Abstract
The vacuole is the main cellular storage pool, where sucrose (Suc) accumulates to high concentrations. While a limited number of vacuolar membrane proteins, such as V-type H+-ATPases and H+-pyrophosphatases, are well characterized, the majority of vacuolar transporters are still unidentified, among them the transporter(s) responsible for vacuolar Suc uptake and release. In search of novel tonoplast transporters, we used a proteomic approach, analyzing the tonoplast fraction of highly purified mesophyll vacuoles of the crop plant barley (Hordeum vulgare). We identified 101 proteins, including 88 vacuolar and putative vacuolar proteins. The Suc transporter (SUT) HvSUT2 was discovered among the 40 vacuolar proteins, which were previously not reported in Arabidopsis (Arabidopsis thaliana) vacuolar proteomic studies. To confirm the tonoplast localization of this Suc transporter, we constructed and expressed green fluorescent protein (GFP) fusion proteins with HvSUT2 and its closest Arabidopsis homolog, AtSUT4. Transient expression of HvSUT2-GFP and AtSUT4-GFP in Arabidopsis leaves and onion (Allium cepa) epidermal cells resulted in green fluorescence at the tonoplast, indicating that these Suc transporters are indeed located at the vacuolar membrane. Using a microcapillary, we selected mesophyll protoplasts from a leaf protoplast preparation and demonstrated unequivocally that, in contrast to the companion cell-specific AtSUC2, HvSUT2 and AtSUT4 are expressed in mesophyll protoplasts, suggesting that HvSUT2 and AtSUT4 are involved in transport and vacuolar storage of photosynthetically derived Suc.
In mature plant cells, the central vacuole occupies 80% to 90% of the cell volume. Vacuoles contain a large number of hydrolytic and biosynthetic enzymes, inorganic ions, soluble carbohydrates, organic acids, amino acids, secondary compounds, and modified xenobiotics (Maeshima, 2001; Martinoia et al., 2002). Based on the potential toxicity of many of these compounds, Matile (1984) suggested that the distance between life and death is 7.5 nm, the thickness of the vacuolar membrane. Plants have only a limited capacity to excrete potentially toxic compounds; therefore, the term internal excretion has also been used (Martinoia et al., 1993) to indicate that, for some classes of compounds, the vacuolar membrane mimics the function and contains homolog transporters of the liver plasma membrane (Kreuz et al., 1996). However, the function of the vacuole is not restricted to the storage of potentially toxic compounds. For optimal function of the metabolic pathways, the concentration of metabolites and ions has to be tightly regulated in the cytoplasm. Metabolites produced in excess are transported into the vacuole, which serves as a temporary storage pool, and released to the cytoplasm when required for metabolism.
Increasing evidence shows that impaired vacuolar deposition or retrieval affects plant metabolism. Catala et al. (2003) showed that the vacuolar calcium-proton exchanger CAX1 is induced during cold treatment and is involved in the regulation of genes responsible for cold tolerance. In the case of the vacuolar AtNRAMP3, Thomine et al. (2003) demonstrated that knockout plants exhibited an increased cadmium tolerance. It was postulated that AtNRAMP3 is an iron exporter and that under iron deficiency the plant is no longer able to mobilize vacuolar iron. The vacuolar malate carrier has recently been identified (Emmerlich et al., 2003). Cellular and vacuolar malate and fumarate contents were strongly reduced in deletion mutants of this transporter, despite the fact that a functional malate channel was still present in the tonoplast (Hurth et al., 2005). Knockout plants also exhibited an increased respiratory coefficient, indicating a shift of the substrates respired from carbohydrates in wild-type plants to mainly organic acids in the mutant. Recently, the Ca2+-dependent Ca2+-release channel TCP1 has been identified and functionally characterized. Deletion mutants lack functional slow channel activity, and are defective in abscisic acid-induced repression of germination and in the response of stomata to extracellular calcium (Peiter et al., 2005). These examples show that a detailed knowledge of vacuolar transporter function and regulation is required to better understand mechanisms regulating cytosolic homeostasis. However, despite some progress in identifying novel vacuolar transporters, our present knowledge of the intrinsic transporters in the tonoplast remains vague. A case in point is that to date one of the most important vacuolar transporters, the Suc transporter (SUT), has not been identified. In leaves, a large proportion of the Suc produced by photosynthesis is transported into the vacuole during the light period for storage. At night, Suc is released and loaded into the phloem for transport to the sink tissues (Kaiser and Heber, 1984; Martinoia et al., 1987). Inhibition of vacuolar Suc transport will increase cytoplasmic Suc concentrations and inhibit photosynthesis. Furthermore, many plants, such as barley (Hordeum vulgare) and wheat (Triticum aestivum), synthesize fructans in leaves (Pollock and Cairns, 1991). Fructan synthesis occurs within the vacuole and requires Suc (Wagner et al., 1983). Fructan accumulation allows such plants to store carbohydrates within the large central vacuole, additionally limiting the increase of osmolarity. Furthermore, fructans may play an important role as cold, drought, and salt stress protectants (Ritsema and Smeekens, 2003).
One possibility to identify new vacuolar transporters is by a proteomic approach. Thus far, vacuolar proteomic approaches have been described using Arabidopsis (Arabidopsis thaliana) vacuoles (Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004). Currently, no data are available for monocotyledonous cereal plants.
In this work, we present data on a vacuolar membrane proteomic approach using highly purified vacuolar membranes isolated from mesophyll cells of the crop plant barley. We identified 101 unique proteins; 45.5% of the detected barley vacuolar proteins have not been reported in the Arabidopsis vacuolar proteome. Among them is the Suc transporter HvSUT2, previously shown to catalyze Suc uptake when heterologously expressed in yeast (Saccharomyces cerevisiae; Weschke et al., 2000). HvSUT2, as well as its Arabidopsis homolog AtSUT4, were localized at the tonoplast as the first vacuolar Suc transporter using green fluorescent protein (GFP) fusion constructs.
RESULTS AND DISCUSSION
To understand the function of the vacuole and the interactions between the vacuole and the metabolic pathways occurring in the cytoplasm, it is important to identify vacuolar transporters that are involved in the export of solutes from the cytosol and the reimport in case of metabolically relevant compounds, such as sugars, organic acids, nitrate, or phosphate. To date, a range of transporters (Maeshima, 2001; Martinoia et al., 2002) have been identified, but, as can be deduced from vacuolar localization and transport studies, a large number remain uncharacterized. To identify novel tonoplast proteins, we isolated highly purified vacuolar membranes from barley mesophyll cells. Barley is an important crop plant closely related to wheat and rice (Oryza sativa). It contains only one type of mesophyll cells, whereas Arabidopsis has two types of mesophyll cells, spongy and palisade cells.
Purification of Barley Vacuoles
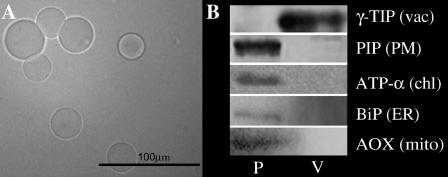
We could overcome the limiting factor of tonoplast proteomic projects, the requirement of highly purified vacuolar membranes in sufficient amounts, by improving a previously described method (Rentsch and Martinoia, 1991) for the isolation of vacuoles from barley leaves. Intact vacuoles were stained with neutral red and the purity was visually checked by bright-field microscopy. No obvious chloroplast or protoplast contaminations were observed (Fig. 1A). As described by Kaiser et al. (1986), the purity of the vacuole preparations was analyzed by the measurement of marker enzymes found in cytoplasm, mitochondria, endoplasmic reticulum (ER), and chloroplast (Table I). Contamination with these organelles was less than 1%, except for the ER (8%).
Figure 1.
Bright-field microscopy of purified vacuoles from barley mesophyll protoplasts (A). Western-blot analysis (B) with compartment-specific antibodies: TIP (dimer 45 kD), PIP (dimer 50 kD), ATP-α (55 kD), BiP (73 kD), and AOX (36 kD). Aliquots (5 μg) of vacuolar membrane proteins (V) and total membrane proteins of protoplasts (P) were loaded on each lane, respectively. vac, Vacuole; PM, plasma membrane; chl, chloroplast; ER, endoplasmic reticulum; mito, mitochondria.
Table I.
Activities of marker enzymes in the vacuolar preparations of barley mesophyll protoplasts
Marker enzyme activities for mitochondria (NAD-malic dehydrogenase), chloroplasts (NADP-gyceraldehydphosphate dehydrogenase, Glc-phosphate isomerase), ER (NADPH-cytochrome-c-reductase), and cytosol (Glc-phosphate isomerase, NAD-malic dehydrogenase) were measured in protoplast and vacuolar fractions. The percentage of contamination was calculated assuming an exclusive localization of α-Mannosidase in the vacuole (Martinoia et al., 1981).
| Marker Enzyme | Activities per 106 Protoplasts | Activities per 106 Vacuoles | % in Vacuole Preparations |
|---|---|---|---|
| nkat | nkat | ||
| α-Mannosidase | 0.15 | 0.147 | 98 |
| NADP-gyceraldehydphosphate dehydrogenase (chloroplast) | 5.7 | 0.009 | 0.16 |
| Glc-phosphate isomerase (cytosol, chloroplast) | 0.35 | 0.003 | 0.96 |
| NAD-malic dehydrogenase (mitochondria, chloroplast, cytosol) | 12.8 | 0.069 | 0.54 |
| NADPH-cytochrome-c-reductase (ER) | 0.32 | 0.27 | 8.4 |
The purity of the tonoplast fractions was further confirmed by western blots. The mitochondrial marker alternative oxidase (AOX), the α-subunit of the chloroplastic ATPase (ATP-α), the endoplasmic marker luminal binding protein (BiP), and the plasma membrane intrinsic protein (PIP) were only detectable in the total membrane protein fraction of protoplasts and not in the tonoplast fraction (Fig. 1B), providing a reliable indicator of the purity of the extraction. The γ-tonoplast intrinsic protein (TIP) was highly concentrated in the tonoplast fraction (Fig. 1B).
Analysis of Tonoplast Proteins by Liquid Chromatography-Tandem Mass Spectrometry
An untreated vacuolar membrane fraction as well as vacuolar membrane fractions washed with either 0.5 m NaOH or 0.3 m KI were analyzed by liquid chromatography-tandem mass spectrometry (MS/MS). Alkaline (NaOH) and saline (KI) treatment reduced the proportion of bound peripheral proteins from 53% in the untreated fraction to 38% in the KI-treated fraction and 34% in the NaOH-treated fraction. Cumulatively, we identified 101 proteins, 49 proteins with at least one transmembrane domain and 52 proteins with no transmembrane domain (Tables II–IV).
Table II.
Characterization of the identified vacuolar proteins
Accession numbers are from NCBI (http://www.ncbi.nlm.nih.gov/). Fractions are indicated by “x” in which each identified protein was present. The number of transmembrane domains (TMDs) is according to the prediction by SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html). MW, Predicted Mr; % Cov., sequence coverage in percent; % Seq., sequence similarity of the closest Arabidopsis homologs; Ref., Arabidopsis homolog(s) found by Carter et al. (2004; a), Shimaoka et al. (2004; b), Szponarski et al. (2004; c), and Sazuka et al. (2004; d).
| Accession No. | Description | Nontreated | KIa | NaOHb | TMDs | MW | pI | % Cov. | ppc | Arabidopsis Homologs | % Seq. | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Known vacuolar proteins | ||||||||||||
| gi|1051258 | V-ATPase A-subunit | x | x | x | 0 | 64 | 5.3 | 77.8 | 1 | At1g78900 | 90 | a, b, c, d |
| gi|167108 | V-ATPase B-subunit (isoform 1) | x | x | x | 0 | 54 | 5.1 | 84.5 | 1 | At1g76030 | 96 | a, b, c |
| At4g38510 | 95 | a, b | ||||||||||
| At1g20260 | 94 | a, b | ||||||||||
| gi|167110 | V-ATPase B-subunit (isoform 2) | x | x | x | 0 | 53 | 5.1 | 81.6 | 1 | At4g38510 | 97 | a, b |
| At1g76030 | 96 | a, b | ||||||||||
| At1g20260 | 95 | a, b | ||||||||||
| gi|12585487 | V-ATPase C-subunit | x | x | x | 0 | 40 | 6.0 | 78.0 | 1 | At1g12840 | 67 | a, b, c |
| gi|50929105 | V-ATPase D-subunit | x | x | x | 0 | 29 | 9.8 | 33.1 | 1 | At3g58730 | 76 | a, b, c |
| gi|4099148 | V-ATPase E-subunit | x | x | x | 0 | 26 | 6.5 | 92.5 | 1 | At4g11150 | 75 | a, b, c |
| At1g64200 | 74 | a, b | ||||||||||
| gi|50916028 | V-ATPase F-subunit | x | x | x | 0 | 14 | 5.6 | 16.9 | 1 | At4g02620 | 84 | a, b |
| gi|50928483 | V-ATPase G-subunit | x | 0 | 12 | 8.0 | 12.7 | 1 | At3g01390 | 72 | a, b | ||
| At4g23710 | 67 | a | ||||||||||
| gi|34898596 | V-ATPase H-subunit | x | x | x | 1 | 51 | 7.1 | 11 | 1 | At3g42050 | 71 | a, b, c |
| gi|18657017 | V-ATPase subunit-a | x | x | x | 7 | 89 | 5.6 | 7.2 | 1 | At2g21410 | 76 | a, b, d |
| At4g39080 | 76 | a, b, c, d | ||||||||||
| gi|52353608 | V-ATPAse subunit-c | x | x | x | 4 | 16 | 8.6 | 10.8 | 1 | At1g19910 | 96 | d |
| At2g16510 | 96 | d | ||||||||||
| At4g34720 | 96 | b, d | ||||||||||
| At4g38920 | 96 | a | ||||||||||
| At1g75630 | 95 | d | ||||||||||
| gi|20160977 | V-ATPAse subunit-d | x | x | 0 | 40 | 4.8 | 27.6 | 1 | At3g28715 | 90 | a, b | |
| At3g28710 | 89 | a, b | ||||||||||
| gi|11527561 | Pyrophosphatase VP1 | x | x | x | 12 | 80 | 5.0 | 27.5 | 1 | At1g15690 | 86 | a, b, c, d |
| gi|18274925 | Pyrophosphatase AVP3 | x | x | x | 12 | 79 | 5.2 | 30.6 | 1 | At1g15690 | 86 | a, b, c, d |
| gi|520936 | γ-TIP-like protein | x | x | x | 6 | 25 | 6.0 | 10.4 | 1 | At2g36830 | 74 | |
| At3g26520 | 74 | a | ||||||||||
| At4g01470 | 68 | |||||||||||
| gi|12278525 | Putative ABC transporter TAP2 | x | x | x | 5 | 69 | 8.5 | 9.3 | 1 | At5g39040 | 77 | a, b |
| gi|22022396 | ABC transporter TaMRP1 | x | 6 | 85 | 5.5 | 10.5 | 1 | At3g62700 | 74 | a, b | ||
| At2g47800 | 72 | a, b, c | ||||||||||
| gi|27263148 | Putative ABC transporter MRP2 | x | x | x | 14 | 183 | 6.3 | 11.8 | 1 | At2g34660 | 72 | b |
| At1g30400 | 70 | a, b, c, d | ||||||||||
| At1g30410 | 69 | |||||||||||
| gi|39545849 | Two-pore calcium channel | x | x | 12 | 85 | 5.4 | 10.4 | 1 | At4g03560 | 59 | a, c | |
| gi|28201131 | Na+/H+ antiporter HvNHX1 | x | 9 | 59 | 7.6 | 8.2 | 0.98 | At3g05030 | 75 | |||
| At5g27150 | 73 | |||||||||||
| gi|50911128 | Putative cytochrome b-561 | x | 6 | 27.6 | 6.6 | 9.1 | 0.97 | At4g25570 | 60 | a, b | ||
| Putative vacuolar transport proteins | ||||||||||||
| gi|7024413 | Suc transporter HvSUT2 | x | x | x | 11 | 54 | 8.9 | 7.4 | 1 | At1g09960 | 62 | |
| gi|26986186 | Hexose transporter HvSTP1 | x | 12 | 79 | 4.8 | 2.0 | 1 | At4g35300 | 71 | a | ||
| At1g20840 | 62 | a | ||||||||||
| gi|55773917 | Putative ABC transporter | x | 10 | 149 | 6.0 | 1.9 | 0.93 | At2g07680 | 59 | |||
| gi|34912536 | Putative ABC transporter (PDR-like) | x | 14 | 162 | 8.0 | 1.5 | 1 | At1g15520 | 68 | |||
| At2g36380 | 60 | |||||||||||
| At1g15210 | 58 | |||||||||||
| gi|34915038 | Putative ABC transporter | x | x | 9 | 162 | 10.7 | 1.7 | 1 | At2g47800 | 50 | a, b, c | |
| At3g62700 | 49 | a, b | ||||||||||
| gi|34898286 | Canalicular multispecific organic anion transporter 2-like protein | x | 15 | 164 | 7.8 | 1.7 | 0.91 | At3g59140 | 65 | |||
| gi|15076661 | Putative peptide transporter | x | x | x | 11 | 65 | 5.1 | 11.3 | 1 | At2g02040 | 69 | a, b |
| At2g02020 | 68 | |||||||||||
| At3g54140 | 62 | |||||||||||
| gi|34015349 | Putative chloride channel (CLC family) | x | 8 | 86 | 8.8 | 3.0 | 0.97 | At5g33280 | 65 | |||
| At5g49890 | 59 | |||||||||||
| gi|47497044 | Amino acid-like transporter | x | 8 | 31 | 9.1 | 4.5 | 0.98 | At2g41190 | 66 | |||
| gi|49388943 | Putative hexose transporter | x | 11 | 80 | 5.2 | 2.1 | 1 | At4g35300 | 72 | a | ||
| gi|51854311 | Putative sugar transporter | x | x | x | 10 | 53 | 9.1 | 4.4 | 1 | At1g75220 | 73 | |
| At1g19450 | 71 | a | ||||||||||
| gi|31433313 | Putative sugar transporter | x | 12 | 79 | 4.9 | 2.2 | 0.96 | At4g35300 | 72 | a | ||
| gi|14275750 | Putative calcium translocating P-type ATPase | x | x | x | 10 | 63 | 4,9 | 8,3 | 1 | At3g57330 | 68 | a, b, c |
| At2g41560 | 66 | a | ||||||||||
| At4g37640 | 64 | |||||||||||
| Putative vacuolar membrane proteins | ||||||||||||
| gi|50726573 | Putative protein kinase Xa21 | x | 2 | 124 | 5.9 | 3.3 | 0.93 | At3g47110 | 41 | |||
| At3g47570 | 41 | |||||||||||
| At3g47090 | 39 | |||||||||||
| gi|8886326 | Vacuolar targeting receptor bp-80 | x | 2 | 69 | 5.4 | 4.8 | 0.97 | At3g52850 | 70 | |||
| At2g14740 | 68 | a | ||||||||||
| At2g14720 | 68 | |||||||||||
| gi|50943395 | Putative syntaxin of plants 52 | x | 1 | 26 | 8.6 | 10.3 | 1 | At1g79590 | 63 | a | ||
| At1g16240 | 62 | a | ||||||||||
| gi|37651973 | Blue copper-binding protein | x | x | 2 | 17 | 5.5 | 13 | 0.99 | At3g27200 | 40 | ||
| At2g32300 | 37 | |||||||||||
| At2g44790 | 36 | |||||||||||
| gi|34913062 | Putative calcium sensor protein | x | 1 | 31 | 4.9 | 2.9 | 1 | At4g33000 | 77 | |||
| gi|41529149 | Putative acid phosphatases | x | x | x | 1 | 29 | 8.6 | 18.4 | 1 | At4g29270 | 45 | |
| At4g29260 | 44 | a | ||||||||||
| At4g25150 | 41 | |||||||||||
| gi|41019551 | Putative Cys protease | x | 1 | 41 | 5.8 | 5.0 | 0.99 | At4g39090 | 67 | |||
| At2g21430 | 64 | a | ||||||||||
| At4g16190 | 62 | |||||||||||
| gi|34904294 | Putative bark storage protein | x | x | 1 | 36 | 5.6 | 5.7 | 1 | At4g24340 | 38 | ||
| gi|53792725 | EREBP-4-like protein | x | x | 1 | 23 | 5.3 | 12.5 | 1 | At1g47740 | 62 | ||
| At4g17486 | 53 | |||||||||||
| At5g47310 | 54 | |||||||||||
| gi|544242 | Endoplasmin homolog precursor | x | 1 | 92 | 4.8 | 6.7 | 1 | At4g24190 | 77 | a | ||
| gi|7288255 | RNase S-like protein precursor | x | x | x | 1 | 27 | 8.8 | 52 | 1 | At1g14220 | 36 | a |
| At2g02990 | 34 | |||||||||||
| At1g26820 | 34 | |||||||||||
| gi|23345048 | Hypersensitive-induced reaction protein 4 | x | 1 | 32 | 5.2 | 27.8 | 1 | At5g51570 | 76 | a, b | ||
| gi|50252990 | SPX domain-containing-like protein | x | x | 10 | 78 | 6.0 | 2.4 | 0.99 | At1g63010 | 68 | ||
| At4g22990 | 68 | |||||||||||
| At4g11810 | 67 | |||||||||||
| gi|50726593 | Putative nodulin-like protein | x | x | 10 | 58 | 8.3 | 9.2 | 1 | At4g34950 | 75 | ||
| At2g16660 | 65 | |||||||||||
| gi|51854288 | Putative integral membrane protein | x | x | x | 10 | 54 | 8.6 | 2.0 | 0.99 | At1g75220 | 71 | |
| At1g19450 | 70 | a | ||||||||||
| gi|51536186 | Translocation related protein | x | 4 | 42 | 9.1 | 3.8 | 0.98 | At3g20920 | 59 | |||
| gi|50948653 | Type 1 membrane-like protein | x | x | 3 | 12 | 4.8 | 8.2 | 1 | At3g24160 | 41 | ||
| gi|50929895 | Unknown protein | x | 2 | 113 | 9.3 | 1.8 | 0.91 | At1g22610 | 58 | |||
| At4g11610 | 52 | |||||||||||
| gi|55771327 | Unknown protein | x | 5 | 152 | 6.0 | 1.7 | 1 | At5g11700 | 66 | a, b | ||
| gi|50926173 | Unknown protein | x | 2 | 47 | 5.2 | 5.4 | 1 | At1g16010 | 78 | a | ||
| At1g80900 | 74 | |||||||||||
| gi|50399936 | Expressed protein | x | 1 | 36 | 6.0 | 4.5 | 1 | At5g06300 | 31 | |||
| At3g53450 | 31 | |||||||||||
| Putative vacuolar membrane-associated proteins | ||||||||||||
| Stress response | ||||||||||||
| gi|53791853 | Putative propyzamide-hypersensitive 1 | x | 0 | 97 | 5.77 | 4.1 | 0.91 | At5g23720 | 59 | |||
| gi|5822249 | Zymogen of a barley vacuolar aspartic proteinase | x | 0 | 51 | 5.2 | 5.4 | 1 | At1g11910 | 72 | a, b | ||
| At1g62290 | 68 | a, b | ||||||||||
| At4g04460 | 64 | |||||||||||
| gi|23345042 | Hypersensitive-induced reaction protein 1 | x | x | 0 | 31 | 5.3 | 45.8 | 1 | At5g62740 | 87 | a, b | |
| At1g69840 | 85 | b | ||||||||||
| gi|18650668 | Temperature stress-induced lipocalin | x | 0 | 22 | 5.5 | 5.3 | 0.99 | At5g58070 | 74 | d | ||
| gi|50912867 | Putative elicitor-inducible protein EIG-J7 | x | 0 | 20 | 6.1 | 4.7 | 0.98 | At4g39730 | 55 | a | ||
| At2g22170 | 52 | a | ||||||||||
| gi|28555904 | NBS-LRR disease-resistance protein homolog | x | 0 | 98 | 7.0 | 2.9 | 0.94 | At5g43470 | 31 | |||
| At1g50180 | 30 | |||||||||||
| At5g48620 | 30 | |||||||||||
| gi|124037 | Bowman-Birk-type trypsin inhibitor | x | 0 | 14 | 8.77 | 10.5 | 0.99 | At5g43060 | 31 | b | ||
| At1g32190 | 30 | |||||||||||
| At1g74670 | 30 | |||||||||||
| gi|50911973 | Ethylene-responsive-like protein | x | 0 | 20 | 5.9 | 6.2 | 0.98 | At5g14680 | 73 | |||
| At3g01520 | 72 | |||||||||||
| Membrane fusion and remodeling | ||||||||||||
| gi|32308080 | α-SNAP | x | x | 0 | 12 | 4.3 | 87.5 | 1 | At3g56190 | 63 | a, b | |
| At3g56450 | 54 | |||||||||||
| gi|47834379 | Cycloartenol synthase | x | x | x | 0 | 86 | 6.1 | 1.8 | 1 | At2g07050 | 74 | a, b |
| gi|50912455 | Putative ADP-ribosylation factor | x | x | 0 | 20 | 8.6 | 12.5 | 1 | At5g37680 | 90 | ||
| At5g67560 | 80 | a | ||||||||||
| gi|50943481 | Putative α-soluble NSF attachment protein | x | 0 | 32 | 5.0 | 20.4 | 1 | At3g56190 | 63 | a, b | ||
| gi|52077324 | Putative GTP-binding protein | x | 0 | 26 | 5.2 | 3.8 | 0.93 | At1g01200 | 65 | |||
| Other | ||||||||||||
| gi|51035342 | Putative carotenoid cleavage dioxygenase 1 | x | x | 0 | 7 | 4.3 | 28.4 | 0.99 | At3g63520 | 77 | a | |
| gi|1167957 | O-Methyltransferase | x | 0 | 30 | 6.4 | 6.6 | 0.99 | At5g54160 | 44 | |||
| At1g33030 | 39 | |||||||||||
| At1g51990 | 38 | |||||||||||
| gi|50900950 | Putative gag-pol precursor | x | 0 | 206 | 8.9 | 3.0 | 1 | |||||
| gi|34901992 | Putative sulfolipid synthase | x | 0 | 46 | 9.0 | 4.6 | 1 | At5g01220 | 71 | |||
| gi|37718792 | Calcineurin B protein | x | 0 | 26 | 4.7 | 12.9 | 1 | At4g26570 | 88 | a | ||
| At5g55990 | 88 | a | ||||||||||
| gi|34904770 | Putative esterase | x | 0 | 38 | 6.0 | 2.5 | 0.99 | At5g23530 | 47 | |||
| At5g27320 | 42 | |||||||||||
| At3g05120 | 39 | |||||||||||
| gi|11360993 | Gly dehydrogenase | x | 0 | 111 | 6.3 | 1.66 | 0.99 | At4g33010 | 81 | |||
| At2g26080 | 80 | |||||||||||
| gi|21954110 | RNase S-like protein | x | 0 | 28 | 6.5 | 21.2 | 1 | At1g26820 | 39 | |||
| At2g02990 | 38 | a | ||||||||||
| At1g14220 | 38 | |||||||||||
| gi|7576892 | Fatty acid α-oxidase | x | 0 | 71 | 7.7 | 5.2 | 0.96 | At3g01420 | 62 | |||
| At1g73680 | 58 | |||||||||||
| gi|51592190 | Nucleotide pyrophosphatase/phosphodiesterase | x | 0 | 39.9 | 5.5 | 3.4 | 0.92 | At4g24890 | 73 | |||
| At5g50400 | 73 | |||||||||||
| At1g13750 | 66 | |||||||||||
| gi|21742152 | Unknown protein | x | 0 | 35 | 9.3 | 11.0 | 1 | At4g13010 | 64 | a | ||
| gi|38345613 | Unknown protein | x | 0 | 33 | 5.0 | 4.4 | 1 | At4g20410 | 67 | |||
| gi|55771325 | Unknown protein | x | 0 | 47 | 5.77 | 1.8 | 0.99 | At5g12010 | 59 | |||
| At4g29780 | 55 | |||||||||||
| gi|42408418 | Hypothetical protein | x | 0 | 16 | 11.5 | 10.8 | 0.99 | |||||
| gi|49389235 | Hypothetical protein | x | 0 | 22 | 11.6 | 13.7 | 0.98 | |||||
| gi|47847977 | Hypothetical protein | x | 0 | 14 | 5.0 | 16.5 | 0.97 | |||||
| gi|50928857 | Unknown protein | x | 0 | 78 | 6.8 | 6.3 | 0.95 | At3g11590 | 44 | |||
| At3g20350 | 42 | |||||||||||
| At1g50660 | 39 | |||||||||||
| gi|27497206 | Unknown protein | x | 0 | 66 | 5.6 | 1.0 | 0.94 | At2g20370 | 67 | |||
| gi|52353607 | Unknown protein | x | 0 | 45 | 8.4 | 4.2 | 1 | |||||
| gi|37533500 | Hypothetical protein | x | 0 | 71 | 6.0 | 3.7 | 0.92 | |||||
Tonoplast fraction washed with 0.3 m KI.
Tonoplast fraction washed with 0.5 m NaOH.
Protein-Prophet scores (pp) above 0.9 were accepted; at this cutoff, the rate of false positive protein identifications is less than 10%.
Table IV.
Soluble known nonvacuolar proteins
Accession numbers are from NCBI. Fractions are indicated by “x” in which each identified protein was present. % Seq., Sequence similarity of the closest Arabidopsis homologs; Ref., indicates Arabidopsis homolog(s) found by Carter et al. (2004; a), Shimaoka et al. (2004; b), Szponarski et al. (2004; c), and Sazuka et al. (2004; d).
| Accession No. | Description | Nontreated | KIa | NaOHb | Arabidopsis Homologs | % Seq. | Ref. |
|---|---|---|---|---|---|---|---|
| gi|1224079 | Rubisco | x | x | rbcL | 91 | ||
| gi|57471706 | Ribosomal protein L13a | x | At5g48760 | 83 | |||
| At4g13170 | 81 | ||||||
| At3g24830 | 81 | ||||||
| gi|729003 | Carbonic anhydrase precursor | x | At5g14740 | 59 | |||
| At1g70410 | 56 | ||||||
| At1g23730 | 54 | ||||||
| gi|41052969 | Putative 60S ribosomal protein L12 | x | At2g37190 | 90 | a | ||
| At3g53430 | 89 | ||||||
| At5g60670 | 87 | ||||||
| gi|1785948 | Cytosolic triosephosphate isomerase | x | At3g55440 | 78 | a | ||
| gi|50917247 | Putative translation elongation factor | x | At3g22980 | 57 | |||
| gi|493591 | Disulfide isomerase | x | At1g21750 | 57 | a | ||
| At1g77510 | 56 | ||||||
| gi|50913205 | Constitutive photomorphogenesis 1 (COP1) | x | At2g32950 | 70 | |||
| gi|7452979 | Allene oxide synthase | x | x | x | At5g42650 | 53 | a |
Tonoplast fraction washed with 0.3 m KI.
Tonoplast fraction washed with 0.5 m NaOH.
Table III.
Known nonvacuolar membrane proteins
Accession numbers are from NCBI. The number of transmembrane domains (TMDs) is according to the prediction by SOSUI. % Seq., Sequence similarity of the closest Arabidopsis homologs; Ref., Arabidopsis homolog(s) found by Carter et al. (2004; a), Shimaoka et al. (2004; b), Szponarski et al. (2004; c), and Sazuka et al. (2004; d).
| Accession No. | Description | TMDs | Arabidopsis Homologs | % Seq. | Ref. |
|---|---|---|---|---|---|
| gi|7649310 | Aquaporin 1 | 4 | At2g37170 | 82 | d |
| At3g53420 | 82 | a, d | |||
| At5g60660 | 80 | ||||
| gi|439586 | Calreticulin | 1 | At1g56340 | 72 | |
| At1g09210 | 72 | a | |||
| gi|19093 | PSI polypeptide | 1 | At1g52230 | 69 | a |
| At3g16140 | 66 | ||||
| gi|37534052 | Putative cell elongation protein DIMINUTO | 1 | At3g19820 | 80 | a, b |
Known Vacuolar Membrane Proteins
All eight subunits of the V1 sector (A–H) and three subunits of the V0 sector (a, c, and d) of the well-characterized V-type H+-ATPase were detected, as well as two isoforms of the V-type H+-pyrophosphatase, indicating the high coverage of vacuolar membrane proteins. Only one aquaporin, a γ-TIP-like protein, was identified. We additionally identified the vacuolar membrane proteins HvTAP2 (Yamaguchi et al., 2002) and TaMRP1 (Theodoulou et al., 2003), homologs of the TCP1 (Peiter et al., 2005), the sodium/proton antiporter NHX (Xia et al., 2002), and the ATP-binding cassette (ABC) transporter MRP2 (Liu et al., 2001), which have been previously shown to reside in the tonoplast (Table II).
We identified 11 of 12 subunits of the V-type H+-ATPase (Sze et al., 2002). In comparison with published Arabidopsis vacuolar proteomic projects, only Carter et al. (2004) and Shimaoka et al. (2004) obtained such a complete set of subunits for the V-type H+-ATPase. Despite this, our proteomic analysis resulted in a limited number of known vacuolar proteins. This discrepancy may be explained by the absence of some known vacuolar transporters in barley mesophyll cells, or barley proteins are not similar enough to permit an unequivocal attribution of peptides during the database searches.
Soluble Proteins and Nonvacuolar Proteins
In our investigation of the tonoplast, we found 52 proteins with no transmembrane domain. Most of these putative membrane-associated proteins are involved in stress response or in membrane fusion and remodeling (Table II).
We identified a few known nonvacuolar proteins, including four membrane proteins (Table III) and nine soluble proteins (Table IV). Soluble proteins can associate with membranes, which might explain their detection in the tonoplast preparation. The view that these proteins were peripherally associated with the membrane is supported by the fact that the proportion of soluble known nonvacuolar proteins was the highest in the nontreated vacuolar membrane fraction (8.8%). However, since the vacuole is a lytic organelle, a proportion of these apparent contaminants may be a result of the ongoing degradation processes. Interestingly, only one plasma membrane protein, aquaporin 1, was detected, demonstrating the high purity of our preparations. Most of the known nonvacuolar proteins (61.5%) were previously reported in one of the Arabidopsis vacuolar proteomic studies (Tables III and IV).
Novel Vacuolar Membrane Proteins
Besides well-known vacuolar membrane proteins, a large number of annotated membrane proteins with an unknown subcellular localization were identified, including several sugar transporters (gi|7024413, gi|26986186, gi|49388943, gi|51854311, and gi|31433313) and ABC proteins (gi|55773917, gi|34912536, and gi|34915038), an amino acid transporter (gi|47497044), a peptide transporter (gi|15076661), an organic anion transporter (gi|34898286), a putative calcium translocating P-type ATPase (gi|14275750), and a putative chloride channel (gi|34015349) of the CLC family (Nakamura et al., 2006). Moreover, there were also nine proteins with at least one transmembrane helix for which a function has yet not been elucidated (Table II).
The closest Arabidopsis homolog of the identified peptide transporter (gi|15076661) is AtPTR2-B (At2g02040), which transports dipeptides and tripeptides and is constitutively expressed in all plant organs (Song et al., 1996). AtPTR2-B was also found by Carter et al. (2004) and Shimaoka et al. (2004) during their Arabidopsis vacuolar proteome analysis. AtPTR1, another peptide transporter of the gene family, has been localized in the plasma membrane (Dietrich et al., 2004). These results indicate that in a multigene family, different members may not be localized at the same membrane. This has also been demonstrated in studies on the subfamily of the MRP-type ABC transporters (Geisler et al., 2004; Klein et al., 2004).
One of the identified sugar transporters (gi|51854311) is a close homolog of a hexose transporter (U43629) in sugar beet (Beta vulgaris), which has previously been postulated to catalyze facilitated diffusion of Glc across the vacuolar membrane of sugar beet (Chiou and Bush, 1996). Facilitated diffusion of Glc has also been reported to occur in barley vacuoles (Martinoia et al., 1987). Therefore, it is highly likely that this hexose transporter is indeed localized at the vacuolar membrane.
Interestingly, an additional sugar transporter identified in our proteomic approach was HvSUT2. HvSUT2 was previously expressed in yeast and shown to catalyze Suc uptake in intact yeast cells (Weschke et al., 2000). The facilitated diffusion of Suc across the vacuolar membrane of barley has already been demonstrated in earlier studies (Martinoia et al., 1987). Although Suc accumulation is one of the key functions of mesophyll vacuoles, the vacuolar transporters responsible for Suc uptake and release are as yet unidentified.
A Comparison of the Arabidopsis and Barley Tonoplast Proteome
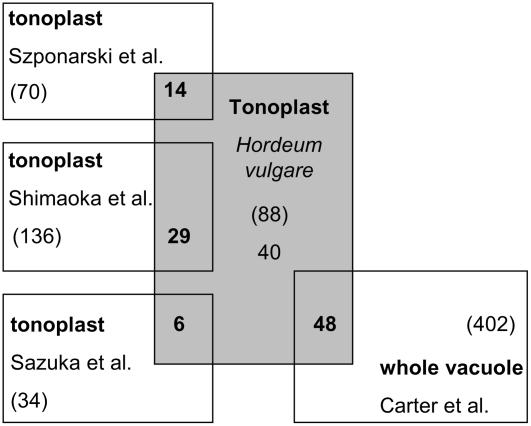
The vacuole possesses specialized functions in different cell types and in different plant species. For example, in Arabidopsis, vacuoles accumulate mainly monosaccharides, whereas in barley, fructans may be synthesized and stored in vacuoles. To identify proteins that are particular for the vacuolar function in barley, we compared the vacuolar proteins found in barley to proteins identified in the four Arabidopsis vacuolar proteomic approaches (Carter et al., 2004; Sazuka et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004). We searched the 88 identified vacuolar proteins for the closest homologs in the Arabidopsis genome. If a similarity of 30% is used as a cutoff, six proteins, including the putative gag-pol precursor and five hypothetical proteins, have no homolog in Arabidopsis (Table II).
Although 34 to 402 vacuolar proteins were detected in the four Arabidopsis proteomic studies, only 48 (54.5%) identified barley vacuolar proteins have also been identified in at least one of the Arabidopsis studies (Table II; Fig. 2). The highest overlap was found with Carter et al. (2004; 48 proteins) and Shimaoka et al. (2004; 29 proteins). However, 40 proteins (45.5%) of 88 detected barley vacuolar proteins were not reported in the Arabidopsis vacuolar proteomic studies, suggesting a higher relative abundance of these proteins in barley leaf mesophyll vacuoles compared to Arabidopsis leaf mesophyll and cell culture vacuoles. Eighteen (43%) of these 40 proteins are membrane proteins, including three ABC transporters (gi|55773917, gi|34912536, and gi|34898286), one amino acid transporter (gi|47497044), one putative CLC-type chloride channel (gi|34015349), six membrane proteins with an unknown function (gi|50252990, gi|50726593, gi|51536186, gi|50948653, gi|50929895, and gi|50399936), and the Suc transporter HvSUT2 (gi|7024413). The detection of HvSUT2 in the vacuolar membrane fraction indicates a high expression of this Suc transporter in barley mesophyll cells. This can be explained by the fact that barley mesophyll cells accumulate large amounts of Suc during the day (Kaiser and Heber, 1984) and that fructan synthesis in barley mesophyll vacuoles requires Suc.
Figure 2.
Comparison between the barley and Arabidopsis vacuolar membrane proteome. Barley tonoplast proteins were BLAST-searched for homologous proteins that were reported in the Arabidopsis tonoplast proteome. Proteins with a similarity of at least 30% were accepted as significant homologs. The number in brackets indicates the total number of identified proteins in each study and the bold numbers represent the number of overlapping proteins. Forty of the detected proteins were only identified in barley, suggesting a significant function at the vacuolar membrane for monocotyledon cereals.
Subcellular Localization of HvSUT2 and AtSUT4
The small number of nonvacuolar proteins that we detected in comparison to the published proteomic approaches using Arabidopsis vacuoles reveals that the purity of our barley tonoplast fractions is clearly higher. However, since the protein content of the vacuolar membrane constitutes less than 1% of the total cellular protein, small contaminations in the range of 2% to 4% have a strong impact on the detected proteins. It should also be taken into account that the plasma membrane contains about double the amount of protein compared to the vacuolar membrane. Furthermore, in mesophyll cells, the surface area of chloroplastic membranes by far exceeds that of the vacuole. Consequently, a contamination of 2% with chloroplasts would result in about 20% plastid proteins in the vacuolar fraction. GFP localization of the newly identified membrane proteins is therefore a prerequisite to confirm their putative localization obtained with a proteomic approach. This effort has not been undertaken in the vacuolar proteomic reports published so far.
Among the potentially novel vacuolar transporters, we were particularly interested in HvSUT2, since the vacuolar Suc transporter, which plays a central role in plant metabolism, still awaits identification. As aforementioned, HvSUT2 exhibited Suc transport activity over the plasma membrane when expressed in yeast (Weschke et al., 2000). However, heterologous expression in yeast does not necessarily provide information about the intracellular localization of the protein in planta.
The high purity of our vacuole preparation encouraged us to test whether HvSUT2 and its Arabidopsis counterpart, AtSUT4, are vacuolar Suc transporters, despite the fact that the closest homologs of HvSUT2 found in tomato (Lycopersicon esculentum; LeSUT4) and potato (Solanum tuberosum; StSUT4) have been immunolocalized in the plasma membrane of sieve elements (Weise et al., 2000).
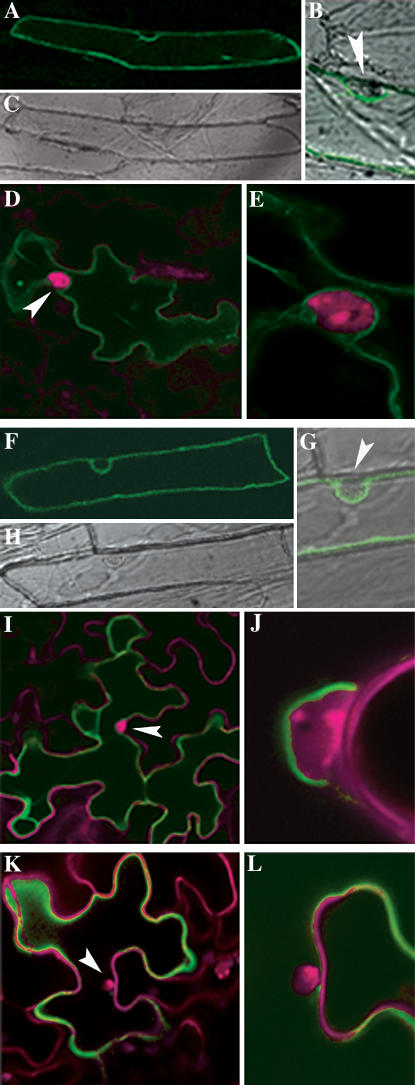
To confirm the tonoplast localization of HvSUT2 and AtSUT4, GFP fusion proteins were transiently expressed in Arabidopsis leaves and onion (Allium cepa) epidermal cells. For both transporters, fluorescence was detected at the vacuolar membrane of Arabidopsis and onion epidermal cells (Fig. 3, A–J). As a control, we cloned AtSUC2, a well-known plasma membrane Suc transporter of the companion cells (Stadler and Sauer, 1996), as a GFP fusion protein. Based on fluorescence intensity, expression of AtSUC2-GFP was similar to AtSUT4-GFP and HvSUT2-GFP. However, unlike AtSUT4 and HvSUT2, AtSUC2 localizes to the plasma membrane (Fig. 3, K and L).
Figure 3.
Subcellular localization of HvSUT2, AtSUT4, and AtSUC2 by transient expression of GFP fusion proteins in onion epidermal cells and Arabidopsis epidermal cells. Cell walls and nuclei of Arabidopsis were stained in red with propidium iodide. Nuclei are indicated by arrowheads. A to E, Tonoplast localization of HvSUT2 in onion epidermal cells (A–C) and Arabidopsis epidermal cells (D and E). Fluorescence of the HvSUT2-GFP fusion protein in onion epidermal cells (A) and transmission picture (C) of the same cell. The tonoplast is located in the internal side of the cytoplasm including the nucleus as shown in the overlay of the detail pictures (B and E). F to J, Localization of the AtSUT4-GFP fusion protein at the tonoplast, transiently expressed in onion epidermal cells (F–H) and Arabidopsis epidermal cells (I and J). As shown in detail pictures (G and J), the green fluorescent tonoplast surrounds the nucleus. K and L, Plasma membrane localization of AtSUC2-GFP in Arabidopsis epidermal cells.
In contrast to this localization study, LeSUT4 and StSUT4, the closest homologs of HvSUT2 and AtSUT4, were immunolocalized in the plasma membrane of enucleate sieve elements, a cell type that does not contain vacuoles. Thus far, SUT4 of Arabidopsis has not been localized at the subcellular level. However, promoter-β-glucuronidase studies showed high expression of AtSUT4 in companion cells (Schulze et al., 2003). High expression in phloem companion cells does not exclude vacuolar localization since these cells contain many small vacuoles. These vacuoles may play a role in temporary storage of Suc that can be used at night as an energy reserve in these energy-demanding cells. The results of our localization study pose the question of whether orthologs might be differently localized or whether the unusual cell structure of sieve elements, cells without nuclei, vacuoles, ribosomes, and Golgi, might entail another subcellular localization of the same protein.
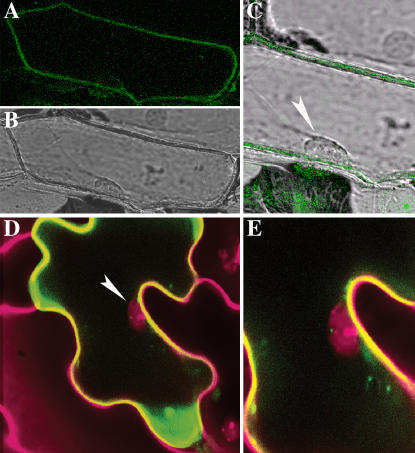
Shimaoka et al. (2004) detected in their Arabidopsis tonoplast proteomic analysis the Suc transporter AtSUC1, which is strongly expressed in Arabidopsis flowers (Stadler et al., 1999). AtSUC1 was also identified in the plasma membrane proteomic approach of Alexandersson et al. (2004). To investigate if AtSUC1 is localized like AtSUT4 at the vacuolar membrane, we expressed AtSUC1-GFP in Arabidopsis leaves and onion epidermal cells. As shown in Figure 4, AtSUC1 is targeted to the plasma membrane as AtSUC2. This observation stresses on the necessity to confirm the localization of novel candidates obtained in proteomic approaches by GFP-fusion proteins.
Figure 4.
Subcellular localization of AtSUC1 by transient expression of GFP fusion proteins in onion epidermal and Arabidopsis leaf cells. AtSUC1-GFP localizes at the plasma membrane of onion epidermal cells (A–C) and Arabidopsis epidermal cells (D and E). Nuclei are indicated by arrows in overlay pictures (C and D). Cell walls and nuclei of Arabidopsis were stained in red with propidium iodide.
Expression of AtSUT4 and HvSUT2 in Mesophyll Cells
AtSUT4 promoter-β-glucuronidase fusion plants exhibited a strong expression in companion cells (Schulze et al., 2003). To investigate whether AtSUT4 is expressed exclusively in companion cells or if transcripts are also present in mesophyll cells, where photosynthetic Suc is stored inside the vacuoles, we performed a reverse transcription (RT)-PCR analysis with isolated leaf mesophyll protoplasts.
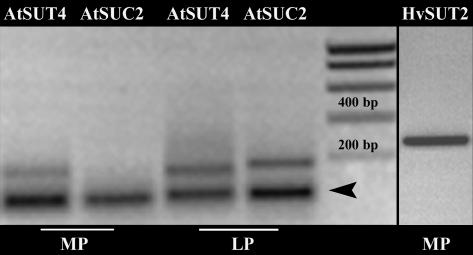
Thirty to 40 mesophyll protoplasts of source leaves were discriminated from smaller companion cell protoplasts, chloroplast-free sieve element protoplasts, and large epidermal protoplasts by bright-field microscopy and collected in a microcapillary. RT-PCR was conducted with primers specific for the amplification of HvSUT2 and AtSUT4. AtSUC2 was used as a marker gene for companion cell contaminations. As illustrated in Figure 5, only AtSUT4 transcripts were detected in the mesophyll protoplast preparation. While in a typical leaf protoplast mixture, both AtSUC2 and AtSUT4 transcripts were present. HvSUT2 transcripts were detected in mesophyll cells (Fig. 5). These results prove that HvSUT2 and AtSUT4 are vacuolar Suc transporters of leaf mesophyll cells.
Figure 5.
Expression analysis of AtSUT4 and HvSUT2 in mesophyll protoplasts. RT-PCR products of AtSUT4 (180 bp) and HvSUT2 (270 bp) are visible in mesophyll protoplast preparations (MP). AtSUC2 RT-PCR products (190 bp), a marker gene for companion cell contaminations, are only detected in the typical leaf protoplast mixture (LP). Actin RT-PCR products (100 bp) are indicated by an arrow.
CONCLUSION
Our results demonstrate that the combination of a high throughput technique, the tonoplast proteomic approach, and subsequent localization by GFP is a powerful approach to identify transporters awaiting identification for a long time. We localized HvSUT2 and the Arabidopsis homolog AtSUT4 as the first Suc transporter at the tonoplast and showed expression of these Suc transporters in leaf mesophyll cells. The results suggest that AtSUT4 and HvSUT2 are involved in the exchange of Suc between vacuole and cytoplasm.
MATERIALS AND METHODS
Plant Material
Barley (Hordeum vulgare) var. Baraka was grown in soil in a controlled environment chamber (16 h light/8 h dark, 300 μE m−2 s−1, 22°C, 60% relative humidity), and Arabidopsis (Arabidopsis thaliana) ecotype Columbia was cultivated in soil in a controlled environment chamber (8 h light/16 h dark, 150 μE m−2 s−1, 18°C, 60% relative humidity).
Tonoplast Isolation
Barley mesophyll vacuoles were isolated from 8-d-old plants according to Rentsch and Martinoia (1991), omitting bovine serum albumin in the isolation medium. Tonoplast vesicles were isolated by sonication of vacuoles and subsequent ultracentrifugation for 1 h at 100,000g. The supernatant was removed and the tonoplast fraction was resuspended in 20 mm HEPES-KOH, pH 7.2, at a concentration of approximately 200 μg protein/mL. The vacuolar membranes were either analyzed directly or treated by washing with 0.3 m KI or 0.5 m NaOH. Proteins were quantified according to Bradford (1976) using the Bio-Rad protein assay (Reinach).
Tonoplast Proteome Analysis and Interpretation of MS Data
Tonoplast proteins were separated using SDS-PAGE (Laemmli, 1970) with 10% acrylamide gels. The SDS gels were cut into 35 sections, and gel slices were immediately subjected to in-gel tryptic digestion (Shevchenko et al., 1996). Tryptic peptides were further fractionated by reverse-phase chromatography coupled online to an LCQ Deca XP ion trap mass spectrometer (Thermo Finnigan). The peptides were analyzed by MS full scan and MS/MS scans of the three most intense parent ions.
MS/MS data sets were interpreted according to the standards of Carr et al. (2004). The SEQUEST software (Thermo Finnigan) was used to search the National Center for Biotechnology Information (NCBI) protein database of the Liliopsida (GI4447) including 139,384 proteins. dta files were created by the SEQUEST software for every MS/MS scan with a total ion count of at least 5 × 104, minimal peak count of 35, and a precursor ion mass in the range of 300 to 2,000 mass-to-charge ratio. Data were searched against the database restricted to tryptic peptides without modifications (except for carboxyamidomethylated Cys 57.0513 and oxidized Met 15.9994), allowing a parent mass error tolerance of 2 D and daughter ion error tolerance of 0.8 D. All SEQUEST data were analyzed by Peptide and Protein-Prophet (Nesvizhskii et al., 2003). Protein-Prophet is based on a statistical model that allows one to assess the reliability of protein identifications on the basis of MS/MS data. The Protein-Prophet score gives a probability estimate for the protein identification. We accepted scores above 0.9; at this cutoff, the rate of false positive protein identifications is less than 10% (Table II). Mostly, we identified the same protein from different biochemical fractions, further compounding the reliability of the identification (Table II).
For the identification of Arabidopsis homologs, we used the BLAST search of the Munich Information Center for Protein Sequences (MIPS) database (http://mips.gsf.de/proj/thal/db/search/search_frame.html). In Tables II, III, and IV, the closest Arabidopsis homologs are listed as well as the second and third homologs if the similarity differed not more than 10% from the closest homolog.
Western Blotting
Western blotting was carried out using antibodies for γ-TIP (Conceicao et al., 1997), ATP-α, luminal BiP (member of the heat shock family), AOX (Elthon et al., 1989), and PIP (Frangne et al., 2001). Secondary antibodies (anti-rabbit or anti-chicken, coupled to alkaline phosphatase [AP] or horse radish peroxidase [HRP]; Promega) were diluted 1:3,000 (AP) or 1:25,000 (HRP) in TBST (0.2 m Tris-HCl, pH 7.5, 0.5 m NaCl, 0.05% Tween 20). AP detection mixture was prepared by adding 6.6 μL of nitroblue tetrazolium (Promega) and 3.3 μL of 5-bromo-4-chloro-3-indolyl phosphate (Promega) to 1 mL of AP buffer (0.1 m Tris-HCl, pH 9.5, 0.5 m MgCl2). The activity of HRP was detected using a Chemiluminescence Blotting Substrate kit (SuperSignal Chemiluminescent Working Solution; Pierce) according to manufacturer's instructions.
Localization Study
To localize HvSUT2 (gi|7024412), AtSUC1 (At1g71880), and AtSUC2 (At1g22710), the respective cDNAs were cloned in frame to the N terminus of GFP into the vector pGFP2 (Haseloff and Amos, 1995). The AtSUT4-GFP fusion construct was cloned into the expression vector pART7 (Gleave, 1992). GFP fusion constructs were transiently expressed in onion (Allium cepa) and Arabidopsis epidermal cells using a Helium Biolistic Particle Delivery system (Bio-Rad). Cell walls and nuclei of Arabidopsis epidermal cells were stained with 1 mm propidium iodide. Fluorescent cells were imaged by confocal microscopy (Leica).
Collection of Mesophyll Protoplasts
The epidermis of Arabidopsis rosette leaves was rubbed off with glass paper P 80. Leaves were transferred into digestion buffer, pH 5.6 (0.5 m sorbitol; 1 mm CaCl2; 10 mm MES) containing 0.75% cellulase YC (w/v) and 0.03% pectolyase Y23 (w/v) (both from Kyowa Chemical Products) and incubated for 1.5 h at 30°C. After digestion, protoplasts were recovered by centrifugation (2,000g for 5 min) and purified by a Percoll gradient. The protoplasts were mixed 3:1 (v/v) with 100% Percoll, pH 6 (0.5 m sorbitol; 1 mm CaCl2; 20 mm MES), overlaid with 25% Percoll, pH 6, and betaine buffer, pH 6 (0.4 m betaine; 30 mm KCl; 20 mm HEPES) followed by centrifugation at 1,500g for 5 min. Protoplasts from the interface between 25% Percoll, pH 6, and betaine buffer were recovered. The leaf protoplast suspension was visualized by bright-field microscopy to distinguish between mesophyll protoplasts and other protoplasts. Thirty to 40 mesophyll protoplasts were drawn up into a microcapillary with a tip opening of approximately 100 μm. Total RNA was extracted from the protoplasts using the PicoPure RNA isolation kit (Arcturus).
RT-PCR
First-strand cDNA was prepared with the DNA-free total RNA using the First-Strand cDNA Synthesis kit (Amersham Biosciences). The following primers were used for RT-PCR: AtSUT4(for) 5′ gtc atc cca cag gta att gtg tct gtt ggc 3′, AtSUT4(rev) 5′ gcg gcc gct cat ggg aga ggg atg gg 3′, AtSUC2(for) 5′ cat tgt cgt ccc tca gat ggt aat atc tg 3′, AtSUC2(rev) 5′ ctc gag atg aaa tcc cat agt agc ttt gaa g 3′, Actin(for) 5′ gga aca gtg tga ctc aca cca tc 3′, Actin(rev) 5′ aag ctg ttc ttt ccc tct acg c 3′, HvSUT2(for) 5′ cac aat ctt agg agc acc tct gtc gat cac g 3′, and HvSUT2(rev) 5′ cat ggg tac ctc gtt ggg tgg ttt tct tct tc 3′.
Acknowledgments
We thank Prof. Masayoshi Maeshima (University of Nagoya) for his kind supply of antibodies to BiP and Prof. Francis Marty (University of Bourgogne) for providing us with antibodies to γ-TIP and AOX.
This work was supported by the Plant Science Center Zurich-Basel (graduate research fellowship), by the project Novel Ion Channels in Plants (grant no. EU HPRN–CT–00245), and by the Deutsche Forschungsgemeinschaft (project no. ME 1955/2).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ulrike G. Schmidt (ulrike.schmidt@botinst.unizh.ch).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.079533.
References
- Alexandersson E, Saalbach G, Larsson C, Kjellbom P (2004) Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol 45: 1543–1556 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A; Working Group on Publication Guidelines for Peptide and Protein Identification Data (2004) The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol Cell Proteomics 3: 531–533 [DOI] [PubMed] [Google Scholar]
- Carter C, Songqin P, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J (2003) Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1996) Molecular cloning, immunochemical localization to the vacuole, and expression in transgenic yeast and tobacco of a putative sugar transporter from sugar beet. Plant Physiol 110: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao ADS, Marty-Mazars D, Bassham DC, Sanderfoot AA, Marty F, Raikhel NV (1997) The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9: 571–582 [PMC free article] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Fluckiger R, Slusarenko AJ, Ward JM, Rentsch D (2004) AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. Plant J 40: 488–499 [DOI] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerlich V, Linka N, Reinhold T, Hurth MA, Traub M, Martinoia E, Neuhaus HE (2003) The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc Natl Acad Sci USA 100: 11122–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangne N, Maeshima M, Schaffner AR, Mandel T, Martinoia E, Bonnemain JL (2001) Expression and distribution of a vacuolar aquaporin in young and mature leaf tissues of Brassica napus in relation to water fluxes. Planta 212: 270–278 [DOI] [PubMed] [Google Scholar]
- Geisler M, Girin M, Brandt S, Vincenzetti V, Plaza S, Kobae Y, Maeshima M, Billion K, Kolukisaoglu UH, Schulz B, et al (2004) Arabidopsis immunophilin-like TWD1 functionally interacts with vacuolar ABC transporters. Mol Biol Cell 15: 3393–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Amos B (1995) GFP in plants. Trends Genet 11: 328–329 [DOI] [PubMed] [Google Scholar]
- Hurth MA, Suh SJ, Kretzschmar T, Geis T, Bregante M, Gambale F, Martinoia E, Neuhaus HE (2005) Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol 137: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser G, Heber U (1984) Sucrose transport into vacuoles isolated from barley mesophyll protoplasts. Planta 161: 562–568 [DOI] [PubMed] [Google Scholar]
- Kaiser G, Martinoia E, Schmitt JM, Hincha DK, Heber U (1986) Polypeptide pattern and enzymic character of vacuoles isolated from barley mesophyll protoplasts. Planta 169: 345–355 [DOI] [PubMed] [Google Scholar]
- Klein M, Geisler M, Suh SJ, Kolukisaoglu HU, Azevedo L, Plaza S, Curtis MD, Richter A, Weder B, Schulz B, et al (2004) Disruption of AtMRP4, a guard cell plasma membrane ABCC-type ABC transporter, leads to deregulation of stomatal opening and increased drought susceptibility. Plant J 39: 219–236 [DOI] [PubMed] [Google Scholar]
- Kreuz K, Tommasini R, Martinoia E (1996) Old enzymes for a new job. Plant Physiol 111: 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Liu G, Sánchez-Fernández R, Li ZS, Rea PA (2001) Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J Biol Chem 23: 8648–8656 [DOI] [PubMed] [Google Scholar]
- Maeshima M (2001) Tonoplast transporters: organization and function. Annu Rev Plant Physiol Plant Mol Biol 52: 469–497 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Grill E, Tommasini R, Kreuz K, Amrhein N (1993) ATP-dependent S-conjugate ‘export’ pump in the vacuolar membrane of plant. Nature 364: 247–249 [Google Scholar]
- Martinoia E, Heck U, Wiemken A (1981) Vacuoles as storage compartments for nitrate in barley leaves. Nature 289: 292–294 [Google Scholar]
- Martinoia E, Kaiser G, Schramm MJ, Heber U (1987) Sugar-transport across the plasmalemma and the tonoplast of barley mesophyll protoplasts: evidence for different transport-systems. Plant Physiol 131: 467–487 [Google Scholar]
- Martinoia E, Klein M, Geisler M, Forestier C, Kolukisaoglu U, Mueller-Roeber B, Schulz B (2002) Multifunctionality of plant ABC transporters: more than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- Matile P (1984) Das toxische Kompartiment Pflanzenzelle. Naturwissenschaften 71: 18–24 [Google Scholar]
- Nakamura A, Fukuda A, Sakai S, Tanaka Y (2006) Molecular cloning, functional expression and subcellular localization of two putative voltage-gated chloride channels in rice (Oryza sativa L). Plant Cell Physiol 47: 32–42 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Peiter E, Maathuis F, Mills L, Knight H, Pelloux J, Hetherington A, Sanders D (2005) The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature 434: 404–408 [DOI] [PubMed] [Google Scholar]
- Pollock CJ, Cairns AJ (1991) Fructan metabolism in grasses and cereals. Annu Rev Plant Physiol Plant Mol Biol 42: 77–101 [Google Scholar]
- Rentsch D, Martinoia E (1991) The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Planta 184: 532–537 [Google Scholar]
- Ritsema T, Smeekens S (2003) Fructan: beneficial for plants and humans. Curr Opin Plant Biol 6: 223–230 [DOI] [PubMed] [Google Scholar]
- Sazuka T, Keta S, Shiratake K, Yamaki S, Shibata D (2004) A proteomic approach to identification of transmembrane proteins and membrane-anchored proteins of Arabidopsis thaliana by peptide sequencing. DNA Res 11: 101–113 [DOI] [PubMed] [Google Scholar]
- Schulze WX, Reinders A, Ward J, Lalonde S, Frommer WB (2003) Interactions between co-expressed Arabidopsis sucrose transporters in the split-ubiquitin system. BMC Biochem 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K, Maeshima M, Yokota A, Tomizawa K, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45: 672–683 [DOI] [PubMed] [Google Scholar]
- Song W, Steiner HY, Zhang L, Naider F, Stacey G, Becker JM (1996) Cloning of a second Arabidopsis peptide transport gene. Plant Physiol 110: 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N (1999) The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. Plant J 19: 269–278 [DOI] [PubMed] [Google Scholar]
- Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109: 299–306 [Google Scholar]
- Sze H, Schumacher K, Muller ML, Padmanaban S, Taiz L (2002) A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H(+)-ATPase. Trends Plant Sci 7: 157–161 [DOI] [PubMed] [Google Scholar]
- Szponarski W, Sommerer N, Boyer JC, Rossignol M, Gibart R (2004) Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics 4: 397–406 [DOI] [PubMed] [Google Scholar]
- Theodoulou FL, Clark IM, He XL, Pallett KE, Cole DJ, Hallahan DL (2003) Co-induction of glutathione-S-transferases and multidrug resistance associated protein by xenobiotics in wheat. Pest Manag Sci 59: 202–214 [DOI] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H (2003) AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J 34: 685–695 [DOI] [PubMed] [Google Scholar]
- Wagner W, Keller F, Wiemken A (1983) Fructan metabolism in cereals: induction in leaves and compartmentation in protoplasts and vacuoles. Z Pflanzenphysiol 112: 359–372 [Google Scholar]
- Weise A, Barker L, Kuehn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low-affinity/high capacity localized in sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Panitz R, Sauer N, Wang Q, Neubohn B, Weber H, Wobus U (2000) Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J 21: 455–467 [DOI] [PubMed] [Google Scholar]
- Xia T, Apse MP, Aharon GS, Blumwald E (2002) Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol Plant 116: 206–212 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) IDI7, a new iron-regulated ABC transporter from barley roots, localizes to the tonoplast. J Exp Bot 53: 727–735 [DOI] [PubMed] [Google Scholar]