Vandenbussche F, Laarhoven LJJ, Dowson-Day MJ, Wang Z-Y, Tobin EM, Harren FJM, Millar AJ, Van Der Straeten D (2004) Circadian rhythms of ethylene emission in Arabidopsis. Plant Physiol 136: 3751–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y (1998) Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol 118: 1517–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun T-P (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XH, Xu ZH, Xue HW (2005) Arabidopsis Membrane Steroid-Binding Protein 1 is involved in inhibition of cell elongation. Plant Cell 17: 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.