Abstract
Isoprenoids are the most diverse and abundant group of natural products. In plants, farnesyl diphosphate (FPP) and geranylgeranyl diphosphate (GGPP) are precursors to many isoprenoids having essential functions. Terpenoids and sterols are derived from FPP, whereas gibberellins, carotenoids, casbenes, taxenes, and others originate from GGPP. The corresponding synthases (FPP synthase [FPPS] and GGPP synthase [GGPPS]) catalyze, respectively, the addition of two and three isopentenyl diphosphate molecules to dimethylallyl diphosphate. Maize (Zea mays L. cv B73) endosperm cDNAs encoding isoprenoid synthases were isolated by functional complementation of Escherichia coli cells carrying a bacterial gene cluster encoding all pathway enzymes needed for carotenoid biosynthesis, except for GGPPS. This approach indicated that the maize gene products were functional GGPPS enzymes. Yet, the predicted enzyme sequences revealed FPPS motifs and homology with FPPS enzymes. In vitro assays demonstrated that indeed these maize enzymes produced both FPP and GGPP and that the N-terminal sequence affected the ratio of FPP to GGPP. Their functionality in E. coli demonstrated that these maize enzymes can be coupled with a metabolon to provide isoprenoid substrates for pathway use, and suggests that enzyme bifunctionality can be harnessed. The maize cDNAs are encoded by a small gene family whose transcripts are prevalent in endosperm beginning mid development. These maize cDNAs will be valuable tools for assessing the critical structural properties determining prenyl transferase specificity and in metabolic engineering of isoprenoid pathways, especially in cereal crops.
With more than 30,000 identified compounds, isoprenoids are the oldest, most abundant, and structurally diverse natural products (Lange et al., 2000; Kharel and Koyama, 2003). In plants, isoprenoids and their derivatives are involved in a number of important processes, including photosynthesis and photoprotection, defense, regulation of growth and development, and attracting animals involved in pollination and seed dispersion (DellaPenna, 1999). Some isoprenoids are substrates for pathways leading to the production of the pharmaceutically relevant taxenes and casbenes. Protein prenylation, a posttranscriptional addition of farnesyl and geranylgeranyl groups, is part of signal transduction, intracellular trafficking, and tissue differentiation in plants (Crowell, 2000). The complexity of isoprenoid biosynthetic pathways is manifest from the scores of genes and proteins reported to date and the large number of genes predicted from analysis of the Arabidopsis (Arabidopsis thaliana) genome (Lange and Ghassemian, 2003).
Isoprenoid biosynthesis proceeds through sequential 1→4′ condensations of isopentenyl diphosphate (IPP; C5) with an allylic acceptor, the first of which is dimethylallyl diphosphate (DMAPP; C5). This reaction is catalyzed by highly conserved prenyl transferases and the elongation stops at discrete chain lengths, depending on enzyme specificity (Ogura and Koyama, 1998). The final size of the isoprenyl product is determined by the bulk of the lateral groups of certain amino acyl residues located in a hydrophobic pocket in the enzyme, as evidenced by x-ray crystallography and kinetic studies of wild-type avian farnesyl diphosphate synthase (FPPS; EC 2.5.1.1 and EC 2.5.1.10) and mutated enzymes in which such amino acyl residues have been substituted (Tarshis et al., 1996). The active site in each one of the two subunits in the FPPS homodimer accommodates an acceptor DMAPP, and two IPP molecules are successively added to form farnesyl diphosphate (FPP; C15). Further elongation of FPP is prevented by aromatic lateral groups from amino acyl residues located at the fourth and fifth positions before the first aspartate-rich motif (FARM: DD(XX)1-2D…RRG). A second aspartate-rich motif (SARM: FQ…DDXXD) also appears to be essential for catalytic function (Koyama, 1999). The sequence preceding a GQ motif may play a role in chain-length determination, at least in some geranygeranyl diphosphate synthases (GGPPS; EC 2.5.1.29; Koyama et al., 2000). In most GGPPS enzymes, the fourth and fifth amino acyl residues before the FARM have small, nonaromatic groups, allowing the elongation to geranylgeranyl diphosphate (GGPP), a C20. Other factors such as subunit composition and steric interactions may contribute to determine isoprenoid product size, as it is in the case of geranyl diphosphate synthase (GPPS) from Mentha piperita (Burke and Croteau, 2002).
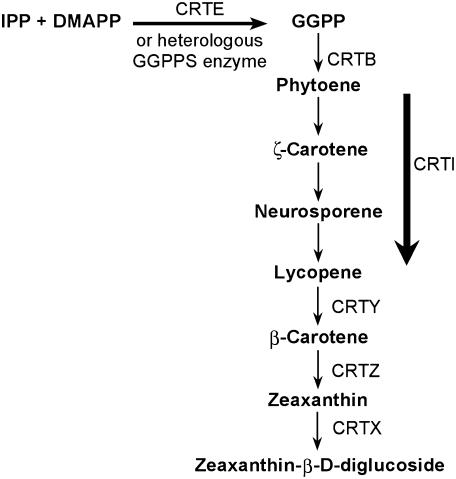
One approach to isolating genes encoding isoprenoid synthases is to couple genes encoding these enzymes with a downstream pathway that acts as a reporter of functional enzyme activity. A valuable reporter system is the carotenoid biosynthetic pathway that converts isoprenoid precursors to easily detected colored products (Cunningham et al., 1993; Gallagher et al., 2003). Several bacterial groups, including Erwinia and Rhodobacter (Armstrong et al., 1990; Misawa et al., 1990; Math et al., 1992), and fungi such as Neurospora crassa (Carattoli et al., 1991; Sandmann et al., 1993) possess a gene cluster, crt, that makes them capable of synthesizing carotenoids. There are six genes in the Erwinia uredovora cluster and most of the encoded enzymes (Fig. 1) have a counterpart in higher plants. When expressed in an appropriate microbial host, this cluster directs synthesis of the yellow carotenoid derivative zeaxanthin-β-d-diglucoside. A gene encoding GGPPS, crtE (Misawa et al., 1990; Math et al., 1992), ensures the supply of GGPP, the direct precursor of carotenoids. In the absence of crtE, carotenoid accumulation is greatly reduced and colonies are colorless (Misawa et al., 1990; Li et al., 1996; Hirschberg et al., 1997; Oh et al., 2000). Complementation of the gene cluster deleted for crtE (Sandmann and Misawa, 1992) facilitated functional identification of maize (Zea mays) cDNAs encoding enzymes with GGPPS activity. Further characterization revealed that these enzymes were bifunctional, having both FPPS and GPPS activities.
Figure 1.
Biosynthetic pathway for zeaxanthin, directed by the Erwinia crt cluster in E. coli, and color complementation strategy. CRTE, Bacterial version of GGPPS; CRTB, phytoene synthase; CRTI, phytoene desaturase; CRTY, lycopene β-cyclase; CRTZ, β-carotene hydroxylase; CRTX, zeaxanthin glucosyl transferase.
RESULTS
Functional Screening to Identify Maize GGPPS cDNAs
As a preliminary step to identify maize GGPPS cDNAs, the E. uredovora carotenoid gene cluster deleted for crtE was used for functional complementation screening of maize B73 inbred endosperm cDNAs. In the absence of GGPPS encoded by crtE, cells were unable to accumulate the pathway end product, zeaxanthin-β-d-diglucoside, unless a plant cDNA encoding a functional enzyme was introduced. Visual screening of approximately 30,000 colonies of Escherichia coli TOP10F′ harboring both pACCAR25ΔcrtE (referred to as ΔcrtE) and library-derived cDNAs in pBluescript II SK (−) resulted in six colonies that accumulated zeaxanthin-β-d-diglucoside, as evidenced by the yellow color not seen in other transformants, and which was confirmed by retransformation. The isolated and related maize cDNAs were 1.42 kb (e.g. pME14D1184) or 1.25 kb (e.g. pME14D1268), either size being sufficient for functional complementation.
Carotenoid Analysis
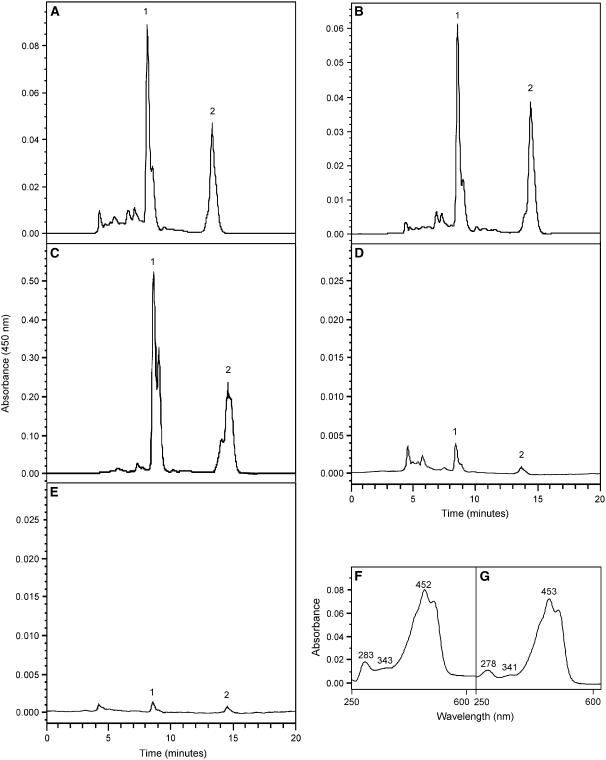
To confirm the identity of the yellow pigment in the ΔcrtE/maize cDNA double transformants as zeaxanthin-β-d-diglucoside, organic extracts from the double transformants and controls were analyzed by reversed-phase HPLC (Fig. 2). Figure 2A shows a chromatogram for pigments accumulating in E. coli TOP10F′ carrying ΔcrtE and maize long clone, pME14D1184. By comparison with carotenoids isolated from pACCAR25 (Misawa et al., 1990) and pACCAR25ΔcrtX cells (Misawa et al., 1990; data not shown), peak 1 corresponds to the pathway end product zeaxanthin-β-d-diglucoside (Sandmann et al., 1998), and peak 2 to free zeaxanthin (Yokoyama et al., 1996), both of which showed predicted spectra (Fig. 2, F and G). The shoulder on the descending slope of peak 1 corresponds to the monoglucoside derivative produced when zeaxanthin becomes limiting (Hundle et al., 1992). Figure 2B also showed zeaxanthin peaks for ΔcrtE cells carrying the shorter maize cDNA pME14D1268, confirming GGPPS activity of its gene product. The total accumulated product for the long clone was 67-fold, and for the short clone 55-fold, compared to the negative control, empty pBluescript II SK (−) vector (Fig. 2E), whose negligible production was due to background GGPP synthesis (Chamovitz et al., 1992; Sandmann and Misawa, 1992; Kainou et al., 2001). For comparison to other prenyl transferases, complementation of ΔcrtE was tested with Gentiana lutea cDNAs previously demonstrated to encode GGPPS (pUCggpps; Zhu et al., 2002) or FPPS (pBS-FPS; C. Zhu and G. Sandmann, unpublished data). As shown in Figure 2C, complementation with the G. lutea GGPPS resulted in combined zeaxanthin/zeaxanthin-glucoside peaks that were 552-fold over the negative control (Fig. 2E) and 8- to 10-fold higher levels compared to the short and long maize clones, respectively. Similar results (data not shown) were also obtained with Hevea brasiliensis GGPPS encoded by pHBGG6 (Takaya et al., 2003). This suggests that there is some variation in either expression or perhaps structure of the encoded maize and bona fide GGPPS enzymes. In contrast, complementation with G. lutea FPPS (pBS-FPS; Fig. 2D) yielded product that was 156-fold lower than that of the bona fide GGPPS and barely above background levels obtained when using the negative control (Fig. 2E). When compared to the G. lutea FPPS, the maize cDNAs yielded 16-fold (short clone) and 19-fold (long clone) more pathway end product. Therefore, while the inferred GGPPS activity of the bona fide GGPPS was about 8- to 10-fold greater than the maize enzyme, this maize enzyme produced as much as 19-fold greater product compared to a known FPPS. Our HPLC data confirmed that similar to a bona fide GGPPS cDNA, the maize cDNAs encoded enzymes yielding GGPP required for zeaxanthin production with activity based on this indirect measurement that was significantly greater than that obtained using a known FPPS.
Figure 2.
HPLC analysis of carotenoids from E. coli TOP10F′ cells containing plant cDNAs and the Erwinia ΔcrtE construct. Besides pACCAR25ΔcrtE, double transformants contained either maize long clone pME14D1148 (A), maize short clone pME14D1268 (B), G. lutea pUCggpps (C), G. lutea pBS-FPS (D), or empty vector pBluescript II SK(−) (E). Absorption spectra and λmax for peaks 1 (zeaxanthin-β-d-diglucoside) and 2 (zeaxanthin) in section A are shown in panels F and G, respectively. Peaks 1 and 2 in all chromatograms displayed the same spectra.
Sequence Analysis of Maize cDNAs
The functional GGPPS endosperm cDNAs from maize were sequenced and found to be virtually identical. Figure 3 shows the nucleotide and predicted amino acid sequences for one long (pME14D1184) and one short cDNA (pME14D1268) that were deposited in GenBank (AF330036 and AF330037, respectively). Excluding the vector-encoded N-terminal β-galactosidase fragment, the peptide encoded by the longer functional GGPPS clone (pME14D1184) was 391 residues (44.1 kD). However, if counting from the first methionine, the predicted peptide was only 350 residues (40.0 kD), which is still greater than the 341 residue (39.4 kD) peptide encoded by the shorter functional clone (pME14D1268).
Figure 3.
Nucleotide sequence of clone pME14D1184 and predicted amino acid sequence. Two DD(XX)1-2D motifs, characteristic of prenyl transferases, are underlined. Nucleotides in the putative polyadenylation signal are in bold and underlined. The GenBank accession number for clone pME14D1184 is AF330036. The [G] at position 10 marks the beginning of a shorter clone (pME14D1268, with GenBank accession number AF330037). The c at position 30 is a t in pME14D1130 but does not affect the translated sequence.
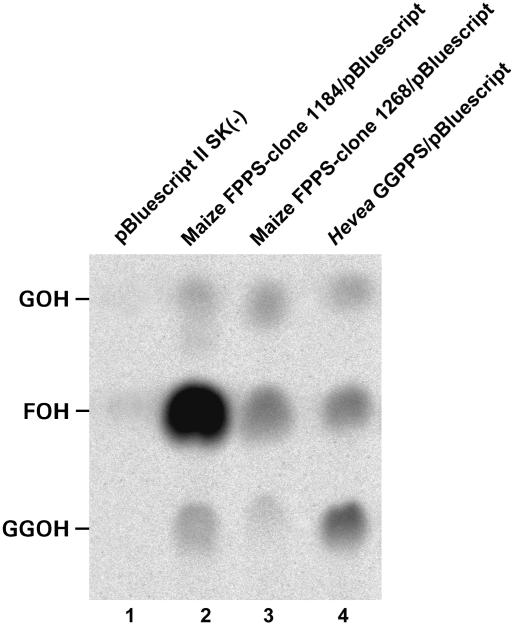
Having employed a GGPPS functional complementation strategy to isolate the maize cDNAs, we predicted homology of the isolated clones to other GGPPS genes. Instead, BLAST searches (Altschul et al., 1997) produced a surprising homology to a truncated but otherwise identical FPPS clone from maize inbred W64A (GenBank L39789; Li and Larkins, 1996). Furthermore, BLAST searches showed that our maize clones, although enabling the synthesis of zeaxanthin-β-d-diglucoside, shared characteristic signatures with other FPPS sequences, including FARM, the chain-length-determining (CLD) region, and the GQ motif. Additionally, the SARM predicted from the maize isoprenoid synthase clones was identical to that in rice (Oryza sativa) and Arabidopsis FPPS enzymes (Fig. 4). Based on characteristics of the CLD region and the sequence preceding the GQ (Hemmi et al., 2003), the polypeptides encoded by the maize clones corresponded to type I FPPS (Wang and Ohnuma, 1999).
Figure 4.
Comparison of CLD regions, GQ motifs, and SARM of predicted amino acid sequences of several plant short-chain prenyl transferases. Zeama FPPS1184, Maize isoprenoid synthase long clone (GenBank AF330036); Zeama FPPS1268, maize short clone (AF330037); Orysa FPPS1J, rice cv Japonica FPPS1 (O04882); Orysa FPPSI, rive cv. Indica FPPS (AAD27558); Arath FPPS1S, Arabidopsis FPPS1-S, short isoform (AAB07264); Arath FPPS1L, Arabidopsis FPPS1-L, mitochondrial long isoform (AAF44787); Arath FPPS2, Arabidopsis FPPS2 (Q43315); Arath GGPPS2, Arabidopsis chloroplastic GGPPS2 (NP_179454); Arath GGPPS4, Arabidopsis endoplasmic reticulum GGPPS4 (AAM15136); Arath GGPPS6, Arabidopsis mitochondrial GGPPS6 (BAA23157); Genlu GGPPS, G. lutea GGPPS (BAB82463); Hevbr GGPPS, H. brasiliensis GGPPS (BAB60678); Erwur CRTE, E. uredovora CRTE (P22873).
In Vitro Isoprenoid Assays
The maize cDNAs shared structural motifs with FPPS enzymes but clearly functioned in the heterologous bacterial system as GGPPS. To directly investigate the reaction products, the novel maize enzymes were further analyzed by in vitro incorporation of [4-14C]IPP in cell-free extracts from transformants of the pBluescript-based constructs. The dephosphorylated, radiolabeled products were identified by reversed-phase thin-layer chromatography (TLC). To ensure that [4-14C]-IPP incorporation results reflected differences in the activities of enzymes tested, but not in their expression levels, cDNAs cloned in vectors with the same promoter [e.g. pBluescript II SK (−) and pUC8] and the same host cells were used when assaying cell extracts. Figure 5 shows that cell extracts containing either the long (1184) or short (1268) maize FPPS cDNAs (lanes 2 and 3, respectively) yielded GGPP (as indicated by the dephosphorylated form, geranylgeraniol [GGOH]), and pathway intermediates GPP and FPP (as indicated by geraniol [GOH] and farnesol [FOH]). Identical products were seen for the bona fide GGPPS from H. brasiliensis (lane 4) but were virtually absent when cells contained only the empty vector, negative control (lane 1). The fact that cell-free extracts containing the maize isoprenoid synthases produced [14C]GGPP (lanes 2 and 3) explains why plasmids pME14D1184 and pME14D1268 could complement ΔcrtE in E. coli. The amounts of GGPP produced by the maize clones, as evidenced by 14C incorporation, were not as large as that produced by the bona fide GGPPS tested (compare with lanes 2 versus 4), but were sufficient to supply GGPP to the enzymes encoded by the ΔcrtE construct in E. coli and therefore to produce zeaxanthin-β-d-diglucoside. The longer maize enzyme (lane 2) yielded a greater proportion of [14C]FPP relative to [14C]GGPP (mean FPP/GGPP = 114, n = 3 representative results) as compared with the shorter maize enzyme (lane 3; mean FPP/GGPP = 10.4, n = 3). Therefore, the truncation of the N terminus increased the relative amount of GGPP produced by almost 11-fold. In comparison, the GGPPS from H. brasiliensis (lane 4) produced significantly more GGPP than FPP (mean FPP/GGPP = 0.59, n = 3). In a two-way ANOVA, mean ratio was significantly different among plants [F (3, 7) = 120, P < 0.0001, R2 = 98.4%] but was no different among experiments [F (2, 7) = 7 × 10−4, P = 0.97]. We concluded that the novel maize cDNA clones code for FPPS that behaves bifunctionally and synthesizes GGPP in addition to the main product, FPP, in a ratio affected by N-terminal sequence.
Figure 5.
In vitro enzymatic activity of recombinant isoprenoid synthases from maize and H. brasiliensis. Cell-free extracts were prepared from E. coli cells containing plasmids encoding prenyl transferases and assayed by incorporation of [4-14C]IPP into short-chain isoprenoids, followed by dephosphorylation, reversed-phase TLC, and development of the chromatography plate in a Phosphorimager. Lane 1, Empty pBluescript II SK (−) vector; lane 2, pME14D1184 (maize long clone); lane 3, pME14D1268 (maize short clone); lane 4, pHBGG6 (H. brasiliensis GGPPS). Authentic isoprenol standards were run along with the enzymatic assay of isoprenol products and developed with p-anysaldehyde reagent.
We proceeded to test if the GGPP produced by the maize enzymes was influenced by the 38-residue N-terminal β-galactosidase (lacZ) sequence encoded by the cloning vector or by the 41-residue N terminus encoded by the 5′-untranslated region (UTR) in the long clone. To this effect, pME14D1184 was subjected to PCR to amplify either the entire insert containing the 5′-UTR that encodes 41 residues upstream of the open reading frame (ORF), or the ORF only (as it starts from nucleotide 123 in Fig. 3 and contains the nine codons missing from the shorter maize clone). The amplification products were subcloned in the expression vector pQE30, which unlike pBluescript II SK (−), does not encode a β-galactosidase (lacZ) N-terminal peptide fusion, but contains instead a 6X-His tag. The 5′-UTR-containing construct was designated pQE1184UTR; the subcloned ORF was named pQE22ORF (see Fig. 6C for construct cartoons).
Figure 6.
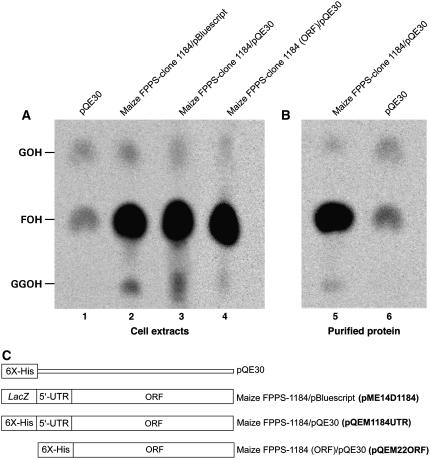
Effect of N terminus on GGPPS activity of maize isoprenoid synthases. Enzymatic assays were performed as described in “Materials and Methods” and Figure 5 legend. A, Cell extracts: lane 1, M15 cells with empty pQE30 vector; lane 2, TOP10F′ cells with pME14D1184; lanes 3 and 4, M15 cells with pQE1184UTR and pQE22ORF, respectively. B, Lane 5, His-tagged recombinant protein encoded by pQE1184UTR (the same construct used in A, lane 3), purified by affinity chromatography; lane 6, same as lane 1. C, Cartoon of constructs used in A and B.
Lane 1 in Figure 6A shows the 14C-incorporation activity in an extract from cells transformed with the empty pQE30 vector. The remaining lanes display the activities of extracts containing protein products of pME14D1184 (contains lacZ, 41-residue maize UTR, and ORF; lane 2; compare with Fig. 5), pQE1184UTR (contains 6X-His tag, 41-residue UTR, and ORF; lane 3), and pQE22ORF (contains 6X-His tag and ORF; lane 4). Polypeptides with UTR-encoded residues produced significant amounts of GGPP, regardless of an N-terminal β-galactosidase fragment (lane 2) or not (lanes 3). This clearly shows that the GGPP production was not a consequence of having a β-galactosidase N-terminal fusion but was a property inherent in the maize enzyme itself. Remarkably, the construct (pQE22ORF) without lacZ and the UTR also produced GGPP (lane 4), and FPP at higher levels then the short clone that is truncated by nine additional codons (Fig. 5, lane 3). To further demonstrate that it was the FPPS enzyme alone that was responsible for GGPPS activity, we also tested the purified protein in the in vitro assay. Lane 5 (Fig. 6B) shows the isoprenoid synthase activity of the purified protein product from pQE1184UTR. This protein, without the β-galactosidase fragment and devoid of bacterial cytoplasmic background, clearly produced GGPP (in addition to FPP and GPP), a phenotype corroborated by pQE1184UTR complementation of the ΔcrtE lesion in E. coli (data not shown). In summary, these results indicate the GGPPS activity associated with the maize FPPS clones was not due to presence of the 38-residue N-terminal β-galactosidase fragment encoded by pBluescript II SK (−). Furthermore, it appears that the 41 amino acids encoded by the 5′-UTR influenced the ratio of FPP/GGPP product associated with the FPPS enzymes. Truncation at the N terminus also interfered with FPPS activity.
Effect of pH Variation on Isoprenoid Synthase Activities
To determine whether GGPP production by the maize enzymes was influenced by pH, we used the in vitro assay at varying pH. Typically, neutral to alkaline pH conditions have been used for in vitro prenyl transferase assays, e.g. FPPS from Arabidopsis and yeast (Saccharomyces cerevisiae) has been assayed at pH 7.0 (Cunillera et al., 1996) and 7.4 (Chambon et al., 1990; Okada et al., 2000). GGPPS from H. brasiliensis shows a broad pH optimum range of 8.0 to 9.0 (Takaya et al., 2003), and the polyprenyl diphosphate synthases from Mycobacterium spp. display an optimum pH near 8 (Crick et al., 2000). The maize isoprenoid synthase (FPPS/GGPPS) was active at pH 6.5 to 10.5, producing both FPP and GGPP, except total isoprenoid synthesis considerably declined at pH 10.5 (data not shown). Besides confirming that the maize isoprenoid synthase was active over a broad pH range, it can be concluded that variations in pH do not affect the product distribution of the prenyl transferase encoded by the maize cDNAs.
Gene Copy Number and Transcript Accumulation in Endosperm
Hybridization of the maize cDNAs to genomic DNA from several maize lines revealed multiple bands and maize B73 bacterial artificial chromosome genomic DNA clones divided into at least three distinct groups (data not shown). Similarly, chromosome mapping data also indicated that the maize isoprenoid synthase gene mapped to three loci, closely linked with bnl117.20 (8L, 69.8), umc15B (3L, 142.6), and npi280 (6L, 144.7).
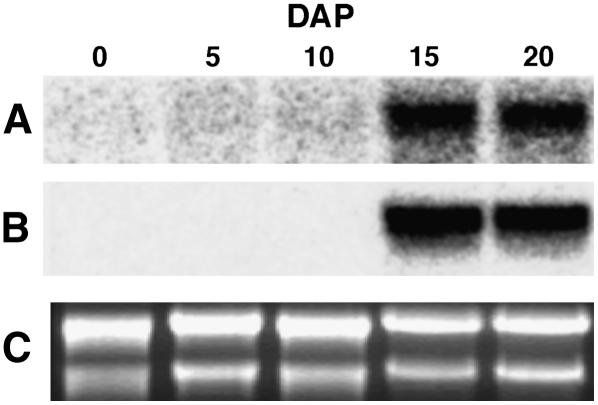
Despite evidence for more than one gene copy in maize, a northern blot of total RNA from a developmental series of maize B73 endosperms showed a single major transcript, approximately 1.5 kb in size (Fig. 7A), a size comparable to that of the endosperm library-derived long cDNA clones. Transcripts were barely detectable until 15 and 20 d after pollination (DAP). The observed transcript levels were consistent with the abundance of cDNA clones isolated from the endosperm cDNA library. Transcript accumulation for Shrunken-1, a gene encoding a starch biosynthetic enzyme, gave a similar profile of temporal expression (Fig. 7B), as previously reported (Wurtzel et al., 1987). Equal amounts of total RNA were loaded, as evidenced by the stained rRNA bands (Fig. 7C).
Figure 7.
Analysis of fpps transcripts from a developmental maize endosperm series. A, A northern-blot prepared from 5 μg/lane of total RNA isolated at the indicated DAP, was probed with the 1.425-kb insert from pME14D1030; or maize Shrunken-1 (B). C, Formaldehyde RNA gel stained with ethidium bromide.
DISCUSSION
The use of a functional complementation strategy as a preliminary screen for plant isoprenoid biosynthesis cDNAs provided new insight into enzyme activity of maize FFPS, which would otherwise not have been predicted from informatic analysis. We demonstrated that the GGPPS activity associated with maize FPPS could be harnessed by coupling to a downstream pathway. While we ruled out the effect of an N-terminal β-galactosidase fusion to elicit this unexpected activity, we showed that the FPPS N terminus impacts both its activity and specificity.
Experimental conversions between FPPS and GGPPS have been achieved using genes from several microorganisms and usually they have been generated by changes in amino acids in the CLD or other regions. The conversion of archaeal GGPPS to FPPS by site-directed mutagenesis identified key residues involved in determining product length specificity, a result that in turn served as the basis for a hypothesis on the evolution of eubacterial and eukaryotic FPPS types from an ancestral GGPPS (Ohnuma et al., 1997). Chemical random mutagenesis was used to determine the conversion of FPPS to GGPPS in Bacillus stearothermophylus (Ohnuma et al., 1996). The mutation responsible for the modified enzymatic activity was found at a position equivalent to crucial residues in other isoprenoid synthases. The use of error-prone PCR for directed evolution of the E. coli FPPS gene (ispA) resulted in GGPPS activity of proteins having novel CLD amino acyl residues (Lee et al., 2004, 2005). These residues are either further upstream from the FARM than others previously identified, in helical regions adjacent to the FARM, or near the SARM. A report in which Streptomyces griseolosporeus mutated GGPPS and FPPS produced FPP and GGPP, respectively, indicates that even amino acid changes far from the FARM can also alter product size (Kawasaki et al., 2003).
Quaternary structure is another factor influencing isoprenoid synthase specificity. For example, the M. piperita GPPS heterotetramer has two regulatory small subunits (GPPS.ssu) and two large catalytic subunits (GGPPS.lsu). When GPPS.ssu is coexpressed with conifer GGPPS in E. coli, it causes the latter to produce GPP instead of GGPP, apparently by constricting the active site. However, GPPS.ssu does not seem to affect FPPS specificity (Burke and Croteau, 2002).
It is conceivable that metabolic context may also impact the maize FPPS enzyme activity, whether the enzyme is expressed by an endogenous gene or as a transgene. Metabolic context has been shown to affect enzyme regiospecificity when subcellular location was altered as in the case of a fatty acid desaturase (Heilmann et al., 2004). The distribution of isoprenoid synthase products in a given organism is also influenced by nonenzymatic factors such as salt and metal ion concentrations, or the available concentration of substrates, conditions that have been studied in vitro (Fujii, 1980; Matsuoka et al., 1991). In vivo, temperature affects final product composition in some archaea, influencing changes in cell membrane archaeols in response to growth temperature (Fujiwara et al., 2004).
Secondary reactions catalyzed by certain enzymes in addition to synthesizing their main products are not a rare occurrence. The term catalytic promiscuity has been applied to this phenomenon (Copley, 2003) that may have an example in the production of 86 sesquiterpenoids from FPP by δ-selinene synthase and γ-humulene synthase in Abies grandis (Steele et al., 1998). Prenylation of aromatic natural products seems to occur promiscuously as well, depending upon factors such as Mg2+ or the availability of aromatic acceptors (Kuzuyama et al., 2005). Further study of this unusual maize FPPS enzyme, including three-dimensional modeling combined with mutagenesis will further expand the understanding of prenyltransferase specificity and activity.
Based on similarity to FPPS2, an Arabidopsis cytoplasmic form (GenBank Q43315) and absence of signatures associated with subcellular targeting (including plastid, mitochondrion, and endoplasmic reticulum; Emanuelsson et al., 2000), the predicted location of the endogenous maize FPPS enzyme is likely cytosolic, where it may play a role in either protein prenylation or in providing biosynthetic precursors to downstream pathways. We do not know if, as for the Arabidopsis fps1 gene, differential transcription leads to multiple isoforms with alternate locations (Cunillera et al., 1997). For the maize long clone pME14D1184 (compare with Fig. 3), the sequence 119-GGCAATGGC-127 displays the properties of a monocot initiation context (Lütcke et al., 1987; Kochetov et al., 1999), and the preceding 118-nucleotide stretch shows characteristics of a monocot 5′-UTR, including a high G + C content (72%; Kozak, 1996). Genomic sequences upstream of the 5′-end of our longest cDNAs (e.g. GenBank CC627299) do not show any in-frame ATG triplets that may indicate alternative translation sites.
The existence of a multigene family, with at least three members, is inferred from genomic DNA analysis and chromosome mapping data. Gene families for isoprenoid biosynthetic enzymes have been reported in other plants. Two cDNAs corresponding to two fpps genes have been reported for Arabidopsis (Lange and Ghassemian, 2003). Two forms of the gene for FPPS have been found for Lupinus albus (Attucci et al., 1995), the rubber-producing shrub Parthenium argentatum (Pan et al., 1996), and rice (Sanmiya et al., 1999). Five cDNAs, encoding GGPPS isoforms targeted to subcellular compartments, have been described in Arabidopsis by functional studies (Okada et al., 2000). Analysis of the Arabidopsis genome sequence predicts 12 genes for GGPPS but only one for GPPS (Lange and Ghassemian, 2003). Several ggpps functional cDNA clones have recently been obtained from maize, confirming that maize has both FPPS and GGPPS (R. Vallabhaneni and E.T. Wurtzel, unpublished data). Therefore, the novel maize FPPS described here does not take the place of a GGPPS but provides an additional GGPPS activity. Interestingly, we did not isolate any of the true GGPPS cDNAs by functional complementation, and instead fortuitously isolated the novel FPPS cDNAs described here. The most likely explanation is that based on transcript profiles, the GGPPS mRNAs are of much lower prevalence (data not shown) and therefore unlikely to be identified by the limited capacity of complementation screening.
To date, metabolic engineering of isoprenoids has sometimes led to unexpected results due to insufficient understanding of metabolic networks (Jørgensen et al., 2005). The future of metabolic engineering for enhancement of plant chemistry rests on investigation of pathway dynamics and plasticity of enzyme function and interaction with natural and artificial metabolons. Further studies on monocot short-chain isoprenoid synthases are necessary if metabolic engineering is to be used in improving the production of isoprenoids in plants. The bifunctional maize clones reported here will be useful tools in such studies.
MATERIALS AND METHODS
Enzymes and Chemicals
All reagents were analytical or HPLC grade. Organic reagents and solvents were purchased from Sigma-Aldrich. Salts and buffers were obtained from Fisher Scientific. Molecular biology reagents, including restriction enzymes, were bought from Invitrogen. Perkin-Elmer was the source of [α-32P]dATP (111TBq/mmol) and [4-14C]IPP (2 mBq/mmol).
Plant Materials
Maize (Zea mays L. cv B73) inbred plants were grown in the field (Pelham Bay, Bronx, NY) and cross pollinated. Endosperms were dissected at various DAP. Tissue was quickly frozen in liquid nitrogen and stored at −80°C until used.
Recombinant DNA Techniques
Cell growth, bacterial transformation, and other methods were performed according to standard protocols (Sambrook et al., 1989; Ausubel et al., 2000).
Construction of Maize Endosperm cDNA Expression Library
Total RNA was isolated from maize B73 endosperm at 14 DAP for production of a unidirectional cDNA library contained in pBluescript II SK (−) (Gallagher et al., 2003). Primary library size was approximately one-million independent clones. Clone representation was estimated by using Shrunken-1 as a hybridization probe on plaque lifts of the lambdoid library and was found to be consistent with transcript prevalence at this developmental stage (Wurtzel et al., 1987).
Library Screening by Color Complementation
The maize B73 endosperm cDNA library was screened by functional complementation of the Erwinia carotenogenic cluster ΔcrtE deletion (Linden et al., 1991; Sun et al., 1996; Hirschberg et al., 1997), according to the strategy depicted in Figure 1. Escherichia coli TOP10F′ cells (Invitrogen) were transformed with the plasmid pACCAR25ΔcrtE (Sandmann and Misawa, 1992) and subsequently transfected with the maize endosperm cDNA library, then diluted, titrated, and plated to 75 to 100 cfu/plate (100 × 15 mm Petri dishes) on Luria-Bertani (LB) medium containing tetracycline, chloramphenicol, and ampicillin. After overnight growth at 37°C and 3 d at room temperature (in the dark), about 30,000 colonies were visually screened. A total of six double-transformant colonies were found to have a yellow hue against a colorless background.
Analysis of Plasmid DNAs
Plasmid DNAs were extracted from the double transformants, after growth in LB medium plus antibiotics, by using the Wizard Midikit (Promega) and separated by agarose gel electrophoresis. Library-derived plasmids were isolated from the agarose gel, purified using Geneclean II (Bio101), and utilized to retransform E. coli TOP10F′ cells containing pACCAR25ΔcrtE, for corroboration of pigment production (Wurtzel et al., 1997), or untransformed E. coli XL1-Blue (Stratagene), for plasmid propagation. Plasmids were digested with EcoRI and XhoI to obtain insert size. Four of the plasmids had inserts of about 1.42 kb and were designated pME14D1030, pME14D1184, pME14D1247, and pME14D1349; the other two had smaller inserts of about 1.25 kb and were named pME14D1268 and pME14D1463.
DNA Sequencing and Analysis
Both strands of cDNAs were initially sequenced using standard primers for pBluescript II SK (−), then by walking, using internal primers, in an automated Applied Biosystems 3730XL capillary apparatus at the DNA Sequencing Facility of the University of Chicago Cancer Research Center. Sequence assembly and analysis were done using BioImage software (Millipore) and Vector NTI Suite v. 8 (InforMax).
Carotenoid Analysis
E. coli TOP10F′ cells containing plasmid pACCAR25ΔcrtE were transformed with either pME14D1184, pME14D1268, pUCggpps (Zhu et al., 2002), pBS-FPS, or pBluescript II SK (−). Double transformants were grown in 100 mL LB medium, plus antibiotics, at 37°C overnight with shaking (200 rpm). Growth was continued for 36 h at room temperature in the dark. After reaching the same culture densities, cells were harvested by centrifugation, followed by pigment extraction through a method adapted from Yokoyama et al. (1995). Cell harvest and subsequent steps were carried out in dim light. Briefly, cell pellets were suspended in 2 mL of acetone, mixed thoroughly, and incubated at 50°C for 15 min. Mixtures were centrifuged for 10 min at 10,000g, and supernatants were recovered into amber vials. The resulting pellets were reextracted as above and supernatants were pooled, dehydrated over Na2SO4, and blown to dryness with nitrogen (g). Pigments were resuspended in injection solvent [acetonitrile:methanol:dichloromethane:hexane (85:10:2.5:2.5, v/v/v/v)], filtered through nylon syringe filters (0.45 μm; Phenomenex), and analyzed on a 250 × 4.6 mm Nucleosil C18 (5 μm particle size) reversed-phase column (Phenomenex) using an Alliance 2690 HPLC system with a 996 photodiode array detector (Waters). Solvent systems A [acetonitrile:methanol (9:1, v/v)] and B [hexane:dichloromethane:methanol (45:45:10, v/v/v)] were used in an isocratic gradient (95% A plus 5% B) for 10 min, followed by a linear gradient (95% to 45% A and 5% to 55% B) for 30 min, at a flow rate of 0.7 mL min−1. Data were acquired using Waters Millennium32 software. Curve points were digitized using the program XYit (Geomatix). Areas under the curves were estimated using Microsoft Excel by the sum of subinterval areas. The data shown are representative of at least three independent experiments.
Preparation of Cell-Free Extracts for Isoprenoid Synthase Assays
E. coli TOP10F′ was used to propagate expression plasmids pME14D1030, pME14D1268, pBS-FPS, pHBGG6 (Takaya et al., 2003), and pUCggpps (Zhu et al., 2002). Extracts were prepared from cells cultured in LB medium with appropriate antibiotics and harvested in mid-log phase (Okada et al., 2000). Cultures of cells carrying pUCggpps were supplemented with 1 mm isopropylthiogalactoside. Protein concentration was determined using the bicinchoninic acid method (Pierce) with bovine serum albumin (BSA) as a standard.
In Vitro Isoprenoid Synthase Assays
A protocol to assay isoprenoid synthase activity was developed from a combination of the procedures of Cunillera et al. (1996), Okada et al. (1998, 2000), and Fujii et al. (1982). Briefly, 100 μL reaction mixtures containing cell-free extract (100 μg of protein) in 100 mm potassium phosphate, pH 7.4, 1 mm EDTA, 1 mm 2-mercaptoethanol, 5 mm MgCl2, 2.2 nmol of DMAPP, and 4.4 nmol of [4-14C]IPP were incubated for 1 h at 37°C. After chilling on ice, isoprenoid diphosphate products were extracted with 200 μL of butanol and dephosphorylated by the addition of 400 μL of methanol, 50 μL of a 5 m potassium acetate (pH 4.7)/10% (w/v) Triton X-100 stock, and 5 mg of potato acid phosphatase (2 u mg−1; Roche Diagnostics). Volume was adjusted to 1 mL with deionized water, and mixtures were incubated for 12 h at 37°C with gentle shaking. Released isoprenols were extracted with 1 mL of hexane. After adding 100 ng each of GOH, FOH, and GGOH to function as carriers, hexane extracts were evaporated to about 25 μL and applied to a 20 cm LKC18 reversed-phase TLC plate (Whatman), along with standard isoprenols (5 nmol each of GOH, FOH, and GGOH). Chromatography was carried out using a methanol:water (95:5, v/v) mobile phase. Isoprenol standards were developed with p-anisaldehyde spray (Dunphy et al., 1967). Radioactive spots were detected on plates after exposure to a Phosphorimager screen as described above. The Volume Report function of the program ImageQuant (Amersham Biosciences) was utilized to estimate the intensity of the radioactive spots on the original scans. Intensity ratios were calculated using Microsoft Excel and JMP v.5.1.2 (SAS) was used for advanced statistical analysis. The data shown are representative of at least three independent experiments.
Subcloning in Expression Vector pQE30
The pME14D1184 insert was amplified using Promega's Master Mix in a Touchgene Gradient thermocycler (Techne). Primers for amplification of insert including the 5′-UTR were 5′-ACTGCAGTCCCTCCCTCCTTCCTTCCTTC-3′ (sense) and 5′-GAAGCTTATCCAAGAGCACCCTACTTCTG-3′ (antisense), which introduced PstI and HindIII restriction sites (underlined) at the product's 5′- and 3′-ends, respectively. To limit amplification to the ORF, the oligonucleotide 5′-ACTGCAGTGGCGGCGGGCGGGAATGG-3′ (sense) was designed. This primer also contains a PstI site (underlined) but the initiation codon was changed to GTG (italicized), as the ATG-containing amplification product did not direct the synthesis of recombinant protein. PCR for both sets was performed with an initial denaturation step at 94°C for 3 min followed by 30 cycles of 45 s at 95°C, 45 s at 60°C, and extension for 2 min at 72°C, and a final extension for 10 min at 72°C. PCR products were digested with PstI and HindIII, gel purified using the Mini-Elute PCR Purification kit (Qiagen), and ligated into the dephosphorylated corresponding sites of expression vector pQE30 (Qiagen).
The resulting sequence-verified constructs, respectively named pQE1184UTR and pQE22ORF, were used to transform E. coli M15 cells. The recombinant protein expressed from these pQE30-derived constructs contains an N-terminal 6XHis tag for purification by affinity chromatography.
Expression and Purification of His-Tagged Isoprenoid Synthase
An extract of E. coli M15 cells containing pQEM1184UTR was prepared as described above, except that cells were washed with and concentrated (40×) in lysis buffer (50 mm sodium phosphate, pH 8.0, 300 mm NaCl, 10 mm imidazole) before sonication. The His-tagged isoprenoid synthase was purified by chromatography of the cell extract in a Ni-NTA Agarose Superflow column, under native conditions, as per the manufacturer's instructions (Qiagen). Protein concentration in column fractions was determined by the bicinchoninic acid method (BCA, Pierce) using BSA dissolved in the corresponding buffers. Concentration of the BSA stock was determined by A280 using an ɛ% = 6.6. Purification to >90% was estimated by SDS-PAGE of samples from the chromatographic steps (data not shown).
In Vitro Isoprenoid Synthase Assays of Purified His-Tagged Enzyme
Activity of the purified recombinant isoprenoid synthase was determined as described above, but using 1 μg of protein.
Transcript Analysis
RNA from maize B73 endosperms at 0, 5, 10, 15, and 20 DAP was isolated as previously described (Burr and Burr, 1981; Wurtzel et al., 1987). Five micrograms from each time point were fractionated by denaturing agarose gel electrophoresis (Sambrook et al., 1989) along with size markers (Promega). The zero time point corresponds to unfertilized ovules and both 0- and 5-DAP endosperms may contain some maternal tissue. RNAs were transferred onto Optitran BA-S 85 supported nitrocellulose membrane. After transfer, membranes were washed with (6×) SSC, UV cross-linked in a Stratalinker 2400 (Stratagene), and prehybridized in 50% (v/v) formamide, (6×) sodium chloride/sodium phosphate/EDTA, (5×) Denhardt's reagent, 0.5% (w/v) SDS, and 100 μg mL−1 denatured salmon sperm DNA. Radiolabeled probe was denatured by boiling and added to prehybridization buffer along with polyethyleneglycol 3000 to 10% (w/v). The membrane was incubated at 42°C for 14 h, and washed twice in (2×) SSC/0.1% (w/v) SDS for 15 min at room temperature, followed by two 15 min washes in (0.2×) SSC/0.1% (w/v) SDS at 50°C, with shaking. Blots were exposed to a phosphor screen and processed as described in a previous section.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF330036 and AF330037.
Acknowledgments
We are grateful to Dr. Norihiko Misawa of the Kamaishi Laboratories, Japan, for supplying the E. uredovora crt constructs. We appreciate suggestions from Dr. Núria Cunillera (Universitat de Barcelona, Spain), who also provided strains and related plasmids, and from Dr. Kazunori Okada (Tokyo Gakugei University, Japan) who provided Arabidopsis GGPPS plasmids. We thank Dr. Tanetoshi Koyama (Tokohu University, Japan) for the Hevea GGPPS plasmid and Dr. Gerhard Sandmann (University of Frankfurt, Germany) for Gentiana cDNAs. We thank Dr. Ben Burr (Brookhaven National Laboratory) for mapping data and the following staff from Lehman College of the City University of New York: Dr. Dwight Kincaid for advice on statistical analysis, David Cain for assistance with maize propagation, and Christina Murillo for technical support.
This work was supported by grants from the National Institutes of Health (grant no. S06 GM08225 to E.T.W. and M.C.C.), the City University of New York Collaborative Incentive Program (to E.T.W. and M.C.C.), and the Professional Staff Congress of the City University of New York (to E.T.W.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eleanore T. Wurtzel (wurtzel@lehman.cuny.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.077008.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong GA, Schmidt A, Sandmann G, Hearst JE (1990) Genetic and biochemical characterization of carotenoid biosynthesis mutants of Rhodobacter capsulatus. J Biol Chem 265: 8329–8338 [PubMed] [Google Scholar]
- Attucci S, Aitken SM, Gulick PJ, Ibrahim RK (1995) Farnesyl pyrophosphate synthase from white lupin: molecular cloning, expression, and purification of the expressed protein. Arch Biochem Biophys 321: 493–500 [DOI] [PubMed] [Google Scholar]
- Ausubel RF, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2000) Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York
- Burke C, Croteau R (2002) Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem 277: 3141–3149 [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA (1981) Controlling-element events at the shrunken locus in maize. Genetics 98: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Romano N, Ballario P, Morelli G, Macino G (1991) The Neurospora crassa carotenoid biosynthetic gene (albino 3) reveals highly conserved regions among prenyltransferases. J Biol Chem 266: 5854–5859 [PubMed] [Google Scholar]
- Chambon C, Ladeveze V, Oulmouden A, Servouse M, Karst F (1990) Isolation and properties of yeast mutants affected in farnesyl diphosphate synthetase. Curr Genet 18: 41–46 [DOI] [PubMed] [Google Scholar]
- Chamovitz D, Misawa N, Sandmann G, Hirschberg J (1992) Molecular cloning and expression in Escherichia coli of a cyanobacterial gene coding for phytoene synthase, a carotenoid biosynthesis enzyme. FEBS Lett 296: 305–310 [DOI] [PubMed] [Google Scholar]
- Copley SD (2003) Enzymes with extra talents: moonlighting functions and catalytic promiscuity. Curr Opin Chem Biol 7: 265–272 [DOI] [PubMed] [Google Scholar]
- Crick DC, Schulbach MC, Zink EE, Macchia M, Barontini S, Besra S, Brennan PJ (2000) Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol 182: 5771–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN (2000) Functional implications of protein isoprenylation in plants. Prog Lipid Res 39: 393–408 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Arró M, Delourme D, Karst F, Boronat A, Ferrer A (1996) Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem 271: 7774–7780 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388 [DOI] [PubMed] [Google Scholar]
- Cunningham FX Jr, Chamovitz D, Misawa N, Gantt E, Hirschberg J (1993) Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of beta-carotene. FEBS Lett 328: 130–138 [DOI] [PubMed] [Google Scholar]
- DellaPenna D (1999) Carotenoid synthesis and function in plants: insights from mutant studies in Arabidopsis. Pure Appl Chem 71: 2205–2212 [Google Scholar]
- Dunphy PJ, Kerr JD, Pennock JF, Whittle KJ, Feeney J (1967) The plurality of long chain isoprenoid alcohols (polyprenols) from natural sources. Biochim Biophys Acta 136: 136–147 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fujii H (1980) Variable product specificity of solanesyl pyrophosphate synthetase. Biochem Biophys Res Commun 96: 1648–1653 [DOI] [PubMed] [Google Scholar]
- Fujii H, Koyama T, Ogura K (1982) Efficient enzymatic hydrolysis of polyprenyl pyrophosphates. Biochim Biophys Acta 712: 716–718 [PubMed] [Google Scholar]
- Fujiwara S, Yamanaka A, Hirooka K, Kobayashi A, Imanaka T, Fukusaki E-i (2004) Temperature-dependent modulation of farnesyl diphosphate/geranylgeranyl diphosphate synthase from hyperthermophilic archaea. Biochem Biophys Res Commun 325: 1066–1074 [DOI] [PubMed] [Google Scholar]
- Gallagher CE, Cervantes-Cervantes M, Wurtzel ET (2003) Surrogate biochemistry: use of Escherichia coli to identify plant cDNAs that impact metabolic engineering of carotenoid accumulation. Appl Microbiol Biotechnol 60: 713–719 [DOI] [PubMed] [Google Scholar]
- Heilmann I, Pidkowich MS, Girke T, Shanklin J (2004) Switching desaturase enzyme specificity by alternate subcellular targeting. Proc Natl Acad Sci USA 101: 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Noike M, Nakayama T, Nishino T (2003) An alternative mechanism of product chain-length determination in type III geranylgeranyl diphosphate synthase. Eur J Biochem 270: 2186–2194 [DOI] [PubMed] [Google Scholar]
- Hirschberg J, Cohen M, Harker M, Lotan T, Mann V, Pecker I (1997) Molecular genetics of the carotenoid biosynthetic pathway in plants and algae. Pure Appl Chem 69: 2151–2158 [Google Scholar]
- Hundle BS, O'Brien DA, Alberti M, Beyer P, Hearst JE (1992) Functional expression of zeaxanthin glucosyltransferase from Erwinia herbicola and a proposed uridine diphosphate binding site. Proc Natl Acad Sci USA 89: 9321–9325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen K, Vinther-Rasmussen A, Morant M, Holm-Nielsen A, Bjarnholt N, Zagrobelny M, Bak S, Lindberg-Møller B (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8: 280–281 [DOI] [PubMed] [Google Scholar]
- Kainou T, Okada K, Suzuki K, Nakagawa T, Matsuda H, Kawamukai M (2001) Dimer formation of octaprenyl-diphosphate synthase (IspB) is essential for chain length determination of ubiquinone. J Biol Chem 276: 7876–7883 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Hamano Y, Kuzuyama T, Itoh N, Seto H, Dairi T (2003) Interconversion of the product specificity of type I eubacterial farnesyl diphosphate synthase and geranylgeranyl diphosphate synthase through one amino acid substitution. J Biochem (Tokyo) 133: 83–91 [DOI] [PubMed] [Google Scholar]
- Kharel Y, Koyama T (2003) Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep 20: 111–118 [DOI] [PubMed] [Google Scholar]
- Kochetov AV, Ponomarenko MP, Frolov AS, Kisselev LL, Kolchanov NA (1999) Prediction of eukaryotic mRNA translational properties. Bioinformatics 15: 704–712 [DOI] [PubMed] [Google Scholar]
- Koyama T (1999) Molecular analysis of prenyl chain elongating enzymes. Biosci Biotechnol Biochem 63: 1671–1676 [DOI] [PubMed] [Google Scholar]
- Koyama T, Gotoh Y, Nishino T (2000) Intersubunit location of the active site of farnesyl diphosphate synthase: reconstruction of active enzymes by hybrid-type heteromeric dimers of site-directed mutants. Biochemistry 39: 463–469 [DOI] [PubMed] [Google Scholar]
- Kozak M (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome 7: 563–574 [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Noel JP, Richard SB (2005) Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435: 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Lange BM, Rujan T, Martin W, Croteau R (2000) Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci USA 97: 13172–13177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Mijts BN, Petri R, Watts KT, Schmidt-Dannert C (2004) Alteration of product specificity of Aeropyrum pernix farnesylgeranyl diphosphate synthase (Fgs) by directed evolution. Protein Eng Des Sel 17: 771–777 [DOI] [PubMed] [Google Scholar]
- Lee PC, Petri R, Mijts BN, Watts KT, Schmidt-Dannert C (2005) Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab Eng 7: 18–26 [DOI] [PubMed] [Google Scholar]
- Li CP, Larkins BA (1996) Identification of a maize endosperm-specific cDNA encoding farnesyl pyrophosphate synthetase. Gene 171: 193–196 [DOI] [PubMed] [Google Scholar]
- Li ZH, Matthews PD, Burr B, Wurtzel ET (1996) Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol 30: 269–279 [DOI] [PubMed] [Google Scholar]
- Linden H, Misawa N, Chamovitz D, Pecker I, Hirschberg J, Sandmann G (1991) Functional complementation in Escherichia coli of different phytoene desaturase genes and analysis of accumulated carotenoids. Z Naturforsch 46c: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Lütcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA (1987) Selection of AUG initiation codons differs in plants and animals. EMBO J 6: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Math SK, Hearst JE, Poulter CD (1992) The crtE gene in Erwinia herbicola encodes geranylgeranyl diphosphate synthase. Proc Natl Acad Sci USA 89: 6761–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Sagami H, Kurisaki A, Ogura K (1991) Variable product specificity of microsomal dehydrodolichyl diphosphate synthase from rat liver. J Biol Chem 266: 3464–3468 [PubMed] [Google Scholar]
- Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172: 6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Koyama T (1998) Enzymatic aspects of isoprenoid chain elongation. Chem Rev 98: 1263–1276 [DOI] [PubMed] [Google Scholar]
- Oh S-K, Kim I-J, Shin DH, Yang J, Kang H, Han K-H (2000) Cloning, characterization and heterologous expression of a functional geranylgeranyl pyrophosphate synthase from sunflower (Helianthus annuus L.). J Plant Physiol 157: 535–542 [Google Scholar]
- Ohnuma S-i, Hirooka K, Ohto C, Nishino T (1997) Conversion from archaeal geranylgeranyl diphosphate synthase to farnesyl diphosphate synthase. J Biol Chem 272: 5192–5198 [DOI] [PubMed] [Google Scholar]
- Ohnuma S-i, Nakazawa T, Hemmi H, Hallberg A-M, Koyama T, Ogura K, Nishino T (1996) Conversion from farnesyl diphosphate synthase to geranylgeranyl diphosphate synthase by random chemical mutagenesis. J Biol Chem 271: 10087–10095 [DOI] [PubMed] [Google Scholar]
- Okada K, Kainou T, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M (1998) Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur J Biochem 255: 52–59 [DOI] [PubMed] [Google Scholar]
- Okada K, Saito T, Nakagawa T, Kawamukai M, Kamiya Y (2000) Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Herickhoff L, Backhaus RA (1996) Cloning, characterization, and heterologous expression of cDNAs for farnesyl diphosphate synthase from the guayule rubber plant reveals that this prenyltransferase occurs in rubber particles. Arch Biochem Biophys 332: 196–204 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Sandmann G, Kuhn S, Boger P (1998) Evaluation of structurally different carotenoids in Eschericia coli transformants as protectants against UV-B radiation. Appl Environ Microbiol 64: 1972–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann G, Misawa N (1992) New functional assignment of the carotenogenic genes crtB and crtE with constructs of these genes from Erwinia species. FEMS Microbiol Lett 90: 253–257 [DOI] [PubMed] [Google Scholar]
- Sandmann G, Misawa N, Wiedemann M, Vittorioso P, Carattoli A, Morelli G, Macino G (1993) Functional identification of al-3 from Neurospora crassa as the gene for geranylgeranyl pyrophosphate synthase by complementation with crt genes, in vitro characterization of the gene product and mutant analysis. J Photochem Photobiol B Biol 18: 245–251 [DOI] [PubMed] [Google Scholar]
- Sanmiya K, Ueno O, Matsuoka M, Yamamoto N (1999) Localization of farnesyl diphosphate synthase in chloroplasts. Plant Cell Physiol 40: 348–354 [DOI] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R (1998) Sesquiterpene synthases from grand fir (Abies grandis): comparison of constitutive and wound-induced activities, and cDNA isolation, characterization, and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem 273: 2078–2089 [DOI] [PubMed] [Google Scholar]
- Sun Z, Gantt E, Cunningham FX Jr (1996) Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271: 24349–24352 [DOI] [PubMed] [Google Scholar]
- Takaya A, Zhang YW, Asawatreratanakul K, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Koyama T (2003) Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyl diphosphate synthase of Hevea brasiliensis. Biochim Biophys Acta 1625: 214–220 [DOI] [PubMed] [Google Scholar]
- Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD (1996) Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA 93: 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ohnuma S (1999) Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem Sci 24: 445–451 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Burr FA, Burr B (1987) DNase I hypersensitivity and expression of the Shrunken-1 gene of maize. Plant Mol Biol 8: 251–264 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Valdez G, Matthews PD (1997) Variation in expression of carotenoid genes in transformed E. coli strains. Bioresearch Journal 1: 1–11 [Google Scholar]
- Yokoyama A, Sandmann G, Hoshino T, Adachi K, Sakai M, Shizuri Y (1995) Thermozeaxanthins, new carotenoid-glycoside-esters from thermophilic eubacterium Thermus thermophilus. Tetrahedron Lett 36: 4901–4904 [Google Scholar]
- Yokoyama A, Shizuri Y, Hoshino T, Sandmann G (1996) Thermocryptoxanthins: novel intermediates in the carotenoid biosynthetic pathway of Thermus thermophilus. Arch Microbiol 165: 342–345 [DOI] [PubMed] [Google Scholar]
- Zhu C, Yamamura S, Koiwa H, Nishihara M, Sandmann G (2002) cDNA cloning and expression of carotenogenic genes during flower development in Gentiana lutea. Plant Mol Biol 48: 277–285 [DOI] [PubMed] [Google Scholar]