Abstract
The domestic apple (Malus domestica; also known as Malus pumila Mill.) has become a model fruit crop in which to study commercial traits such as disease and pest resistance, grafting, and flavor and health compound biosynthesis. To speed the discovery of genes involved in these traits, develop markers to map genes, and breed new cultivars, we have produced a substantial expressed sequence tag collection from various tissues of apple, focusing on fruit tissues of the cultivar Royal Gala. Over 150,000 expressed sequence tags have been collected from 43 different cDNA libraries representing 34 different tissues and treatments. Clustering of these sequences results in a set of 42,938 nonredundant sequences comprising 17,460 tentative contigs and 25,478 singletons, together representing what we predict are approximately one-half the expressed genes from apple. Many potential molecular markers are abundant in the apple transcripts. Dinucleotide repeats are found in 4,018 nonredundant sequences, mainly in the 5′-untranslated region of the gene, with a bias toward one repeat type (containing AG, 88%) and against another (repeats containing CG, 0.1%). Trinucleotide repeats are most common in the predicted coding regions and do not show a similar degree of sequence bias in their representation. Bi-allelic single-nucleotide polymorphisms are highly abundant with one found, on average, every 706 bp of transcribed DNA. Predictions of the numbers of representatives from protein families indicate the presence of many genes involved in disease resistance and the biosynthesis of flavor and health-associated compounds. Comparisons of some of these gene families with Arabidopsis (Arabidopsis thaliana) suggest instances where there have been duplications in the lineages leading to apple of biosynthetic and regulatory genes that are expressed in fruit. This resource paves the way for a concerted functional genomics effort in this important temperate fruit crop.
Apples are recognized by consumers for their flavor, health, and nutritional attributes (Harker et al., 2003). Because of this, they have become the major temperate horticultural fruit crop and a significant component of fresh fruit traded internationally (Zohary and Hopf, 2000). The domestic apple (Malus domestica; also known as Malus pumila Mill.) belongs to the family Rosaceae. Together with other commercial fruit and ornamental species, it forms the subfamily Maloideae (Challice, 1974), which is thought to have evolved by hybridization from the families Spiraeoideae (x = 9) and Prunoideae (x = 8; Lespinasse et al., 2000). The resulting allopolyploid has a basic haploid number of x = 17 and an estimated genome size of 743 to 796 Mb (Arumuganathan and Earle, 1991).
Apple has become a model for understanding important traits in fruiting tree crops. The ability to graft scions to speed propagation and mass produce a genetically uniform fruit from an outbreeding plant has contributed to the success of apple and many other horticultural crops. Also, other important traits, including dwarfing and some insect resistance traits, can be conferred by rootstocks (Ferree and Carlson, 1987). Compounds in the skin and flesh of the fruit confer flavor, taste, and health benefits that are important consumer traits in apple. Presumably, these compounds evolved as attractants and bribes for their seed dispersers. Flavor compounds increase substantially during fruit ripening, which takes place toward the end of 20 to 21 weeks of fruit development. This increase in flavor is caused by an autocatalytic burst of ethylene production late in fruit development, characteristic of all climacteric fruit (Fellman et al., 2000). Also triggered by ethylene are a marked increase in cell wall and starch breakdown and a general progression through ripening, senescence, and breakdown (Giovannoni, 2001).
The genes involved in many of the aforementioned traits are yet to be identified in apple. However, with the advent of high-throughput sequencing, isolation of genes potentially involved in such traits is now more readily attainable. One approach is the single-pass sequencing of cloned cDNAs representing RNA transcripts (mRNAs). These are otherwise known as expressed sequence tags (ESTs) and have become an established method for rapidly developing gene databases (Adams et al., 1993). By sequencing numbers of clones from cDNA libraries derived from RNA from different source tissues, the total set of genes sampled from the genome can be maximized. Bioinformatic sorting and clustering of the resulting sequences yield databases of putative genes that form the basis of a functional genomics program. Gene mining of these databases, aided by techniques such as microarrays, can be used to select candidate genes that are implicated in particular crop traits. In addition, ESTs have been identified as useful sources of both simple sequence repeats (SSRs) and single-nucleotide polymorphisms (SNPs), both useful markers for creating genetic maps in plants (Morgante et al., 2002; Rafalski, 2002).
ESTs have been collected for many plant species. The most comprehensively surveyed are Arabidopsis (Arabidopsis thaliana; 418,563 in GenBank) and rice (Oryza sativa; 406,624 in GenBank), both of which have also had their entire genome sequenced (Arabidopsis Genome Initiative, 2000; International Rice Genome Sequencing Project, 2005). Whereas fruit crops have been less extensively surveyed using an EST approach, recently there have been a number of reports on fruit EST projects. There is an extensive EST collection available from tomato (Lycopersicon esculentum; Van der Hoeven et al., 2002), and genes likely to be involved in the ripening process have been identified by virtual northern analysis (Fei et al., 2004). ESTs of strawberry (Fragaria ananassa) fruit have been analyzed by microarray technology (Aharoni et al., 2000) also during ripening (Aharoni and O'Connell, 2002). In an EST collection from the fruit of pineapple (Ananas comosus), Moyle et al. (2005) found a very high abundance of metallothione gene transcripts, whereas reports from grape (Vitis vinifera) identify many new SSRs useful for grape mapping (Moser et al., 2005) and many candidate genes involved in fruit development traits (Goes da Silva et al., 2005). The only other Rosaceae species to have a significant number of ESTs described is apricot (Prunus armeniaca; Grimplet et al., 2005).
Here we describe the first EST sequencing project in apple. We report the collection and analysis of 151,687 high-quality apple ESTs, largely from the commercial apple cultivar Royal Gala. From this sequence information, we put sequences into functional categories in preparation for functional genomics programs and describe SSRs and SNPs in the sequence data that will be useful in marker-assisted breeding programs. In addition, we show that there are many ESTs that potentially encode enzymes of important flavor and health compound biosynthetic pathways, and explore whether there has been an expansion of the number of genes from gene families involved in secondary metabolite biosynthesis and regulation that are expressed in fruit tissues.
RESULTS
EST Sequencing and Clustering
cDNA libraries were constructed from a range of different tissues and developmental time points using material from the apple cultivars Royal Gala, Pinkie, Pacific Rose, and the dwarfing rootstock M9. Libraries were also constructed from some tissues, plants, and cell lines that were subjected to biotic and abiotic stresses. The libraries were sequenced to varying depths (Table I), depending on library quality and novelty. Over the 43 cDNA libraries sequenced, 151,687 good-quality sequences were recovered. The average edited length of the sequences was 468 bases.
Table I.
Summary of apple ESTs
| Library Code | Library Description | Minimum Sequence Length | Average Sequence Length | Maximum Sequence Length | Total No. ESTs | No. ESTs Assigned to TCa | No. Singletons | Percentage of Singletons per Library |
|---|---|---|---|---|---|---|---|---|
| AAAA | Royal Gala 59 DAFBb fruit, seeds removed | 50 | 425 | 767 | 7,018 | 5,495 | 1,523 | 21.7 |
| AAFA | Royal Gala apple skin peel, tree-ripened fruit 150 DAFB | 47 | 382 | 821 | 6,184 | 5,389 | 795 | 12.9 |
| AAFB | Royal Gala apple skin peel, tree-ripened fruit 150 DAFB | 50 | 331 | 682 | 1,556 | 1,058 | 498 | 32.0 |
| AAGA | Pinkie expanding leaf, normalized | 51 | 342 | 702 | 530 | 418 | 112 | 21.1 |
| AAKA | Pinkie expanding leaf, normalized | 51 | 365 | 687 | 516 | 389 | 127 | 24.6 |
| AALA | Royal Gala 150 DAFB fruit cortex | 50 | 348 | 721 | 5,282 | 4,130 | 1,152 | 21.8 |
| AALB | Royal Gala 150 DAFB fruit cortex | 51 | 341 | 739 | 1,275 | 933 | 342 | 26.8 |
| AAMA | Royal Gala spur bud autumn | 51 | 374 | 668 | 988 | 679 | 309 | 31.3 |
| AANA | Royal Gala partially senescing leaf | 50 | 318 | 729 | 1,888 | 1,453 | 435 | 23.0 |
| AAOA | Royal Gala phloem | 50 | 371 | 734 | 4,519 | 3,059 | 1,460 | 32.3 |
| AAPA | Royal Gala 24 DAFB fruit | 50 | 482 | 771 | 2,649 | 2,003 | 646 | 24.4 |
| AARA | Royal Gala partially senescing leaf | 50 | 451 | 770 | 8,838 | 6,412 | 2,426 | 27.4 |
| AASA | Royal Gala 10 DAFB fruit | 51 | 553 | 767 | 4,860 | 4,169 | 691 | 14.2 |
| AASB | Royal Gala 10 DAFB fruit | 187 | 562 | 777 | 4,240 | 3,945 | 295 | 7.0 |
| AASC | Royal Gala 10 DAFB fruit | 200 | 440 | 697 | 405 | 378 | 27 | 6.7 |
| AAUA | Royal Gala 87 DAFB fruit cortex | 182 | 511 | 748 | 2,458 | 2,361 | 97 | 3.9 |
| AAWA | Royal Gala 59 DAFB seeds | 101 | 520 | 766 | 5,472 | 4,769 | 703 | 12.8 |
| AAXA | Royal Gala 126 DAFB fruit core | 195 | 514 | 766 | 5,092 | 4,655 | 437 | 8.6 |
| AAYA | Royal Gala 126 DAFB fruit cortex | 118 | 521 | 742 | 4,481 | 3,946 | 535 | 11.9 |
| AAZA | M9 xylem | 97 | 506 | 756 | 5,004 | 3,914 | 1,090 | 21.8 |
| ABAA | Pacific Rose spur buds | 139 | 512 | 736 | 789 | 600 | 189 | 24.0 |
| ABAB | Pacific Rose spur buds | 105 | 589 | 751 | 486 | 378 | 108 | 22.2 |
| ABBA | Pacific Rose spur buds | 201 | 524 | 740 | 1,078 | 886 | 192 | 17.8 |
| ABBB | Pacific Rose spur buds | 226 | 591 | 753 | 495 | 403 | 92 | 18.6 |
| ABCA | Royal Gala fruit stored at 0.5°C for 24 h | 186 | 523 | 756 | 4,693 | 4,215 | 478 | 10.2 |
| ABDA | Royal Gala fruit stored for 24 h under low oxygen/high CO2 | 165 | 552 | 741 | 4,808 | 4,616 | 192 | 4.0 |
| ABEA | Royal Gala seedling leaves infected with V. inaequalisc | 170 | 564 | 775 | 4,798 | 3,967 | 831 | 17.3 |
| ABEB | Royal Gala seedling leaves infected with V. inaequalisc | 198 | 521 | 766 | 562 | 395 | 167 | 29.7 |
| ABKA | Royal Gala temperature stress leaves | 128 | 539 | 770 | 1,075 | 927 | 148 | 13.8 |
| ABLC | Braeburn cell culture 3 d after subculture | 72 | 573 | 764 | 4,715 | 4,082 | 633 | 13.4 |
| ABMA | M9 phloem | 108 | 557 | 781 | 4,900 | 4,383 | 517 | 10.6 |
| ABNB | Braeburn cultured fruit cells, boron exposed | 209 | 585 | 754 | 944 | 777 | 167 | 17.7 |
| ABPB | M9 root tips | 86 | 562 | 760 | 4,813 | 3,963 | 850 | 17.7 |
| ABQA | Royal Gala flower | 199 | 504 | 749 | 926 | 831 | 95 | 10.3 |
| AEAA | Aotea expanding leaf | 51 | 349 | 758 | 1,665 | 1,524 | 141 | 8.5 |
| AELA | Royal Gala young expanding leaf | 50 | 318 | 712 | 5,457 | 4,388 | 1,069 | 19.6 |
| AENA | Northern Spy expanding leaf | 52 | 428 | 759 | 1,222 | 918 | 304 | 24.9 |
| AEPA | Pinkie expanding leaf | 50 | 370 | 828 | 4,786 | 3,891 | 895 | 18.7 |
| AFBC | Royal Gala preopened floral bud | 50 | 350 | 877 | 4,959 | 3,933 | 1,026 | 20.7 |
| AOFA | Royal Gala 24 DAFB fruit | 51 | 344 | 777 | 1,077 | 782 | 295 | 27.4 |
| AVBB | Royal Gala young shoot | 50 | 296 | 737 | 5,572 | 4,511 | 1,061 | 19.0 |
| AVBC | Royal Gala young shoot | 57 | 544 | 790 | 17,967 | 15,874 | 2,093 | 11.6 |
| AYFB | Royal Gala 10 DAFB fruit | 54 | 304 | 693 | 645 | 410 | 235 | 36.4 |
| Total for all libraries combined | 47 | 468 | 877 | 151,687 | 17,460d | 25,478 | ||
| Total no. of NR sequences 43,938 | ||||||||
Number of ESTs from each library that have been assigned to a TC sequence (contig).
Days after full bloom.
These libraries also will contain fungal sequences derived from the pathogen V. inaequalis (K. Plummer, W. Cui, and M. Templeton, unpublished data).
Total number of unique TC sequences in the entire database (i.e. this figure is not additive).
Clustering of the sequences using a 95% threshold yielded 17,460 tentative consensus (TC) sequences with 25,478 sequences remaining unclustered (singletons). TC sequences range in length from 66 to 6,145 bases with an average of 745 bases, whereas singletons range in size from 47 to 790 bases with an average of 394 bases. The GC ratio of singletons ranged from 13% to 78%, with an average of 44%, whereas that for TC sequences ranged from 14% to 69%, also with an average of 44%. Together, the TC sequences and singletons yielded an apple EST dataset of 42,938 sequences. Hereafter, the singletons and TC sequences are collectively referred to as the nonredundant (NR) gene set. Clustering using a threshold of 90% generated fewer TC sequences (16,756) and singletons (17,858). However, using this lower threshold increased the number of instances of paralogs being incorporated into the same cluster and was therefore not used subsequently.
Codon usage was assessed on a set of 545 apple cDNA sequences predicted to contain full-length coding regions. These cDNAs were checked by manual inspection of BLASTx versus NRDB90 reports to make sure they are devoid of introns and frameshift errors. From these data, the open reading frames were defined and a codon usage table created from the 203,267 codons (Table II). All codons are found in the full-length cDNA dataset, with the least frequent codon represented over 100 times. The GC content at the third position of the codon is 52%.
Table II.
Codon usage calculated using 545 full-length apple cDNA sequencesa
| Codon | Amino Acid | Fractionb | Per 1,000c | No. |
|---|---|---|---|---|
| GCA | A | 0.27 | 18.92 | 3,846 |
| GCC | A | 0.26 | 18.42 | 3,745 |
| GCG | A | 0.14 | 9.82 | 1,997 |
| GCT | A | 0.33 | 23.36 | 4,748 |
| TGC | C | 0.59 | 10.38 | 2,110 |
| TGT | C | 0.41 | 7.20 | 1,464 |
| GAC | D | 0.44 | 23.91 | 4,860 |
| GAT | D | 0.56 | 29.98 | 6,095 |
| GAA | E | 0.45 | 28.70 | 5,834 |
| GAG | E | 0.55 | 34.67 | 7,047 |
| TTC | F | 0.53 | 22.14 | 4,500 |
| TTT | F | 0.47 | 19.95 | 4,056 |
| GGA | G | 0.29 | 19.85 | 4,034 |
| GGC | G | 0.23 | 16.09 | 3,270 |
| GGG | G | 0.22 | 15.40 | 3,130 |
| GGT | G | 0.25 | 17.43 | 3,542 |
| CAC | H | 0.51 | 13.34 | 2,712 |
| CAT | H | 0.49 | 12.67 | 2,575 |
| ATA | I | 0.21 | 9.98 | 2,028 |
| ATC | I | 0.37 | 17.49 | 3,556 |
| ATT | I | 0.42 | 19.74 | 4,012 |
| AAA | K | 0.39 | 23.77 | 4,832 |
| AAG | K | 0.60 | 36.10 | 7,337 |
| CTA | L | 0.09 | 7.77 | 1,579 |
| CTC | L | 0.21 | 18.83 | 3,828 |
| CTG | L | 0.17 | 15.33 | 3,116 |
| CTT | L | 0.21 | 18.76 | 3,813 |
| TTA | L | 0.09 | 8.09 | 1,644 |
| TTG | L | 0.23 | 21.11 | 4,291 |
| ATG | M | 1 | 24.87 | 5,056 |
| AAC | N | 0.51 | 23.02 | 4,679 |
| AAT | N | 0.49 | 21.98 | 4,467 |
| CCA | P | 0.30 | 17.37 | 3,530 |
| CCC | P | 0.20 | 11.60 | 2,358 |
| CCG | P | 0.21 | 11.84 | 2,406 |
| CCT | P | 0.29 | 16.37 | 3,328 |
| CAA | Q | 0.48 | 19.08 | 3,879 |
| CAG | Q | 0.52 | 20.52 | 4,172 |
| AGA | R | 0.26 | 13.02 | 2,646 |
| AGG | R | 0.27 | 13.60 | 2,764 |
| CGA | R | 0.11 | 5.28 | 1,073 |
| CGC | R | 0.12 | 6.17 | 1,254 |
| CGG | R | 0.12 | 6.17 | 1,255 |
| CGT | R | 0.10 | 5.14 | 1,044 |
| AGC | S | 0.16 | 14.65 | 2,977 |
| AGT | S | 0.14 | 12.18 | 2,476 |
| TCA | S | 0.19 | 16.63 | 3,381 |
| TCC | S | 0.19 | 16.99 | 3,453 |
| TCG | S | 0.13 | 11.21 | 2,279 |
| TCT | S | 0.19 | 17.28 | 3,512 |
| ACA | T | 0.26 | 12.97 | 2,636 |
| ACC | T | 0.30 | 15.15 | 3,079 |
| ACG | T | 0.14 | 7.23 | 1,469 |
| ACT | T | 0.29 | 14.68 | 2,985 |
| GTA | V | 0.11 | 7.27 | 1,478 |
| GTC | V | 0.23 | 14.47 | 2,942 |
| GTG | V | 0.33 | 20.81 | 4,231 |
| GTT | V | 0.33 | 20.57 | 4,181 |
| TGG | W | 1 | 14.01 | 2,847 |
| TAC | Y | 0.57 | 14.83 | 3,015 |
| TAT | Y | 0.43 | 11.16 | 2,269 |
| TAA | * | 0.34 | 0.91 | 185 |
| TAG | * | 0.25 | 0.66 | 135 |
| TGA | * | 0.41 | 1.11 | 225 |
Codon usage calculated using the CUSP program from EMBOSS (Rice et al., 2000).
Proportion of usage of a given codon among its redundant set (i.e. the set of codons that code for this codon's amino acid).
Codon frequency normalized per 1,000 bases.
SSRs and SNPs
cDNA sequences are a useful source of microsatellites or SSRs in plants. SSRs are particularly common in the 5′-untranslated region (UTR) and, to a lesser extent, in the 3′-UTR of transcribed plant sequences (Morgante et al., 2002). We analyzed the nature of the perfect SSRs in the apple sequence dataset. Approximately 17% of the apple sequences contained one or more di-, tri-, or tetranucleotide SSRs. The relative frequency of di- and trinucleotide repeats is similar (4,018 versus 4,010, respectively; Table III). Just over one-half (57%) of the repeats were between 12 and 14 bases in length and only 17% of the di-, tri-, and tetranucleotide repeats were longer than 20 nucleotides in length. We compared the relative frequency of repeats with different dinucleotide compositions and found there to be a striking bias to one of four possible repeat classes (Table III). AG repeats were by far the most common dinucleotide repeat, constituting nearly 88% of dinucleotide repeats. A similar bias to AG repeats has been found in Arabidopsis (83%) and indeed other plants (Zhang et al., 2004). AT repeats were the next most common at 7.6% in apple compared with 8.8% for Arabidopsis, followed by AC repeats at 4% in apple compared with 8% in Arabidopsis. CG repeats are very infrequent in plants at 0.05% in apple and 0.14% in Arabidopsis.
Table III.
Summary of microsatellites in apple ESTs and comparison with Arabidopsis
| Dinucleotide Repeat Composition | No. NR Sequences | Percentage of Apple Di Repeats | Apple Rank | Percentage of Arabidopsis Di Repeats |
|---|---|---|---|---|
| AC/CA/GT/TG | 162 | 4.0 | 3 | 8.0 |
| AG/GA/CT/TC | 3,548 | 88.3 | 1 | 83.0 |
| AT/TA | 306 | 7.6 | 2 | 8.8 |
| CG/GC | 2 | 0.1 | 4 | 0.14 |
| Totals | 4,018 | 100 | – | 100 |
| Trinucleotide Repeat Composition | No. NR Sequences | Percentage of Apple Tri Repeats | Apple Rank | Arabidopsis Rank |
| AAC/ACA/CAA/GTT/TGT/TTG | 223 | 5.6 | 7 | 3 |
| AAG/AGA/GAA/CTT/TCT/TTC | 918 | 22.9 | 1 | 1 |
| AAT/ATA/TAA/TTA/TAT/ATT | 126 | 3.1 | 9 | 5 |
| ACC/CAC/CCA/GGT/GTG/TGG | 635 | 15.8 | 3 | 4 |
| ACG/CGA/GAC/CGT/GTC/TCG | 202 | 5.0 | 8 | 9 |
| ACT/CTA/TAC/AGT/TAG/GTA | 52 | 1.3 | 10 | 8 |
| AGC/CAG/GCA/TGC/CTG/GCT | 544 | 13.6 | 4 | 7 |
| AGG/GGA/GAG/TCC/CTC/CCT | 752 | 18.8 | 2 | 6 |
| ATC/CAT/TCA/GAT/ATG/TGA | 291 | 7.3 | 5 | 2 |
| CCG/CGC/GCC/GGC/GCG/CGG | 267 | 6.7 | 6 | 10 |
| Totals | 4,010 | 100% | – | – |
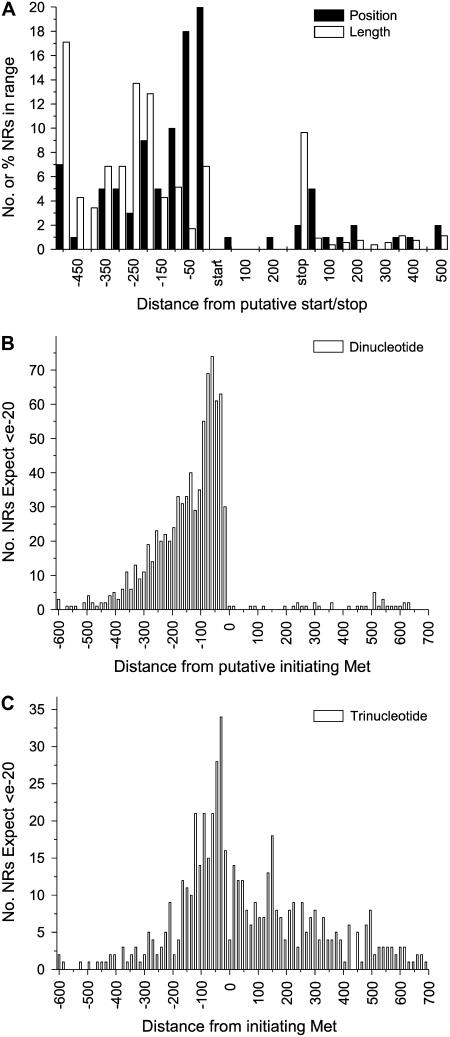
Next we investigated the position of the SSRs in relation to putative initiation (Met) and stop codons within the apple sequence dataset. First, we identified sequences containing a dinucleotide repeat with more than 100 bp of flanking DNA and ranked the sequences in order of significance of match to public sequences using BLASTx (Altschul et al., 1990). This ensured that we had identified the correct open reading frame and, therefore, the start and stop codons accurately (Fig. 1A). Of the top 100 in this ranking, we found that 83% contained dinucleotide repeats in the putative 5′-UTR, 2% in the putative coding region, and 15% in the putative 3′-UTR. These figures are similar for the Arabidopsis genome (5′-UTR 83%, coding region 0.4%, and 3′-UTR 16% as deduced from relative frequencies per Megabase pair given by Zhang et al. [2004]). These data suggested that repeats in the 5′-UTR are disproportionately high within 100 bp of the translation start site. We then analyzed all dinucleotide (Fig. 1B) and trinucleotide (Fig. 1C) repeats longer than six repeats present in the entire apple database using a BLASTx E-value significance cutoff criterion of e-20 to identify all sequences with a reasonable protein match in GenBank on which to base putative start sites. At least for the dinucleotide repeats, this pattern seen on the global data is consistent with that manually collected for the top 100 ranked genes used above. Both datasets show a consistent pattern, with SSRs clustered in the 5′-UTR closest to the start codon.
Figure 1.
Microsatellite (SSR) positions relative to predicted coding regions of the apple NR sequences. A, Manual analysis of the number of NR sequences containing dinucleotide repeats and their distance from the putative initiating Met (5′-SSR) or stop codon (3′-SSR) in 50-base bin sets. NR sequences were ranked in order of most significant to least significant BLASTx match to public domain databases to decrease the influence of incorrect putative initiating Met and stop codon identifications. The 100 top-ranked NR sequences with a BLASTx match more significant than e-38 were manually inspected for repeat position. Also shown are the lengths of the UTRs (% of total for the same dataset) that fit into the same bin set. The stop and start positions are shown and the numbering in between these two indicates the distance from the start codon. Repeats more than 500 bases from the putative start and stop sites (and UTRs longer than 500) have been combined into one bin set at the beginning and end of the range, respectively. B, Position of dinucleotide repeats in relation to the putative initiating Met from an automated analysis, including all NR sequences with a BLASTx match more significant than e-20 in 15-base bin sets. Note that, in contrast to the manual analysis in A, the stop sites have not been predicted with this automated analysis. C, Position of trinucleotide repeats in relation to the putative initiating Met from an automated analysis including all NR sequences with a BLASTx match more significant than e-20 in 15-base bin sets. As for the automated analysis in B, the stop sites have not been predicted with this automated analysis.
In addition to SSRs, EST sequences are also a useful source of SNPs, which can also be used in mapping and marker-assisted breeding. The major cultivar sequenced in this study was Royal Gala (78.9%). However, some sequences were also from other apple cultivars, including M9 (9.7%), Pinkie (3.8%), Braeburn (3.7%), Pacific Rose (1.9%), Aotea (1.1%), and Northern Spy (0.8%). Apple is also an outbreeder, which will increase levels of heterozygosity within cultivars. Together, these factors should increase the instances of SNPs in the apple EST data. This seems to be the case with evidence for 18,408 bi-allelic SNPs confirmed by more than one sequence per allele from the 13.0 Mb of aligned NR sequences analyzed. Bi-allelic SNPs are therefore found, on average, every 706 bp of transcribed DNA. Transitions were more common than transversions. There were 4,592 AG and 5,112 CT transitions compared with 2,032 AC, 2,372 AT, 2,228 CG, and 2,072 GT transversions (Table IV). Furthermore, one or more restriction endonuclease cleavage site polymorphisms were revealed with candidate SNPs in approximately 82% of NR sequences with predicted SNPs.
Table IV.
SNP analysis of apple NR sequences
| Cumulative length of analyzed NR sequence entries | 13.0 Mb |
| Total no. bi-allelic SNPs predicted from 42,938 NR sequences (25,478 TC sequences + 17,460 singletons) | 18,408 |
| Average occurrence of predicted bi-allelic SNPs | 1 in 706 bp |
| Average occurrence of predicted bi-allelic SNPs + sequences with one polymorphic base | 1 in 144 bp |
| Contig NR sequences with predicted bi-allelic SNPs | 20.61% |
| Average no. predicted bi-allelic SNPs per NR sequence | 1.05 |
| Transitions and transversions | |
| AG transitions | 4,592 |
| CT transitions | 5,112 |
| Total transitions | 9,704 |
| AC transversions | 1,516 |
| AT transversions | 3,508 |
| CG transversions | 2,726 |
| GT transversions | 2,570 |
| Total transversions | 8,704 |
| Total | 18,408 |
Functional Categorization
Annotation of the apple sequences was based on similarity to Arabidopsis genes and transfer of their annotation to apple sequences. BLASTx comparisons to predicted proteins from Arabidopsis were used to assign apple NR sequences into 21 functional categories based on functional annotations available for the Arabidopsis proteins following the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de) Functional Catalogue (FunCat) schema (Ruepp et al., 2004). Only 5.82% of the apple NR sequences did not have a match in Arabidopsis. Of those that do have a match, 72.79% are most similar to unclassified proteins in Arabidopsis (83.39%). Of the classified proteins in apple, the category Metabolism (5.39%) contains the most genes, as it does in Arabidopsis (4.09%; Table V).
Table V.
MIPS FunCat analysis of apple NR sequences compared with Arabidopsis
| No. | Functional Category | Apple NR Sequences | Arabidopsis |
|---|---|---|---|
| % | % | ||
| 01 | Metabolism | 5.39 | 4.09 |
| 02 | Cell fate | 2.13 | 0.26 |
| 04 | Storage protein | 0.13 | 0.07 |
| 10 | Cell cycle and DNA processing | 0.47 | 0.44 |
| 11 | Transcription | 2.47 | 2.29 |
| 12 | Protein synthesis | 1.17 | 0.64 |
| 14 | Protein fate | 2.30 | 0.93 |
| 16 | Protein with binding function or cofactor requirement | 0.42 | 0.41 |
| 18 | Protein activity regulation | 0.03 | 0.02 |
| 20 | Cellular transport | 2.19 | 1.39 |
| 30 | Cellular communication/signal transduction mechanism | 2.07 | 1.19 |
| 32 | Cell rescue, defense, and virulence | 2.02 | 1.02 |
| 34 | Interaction with the cellular environment | 0.10 | 0.03 |
| 36 | Interaction with the environment | 0.16 | 0.07 |
| 38 | Transposable elements, viral and plasmid proteins | 0.01 | 0.48 |
| 40 | Cell fate | 0.24 | 0.11 |
| 41 | Development | 0.31 | 0.11 |
| 42 | Biogenesis of cellular components | 1.36 | 0.40 |
| 70 | Subcellular localization | 1.75 | 0.31 |
| 98 | Classification not yet clear cut | 2.49 | 2.36 |
| 99 | Unclassified proteins | 72.79 | 83.39 |
The representation of protein families, domains, and functional sites within the apple sequence dataset was determined by comparison to the Inter-Pro (Zdobnov and Apweiler, 2001; Mulder et al., 2003) protein family database. In total, matches to 2,692 Inter-Pro families were found. The Inter-Pro families with the most frequent representation in the apple sequence dataset are presented in Table VI. Protein kinases are the most abundant family (IPR000719), with 801 NR sequences identified from apple. We used automated predictions based on comparisons to the Inter-Pro database to analyze transcription factors in greater detail and identified the most common transcription factor families in the apple sequences and compared the rankings of these with Arabidopsis (Table VII). The MYB transcription factor family is the most common within the apple NR sequences.
Table VI.
Fifty most common Inter-Pro families represented within the apple NR sequences
| Inter-Pro No. | Description | Frequency |
|---|---|---|
| IPR000719 | Protein kinase | 801 |
| IPR002290 | Ser-Thr protein kinase | 359 |
| IPR001611 | LRR | 346 |
| IPR008271 | Ser-Thr protein kinase, active site | 274 |
| IPR001245 | Tyr protein kinase | 269 |
| IPR000504 | RNA-binding region RNP-1 (RNA recognition motif) | 202 |
| IPR001680 | G-protein β WD-40 repeat | 193 |
| IPR007090 | LRR, plant specific | 170 |
| IPR001128 | Cytochrome P450 | 159 |
| IPR001841 | Zinc finger, RING | 156 |
| IPR001005 | Myb DNA-binding domain | 133 |
| IPR001806 | Ras GTPase superfamily | 124 |
| IPR000626 | Ubiquitin | 124 |
| IPR002048 | Calcium-binding EF-hand | 118 |
| IPR002885 | PPR repeat | 117 |
| IPR002110 | Ankyrin | 111 |
| IPR001810 | Cyclin-like F-box | 106 |
| IPR006662 | Thioredoxin-type domain | 100 |
| IPR000608 | Ubiquitin-conjugating enzymes | 98 |
| IPR001410 | DEAD/DEAH box helicase | 96 |
| IPR001440 | TPR repeat | 95 |
| IPR002401 | E-class P450, group I | 95 |
| IPR001932 | Protein phosphatase 2C-like | 83 |
| IPR001993 | Mitochondrial substrate carrier | 81 |
| IPR001623 | Heat shock protein DnaJ, N terminus | 78 |
| IPR001471 | Pathogenesis-related transcriptional factor and ERF | 76 |
| IPR002198 | Short-chain dehydrogenase/reductase SDR | 73 |
| IPR001087 | Lipolytic enzyme, G-D-S-L | 65 |
| IPR001344 | Chlorophyll a/b-binding protein | 63 |
| IPR001356 | Homeobox | 63 |
| IPR007087 | Zinc finger, C2H2 type | 61 |
| IPR002130 | Peptidyl-prolyl cis-trans isomerase, cyclophilin type | 60 |
| IPR003439 | ABC transporter | 60 |
| IPR002347 | Glucose/ribitol dehydrogenase | 60 |
| IPR001878 | Zinc finger, CCHC type | 57 |
| IPR000157 | TIR | 56 |
| IPR000795 | Protein synthesis factor, GTP binding | 54 |
| IPR000425 | Major intrinsic protein | 53 |
| IPR002016 | Haem peroxidase, plant/fungal/bacterial | 52 |
| IPR001395 | Aldo/keto reductase | 51 |
| IPR000008 | C2 domain | 49 |
| IPR002423 | Chaperonin Cpn60/TCP-1 | 48 |
| IPR004087 | KH | 46 |
| IPR000916 | Bet v I allergen | 45 |
| IPR001092 | Basic helix-loop-helix dimerization domain bHLH | 43 |
| IPR001023 | Heat-shock protein Hsp70 | 42 |
| IPR000571 | Zinc finger, C-x8-C-x5-C-x3-H type | 41 |
| IPR000217 | Tubulin | 40 |
| IPR001938 | Thaumatin, pathogenesis-related | 39 |
| IPR000823 | Plant peroxidase | 38 |
Table VII.
The 10 most common transcription factor (TF) families in apple identified by searches of automated predictions using Inter-Pro
| Top 10 TF Family Descriptions | No. Apple NR Sequences | Inter-Pro Accessions Nos. | TF Family Rank
|
||
|---|---|---|---|---|---|
| Apple | Arabidopsisa | Riceb | |||
| MYB | 138 | IPR001005, | 1 | 1, 11, 14 | 1, 9 |
| IPR006447, | |||||
| IPR000818, | |||||
| Pathogenesis related | 76 | IPR001471 | 2 | 2 | 2 |
| C2C2 Zn finger | 74 | IPR002991, | 3 | 6 | 7, 8, 10 |
| IPR000315, | |||||
| IPR000679, | |||||
| IPR003851, | |||||
| IPR006780 | |||||
| Homeobox | 66 | IPR001356, | 4 | 7 | NDc |
| IPR003106, | |||||
| IPR000047 | |||||
| C2H2 Zn finger | 64 | IPR007087, | 5 | 5 | 3 |
| IPR003656 | |||||
| NAC | 52 | IPR008917, | 6 | 4 | ND |
| IPR003441 | |||||
| Basic helix-loop-helix | 43 | IPR001092 | 7 | 3 | ND |
| C3H-type 1 Zn finger | 41 | IPR000571 | 8 | 18 | ND |
| WRKY | 40 | IPR003657 | 9 | 10 | 4 |
| bZip | 36 | IPR004827 | 10 | 9 | 5 |
| Total No. TFs | 952 | – | 1,470 | 1,306 | |
Based on data from Riechmann et al. (2000).
Based on data from Goff et al. (2002).
Family not determined by Goff et al. (2002).
Genes Encoding Important Traits in Apple Fruit
This collection of ESTs contains signatures of many genes involved in important traits in apple. Whereas much of primary metabolism and basic plant physiological processes are not peculiar to apple, some elements of the biology of apple are unique to the species, or at least members of the Rosaceae or other climacteric fruit.
Fruit Ripening
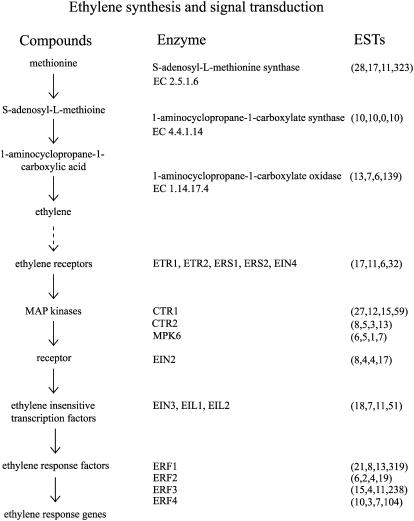
Apples are a climacteric fruit, displaying a rapid increase in respiration at the onset of ripening simultaneous with an increase in the production of the hormone ethylene (Knee, 1993). This process alters the biochemistry and physiology of the fruit to produce the attributes we associate with fruit that are ready to eat, including color, texture, flavor, and nutritional content (Fellman et al., 2000). Many of these processes are under the control of ethylene, the synthesis of which is autocatalytic (McKeon and Yang, 1987; Fig. 2). In the first biosynthetic step, Met is converted to S-adenosyl-l-Met (SAM) by S-adenosyl-l-Met synthetase (EC 2.5.1.6), represented by 28 NR sequences in the apple sequence dataset. Next, SAM is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (EC 4.4.1.14; 10 NR sequences) in what is the rate-limiting step in the pathway. Finally, ethylene is synthesized by ACC oxidase (EC 1.14.17.4; 13 NR sequences). In apple, an ACC synthase and an ACC oxidase gene have each been silenced in transgenic lines, revealing that many of the flavor and texture traits are under ethylene control (Dandekar et al., 2004).
Figure 2.
Ethylene synthesis and signal transduction as inferred from Giovannoni (2001) and Adams-Phillips et al. (2004). Apple sequences encoding enzymes and signal transduction proteins were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003) or sequences quoted in Adams-Phillips et al. (2004). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively.
Proteins involved in the perception of ethylene have been isolated almost exclusively through the positional cloning of genetic mutants in tomato and Arabidopsis (Giovannoni, 2001; Adams-Phillips et al., 2004). Using these sequences, many of the apple representatives can be found in the apple sequence dataset (Fig. 2). Ethylene receptors are members of the His kinase receptor class of which there are 17 NR sequences in the apple sequence dataset. These receptors transduce their signal through a mitogen-activated protein (MAP) kinase cascade. MAP kinase kinase kinases are negative regulators of the receptors with mutants for these genes showing constitutive activation of the pathway. Two representative families have relatives in the apple sequence dataset (CTR1, 27 NR sequences; CTR2, eight NR sequences). Alternatively, an ethylene-inducible MAP kinase, MPK6, is represented by six NR sequences. Downstream of these are a membrane-bound receptor ethylene insensitive-2 (EIN2; eight NR sequences) and then two sets of transcription factors, including the ethylene-insensitive-like (EIL) family (18 NR sequences) and the ethylene-response factors (ERF1, 21 NR sequences; ERF2, six NR sequences; ERF3, 15 NR sequences; ERF4, 10 NR sequences). The targets of the ERFs are likely to include genes involved in flavor biosynthesis and texture modification in ripening fruit.
Flavor Biosynthesis
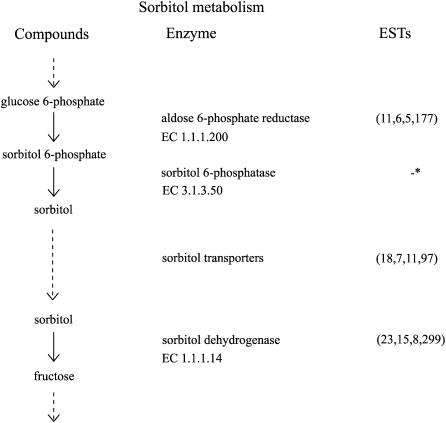
Sugars are important contributors and modulators of flavor in fruit. Whereas in most fruit species Suc is the major transported photosynthate, in members of the Rosaceae, including apple, sorbitol accounts for more than 50% of the fixed carbon and the carbon exported from the leaves (Bieleski, 1982). The enzymatic steps required for the synthesis of sorbitol in source tissues and its metabolism in sink tissues are known. Genes encoding these enzymes are well represented in this apple NR set (Fig. 3). In source leaves, sorbitol is derived from the same hexose phosphate pool as Suc. The enzyme aldose 6-P reductase (EC 1.1.1.200; 11 NR sequences) is the rate-limiting step for the conversion to sorbitol from the hexose phosphate pool (Negm and Loescher, 1981), synthesizing sorbitol 6-P from Glc 6-P. Antisense suppression of the aldose 6-P reductase transcript to 15% to 30% that of control apple plants results in a switch from sorbitol production to starch synthesis with no overall effect on the amount of CO2 fixation (Cheng et al., 2005). Sorbitol is then produced through dephosphorylation of sorbitol 6-P by an as yet unidentified 61-kD phosphatase (Zhou et al., 2003; EC 3.1.3.50). The sorbitol is then transported to sink tissues by a family of specialized sorbitol transporters (18 NR sequences) for unloading (Watari et al., 2004). Once in the sink tissue such as fruit, sorbitol dehydrogenase (EC 1.1.1.14; 23 NR sequences) converts sorbitol to Fru (Loescher et al., 1982), thereby reentering the pool of available sugars.
Figure 3.
The sorbitol metabolism pathway in apple. Apple sequences encoding enzymes involved in sorbitol metabolism were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively. *, No PIR reference gene currently available for EC 3.1.3.50 because the gene has yet to be isolated.
Upon ripening, apple fruit produce large quantities of volatiles presumably to attract and provide a taste reward for seed dispersers (Yahia, 1994; Fellman et al., 2000). In addition, other parts of the plant, such as the leaves, also produce volatiles that can act as attractants for insects (Bengtsson et al., 2001). The volatiles produced by apple, including alcohols, aldehydes, esters, terpenes, terpenoids, and polyphenolics, are derived from secondary metabolite pathways. The apple terpenes (E,E)- and (Z,E)-α-farnesene are produced via the mevalonate pathway (Ju and Curry, 2000; Fig. 4). In addition to being important flavor compounds in apple, (E,E)- and (Z,E)-α-farnesene are attractants for codling moth (Bengtsson et al., 2001) and have been implicated in the storage disorder scald (Pechous et al., 2005). All the enzymes involved in the mevalonate pathway are represented in the apple sequence dataset. Initial steps from mevalonate include the enzymes mevalonate kinase (EC 2.7.1.36; three NR sequences), phosphomevalonate kinase (EC 2.7.4.2; one NR sequence), mevalonate diphosphate decarboxylase (EC 4.1.1.33; four NR sequences), and isopentyl-diphosphate δ-isomerase (EC 5.3.3.2; four NR sequences). The progenitors of the terpenoids, geranyl diphosphate, farnesyl diphosphate, and geranylgeranyl diphosphate, are synthesized by polyisoprene synthases (EC 2.5.1.x; 12 NR sequences). The sesquiterpenes (E,E)- and (Z,E)-α-farnesene are produced from farnesyl diphosphate by the enzyme α-farnesene synthase. The gene encoding α-farnesene synthase has been isolated and shown to be up-regulated in fruit during ripening (Pechous and Whitaker, 2004). The α-farnesene synthase gene is represented by three NR sequences in the apple dataset. Other sesquiterpenes (e.g. β-caryophyllene, β-farnesene, germacrene D) and monoterpenes (e.g. ocimene, linalool) are produced by apple (Bengtsson et al., 2001); however, the terpene synthases responsible for their biosynthesis are yet to be identified.
Figure 4.
The terpene biosynthesis pathway via the mevalonate pathway. Apple sequences encoding enzymes involved in the mevalonate pathway were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively.
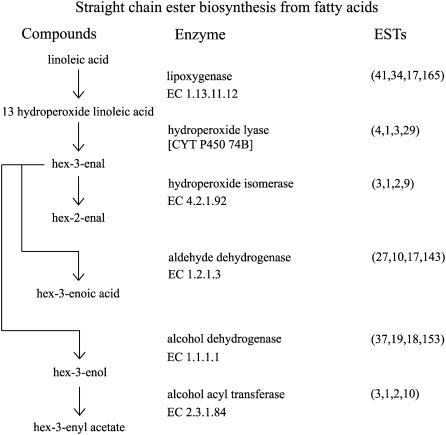
The major group of compounds produced from ripe fruit of apple cultivars, such as Royal Gala, is esters (Young et al., 1996, 2004), including straight-chain esters derived from fatty acids (Rowan et al., 1999) and branched-chain esters derived from branched-chain amino acids (Rowan et al., 1996). Of the straight-chain esters, C-6 constituents are thought to be derived via the lipoxygenase pathway from linoleic acid (Fig. 5). The first committed step is performed by members of the lipoxygenase family (EC 1.13.11.12), which produces 13-hydroperoxide linoleic acid from linoleic acid. A large number of candidate lipoxygenases have been found in the apple sequence dataset (41 NR sequences); however, not all of these will necessarily be involved in ester biosynthesis in fruit. In tomato, at least five lipoxygenase genes have been identified, but only one of these has been directly implicated in the production of flavor compounds (Chen et al., 2004). From 13-hydroperoxide linoleic acid, the cytochrome P450, hydroperoxide lyase, is responsible for the conversion to the aldehyde, hex-3-enal (four NR sequences), which can be converted to hex-2-enal by another cytochrome P450, hydroperoxide lyase (EC 4.2.1.92; three NR sequences). Alcohol dehydrogenases can reduce the aldehydes to alcohols (EC 1.1.1.1; 37 NR sequences). To date, one alcohol dehydrogenase has been identified from apple and shown not to be under the control of ethylene (Defilippi et al., 2005b). In addition to alcohols, aldehydes can be converted to acids by aldehyde dehydrogenases (EC 1.2.1.3; 27 NR sequences). Alcohols are then able to be esterified with CoA acids by alcohol acyl transferases (EC 2.3.1.84; three NR sequences). For example, the common apple ester, hexyl acetate, is synthesized from hexanol and acetyl CoA. One apple alcohol acyl transferase (MpAAT1) has been well characterized. MpAAT1 is up-regulated during ripening in response to ethylene (Defilippi et al., 2005b; Souleyre et al., 2005). The MpAAT1 enzyme is able to synthesize a wide range of esters, including many found in Royal Gala fruit (Souleyre et al., 2005). It is possible that the other alcohol acyl transferases found in the apple sequence dataset may also be contributing to the production of volatile esters.
Figure 5.
The straight-chain ester biosynthetic pathway from fatty acids. Apple sequences encoding enzymes involved in straight-chain ester biosynthesis were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively.
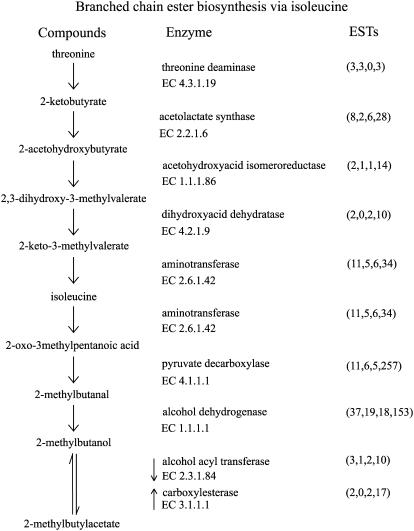
Branched-chain esters are characteristic of many cultivars of apple, including Royal Gala (Young et al., 1996, 2004). These are produced from Ile (Rowan et al., 1996), which increases in quantity in apple fruit skin during ripening (Defilippi et al., 2005a). Ile is synthesized from the amino acid Thr via five enzymatic steps. NR sequences representing each of these steps are present in the apple sequence dataset (Fig. 6). Ile is then metabolized first by branched-chain aminotransferases (EC 2.6.1.42; 11 NR sequences) and then pyruvate decarboxylase (EC 4.1.1.1; 11 NR sequences) to derive aldehydes available for alcohol dehydrogenases and alcohol acyl transferases to form esters. In addition to contributing to the pool of alcohols, branched hydrocarbons derived from Ile can also form CoA acids. For example, in addition to the branched-chain ester 2-methyl butyl acetate, the same branched chain can form ethyl-2-methyl butanoate, the predominant branched-chain ester found in Granny Smith apples (Rowan et al., 1996). Once produced, esters are available to be hydrolyzed by esterases (EC 3.1.1.1; two NR sequences). Such esterases are perhaps responsible for the large quantities of alcohols found in very ripe apple fruit and apple juice.
Figure 6.
Branched-chain ester biosynthesis via Ile. Apple sequences encoding enzymes involved in branched-chain ester biosynthesis were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively.
Color and Health-Related Compound Biosynthesis
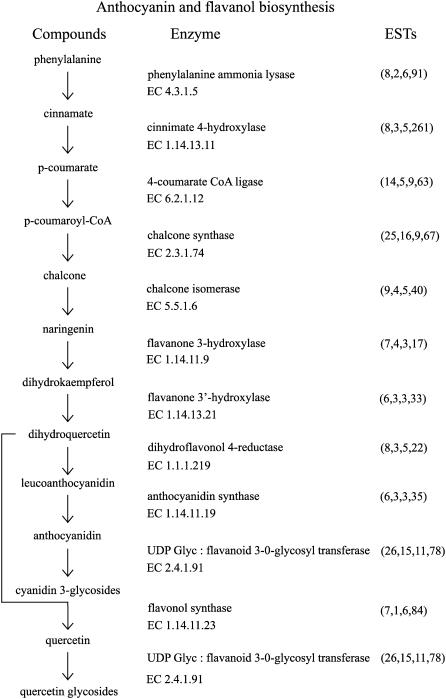
Flavonoids, including anthocyanins and flavanols, are a class of secondary metabolites, derived from the amino acid Phe, that impart important beneficial health attributes probably through their antioxidant activity (Wolfe et al., 2003; Boyer and Liu, 2004). Representative anthocyanins and flavanols are found in apple (McGhie et al., 2005). The major anthocyanins in apple are glycosylated cyanidins, which produce the red color observed in the fruit of many cultivars, including Royal Gala. The flavanol quercetin is found in apple fruit and has also been associated with health benefits. Representatives of the genes involved in flavonoid biosynthesis are present in the apple sequence dataset (Fig. 7). There are multiple NR sequences for all the genes in the pathway, including Phe ammonia lyase (EC 4.3.1.5; eight NR sequences), cinnamate 4-hydroxylase (EC 1.14.13.11; eight NR sequences), 4-coumarate CoA ligase (EC 6.2.1.12; 14 NR sequences), chalcone synthase (EC 2.3.1.74; 25 NR sequences), chalcone isomerase (EC 5.5.1.6; nine NR sequences), flavanone 3-hydroxylase (EC 1.14.11.9; seven NR sequences), and flavanone 3′-hydroxylase (EC 1.14.13.21; six NR sequences). From dihydroquercetin to the production of anthocyanins, the pathway proceeds to leucoanthocyanidins produced by dihydroflavonol reductase (EC 1.1.1.219; eight NR sequences) then to anthocyanidins by anthocyanidin synthase (EC 1.14.11.19; six NR sequences). Finally, the red-colored cyanidin 3-glycosides are formed through the transfer of a sugar onto a hydroxyl group by a glycosyl transferase (EC 2.4.1.91; 26 NR sequences). Also, from dihydroquercetin, quercetin can be synthesized by flavonol synthase (EC 1.14.11.23; seven NR sequences), which in turn can be glycosylated by glycosyl transferases. Some members of these gene families that are expressed in the apple skin have been isolated and shown to be inducible by UV and coordinately up-regulated in the skins of red apple varieties (Kim et al., 2003; Ben-Yehudah et al., 2005). In addition, members of the MYB transcription factor family have been identified that can interact with promoters of these genes (Hellens et al., 2005). Furthermore, one MYB (MdMYB10; one NR sequence) has been identified that up-regulates this pathway in apple skin (R.V. Espley, R.P. Hellens, J. Putterill, and A.C. Allan, personal communication).
Figure 7.
Anthocyanin and flavanol biosynthesis in apple. Apple sequences encoding enzymes involved in anthocyanin and flavanol biosynthesis were identified by BLASTx (e-05 cutoff) using the PIR NREF database (Wu et al., 2003). Numbers in parentheses under ESTs refer to the number of apple NR sequences, singletons, TC sequences, and total number of ESTs, respectively.
Gene Family Evolution
Within the apple sequence dataset, there are representatives of many large gene families involved in the biosynthesis of phytochemicals, such as the flavor and health compounds described above. Such multigene families include the acyl transferases, methyl transferases, glycosyl transferases, and cytochrome P450s. We have compared the predicted amino acid sequences of members of selected biosynthetic gene families from Arabidopsis and apple using phylogenetic methods to identify clades where apple genes may have expanded in number, presumably by gene duplication. An example of this type of analysis is shown for the acyl transferases (Fig. 8), a gene family that contains members that are involved in ester biosynthesis in apple (Souleyre et al., 2005). For this gene family, there is at least one clade with more representatives from apple than Arabidopsis. Expansions of gene number in apple are frequent in genes that are found in fruit cDNA libraries. For example, there have been more expansions of clades in apple P450s that contain representative ESTs from fruit libraries over duplicated genes containing only ESTs from exclusively nonfruit libraries (15 versus four duplicated genes). Comparisons of numbers of nonduplicated P450 genes expressed in fruit compared with genes that contained ESTs exclusively from nonfruit were 13 versus 11 nonduplicated genes. We also examined gene families involved in gene regulation because presumably additional transcription factor families and control genes might be required to regulate new biosynthetic pathways. Transcription factor gene families also show an expansion of clades of apple members of these families that are found in cDNA libraries made from fruit tissues. For example, the bZIP transcription factors contain eight duplicated genes that were expressed in fruit tissues compared to three that were not, whereas the MYB transcription factors contain nine expressed in fruit compared to six that are not. Comparisons for orthologous genes between fruit- and nonfruit-expressed genes for the bZIPs were four versus eight and for the MYBs were 16 versus 10.
Figure 8.
Phylogenetic tree of Arabidopsis (AtATs) and apple (numbers) members of the acyl transferase family (AT). Apple ATs that have duplicated in the apple lineage are colored green, orthologous apple ATs are colored red, and apple ATs for which assignment is ambiguous are colored blue. The whole-apple image on the right of the apple ATs identifies those that include ESTs from fruit tissue cDNA libraries, whereas the crossed apple image identifies apple ATs that do not have EST representatives from fruit libraries.
DISCUSSION
We report here a significant sample of transcripts taken from 43 apple cDNA libraries. We constructed cDNA libraries from various tissue types, but with a bias toward fruit tissues. A staged series of developing and then ripening Royal Gala fruit were sampled for ESTs (53,620 ESTs). This series included flower, whole fruit, fruit cortex, skin, and seed samples. Such a series will become a useful resource of genes for experiments aimed at understanding important processes and transformations in fruit development, such as early cell proliferation, cell expansion, and, finally, ripening. Also, from these libraries will come genes encoding enzymes and transcription factors involved in the biosynthesis of health and flavor compounds from apple fruit. Other major plant tissues were also sampled, including buds, shoots, leaves, roots, phloem, and xylem (76,472 ESTs). Finally, many genes are only expressed in response to external effects. We therefore sampled ESTs from various tissues that had been exposed to biotic and abiotic stresses. These included harvested fruit stored at low temperature and altered storage atmospheric conditions, leaves that had been infected with the fungal pathogen Venturia inaequalis and exposed to high temperature, and fruit cell lines that had been exposed to boron (21,595 ESTs).
As is typical for EST gene-sampling strategies, there is a high degree of redundancy in the sequences collected. Clustering of the sequences reduced the number of sequences to 43,938 NR sequences composed of 17,460 TC sequences and 25,478 singletons. The proportion of singletons compared to the total number of ESTs can provide a measure of the overall contribution of the library to the dataset. No single library contained more than 8% of the total number of singletons, indicating that much of the diversity is derived by sequencing different sources of tissue. The AARA library contained the greatest proportion of singletons per total number of apple ESTs (7.8%), with the next highest being the library sequenced to greatest depth, the leaf library AVBC (6.7% of singletons) that contained 11.8% of all apple sequences. Sequencing a number of different genotypes is also a good strategy for identifying new genes. The extreme of this is illustrated by a comparison of the NR clusters shared between the two largest expanding leaf libraries from the cultivars Royal Gala (AELA, 2,629 NR sequences) and Pinkie (AEPA, 2,074 NR sequences). These two libraries only share 14 NR sequences between them, which comprises only 0.3% of the total NR sequences represented in the combined dataset of the two libraries (4,689). These differences are not solely due to genotype-specific expression profiles, but also will include differences introduced by the two separate cloning procedures involved with making the libraries. Tissues where further sequencing would be useful are indicated by the percentage of singletons by library figures. High percentages of singletons in libraries such as AYFB (36.4%), AAFB (32%), AAMA (31.3%), and AAOA (32.3%) suggest that these libraries could be targeted for sampling for further genes from apple.
Overall, it is expected that 43,938 NR sequences is an overestimate of the number of protein-coding transcripts (protein-coding genes) represented in apple and that more sequencing, both of the cDNAs sampled here and novel cDNAs from apple, would reduce this number of NR sequences. Other EST projects undertaken in fruit crops of a similar size in terms of total number of ESTs collected have reported lower numbers of NR sequences. For example, a study of 152,635 tomato ESTs produced 31,012 NR sequences (Fei et al., 2004), whereas a collection of 146,075 grape ESTs rendered 25,746 NR sequences (Goes da Silva et al., 2005). This is likely due to the higher clustering threshold (95%) used in this study compared to the tomato and grape studies (90%). If the apple EST dataset is analyzed using a 90% clustering threshold, similar numbers of NR sequences are attained in the other fruit EST studies (34,614 NR sequences composed of 16,756 TC sequences and 17,858 singletons). However, even this lower number of NR sequences is likely to be an overestimate of the number of genes in the apple genome. The Arabidopsis unigene set estimated from all Arabidopsis ESTs produces a 35% overestimate of the actual number of protein-coding genes estimated from the genome sequence. Using this figure, the predicted actual number of apple genes may be approximately 27,000. However, when data from full cDNA sequences are incorporated and homology to Arabidopsis genes is taken into account, it is likely the apple NR set presented here represents approximately one-half the number of expressed genes found in apple.
A common feature of the cDNA sequences obtained from apple, and indeed other plants (Morgante et al., 2002), is the high frequency of SSRs contained within them, with 8,028 of the 43,938 apple NR sequences (19%) containing di- or trinucleotide repeats. Dinucleotide repeats were most frequent in the 100 bp immediately 5′ of the presumptive start AUG, whereas trinucleotide repeats were more common in the coding region. By far the most common class of dinucleotide repeat were AG repeats, making up 88.3% of all dinucleotide repeats. Least frequent were CG repeats at 0.1%. This bias toward AG and/or against CG repeats may be due to the tendency of CpG sequences to be methylated (Finnegan et al., 1998), which potentially might inhibit transcription. Another interesting feature of apple SSRs is the difference between the relative frequency of AG to the other dinucleotide repeat types in transcribed sequences compared with those found in genomic DNA (Guilford et al., 1997). For example, in apple genomic DNA, the AG repeats are approximately 60% more common than AC repeats, whereas in transcribed sequences AG repeats are almost 22 times (i.e. 2,200%) more common than AC repeats. A similar bias is found in Arabidopsis (Zhang et al., 2004). One possible explanation for this phenomenon is that there is an active role being played by these AG repeats in plant species. This could also account for why they are so common in transcribed sequences compared to other repeats. Factors that bind AG repeats in regulatory regions are known in both animals and plants (Epplen et al., 1996; Sangwan and O'Brian, 2002; Iglesias et al., 2004). Other potential ways SSRs affect regulation is by hypermethylation and/or secondary structure (Jacobsen et al., 2000).
Numerous SNPs were detected in the apple sequence dataset. From a cumulative length of 13.0 Mb of contiguous NR sequences sampled, 18,408 bi-allelic SNPs were detected. Bi-allelic SNPs occur with a frequency of one in every 706 bp of sequence. This is a relatively high level of variation probably due to two factors. The apple NR sequences, while predominantly from the cultivar Royal Gala, also contain sequences from six other cultivars, including Aotea, Braeburn, Pacific Rose, Pinkie, M9, and Northern Spy. Also, apple utilizes a strong incompatibility system selecting against self-crosses. Therefore, high levels of heterozygosity are expected. The ratio of transitions to transversions in the apple bi-allelic SNPs is close to 1:1, with 52.7% transitions. Similarly, in a SNP analysis of a comparison of the Columbia and Landsberg erecta accessions of Arabidopsis, 52.8% of the SNPs were transitions (Jander et al., 2002). With the advent of high-throughput detection systems, these SSRs and SNPs will form a large resource for mapping and marker-assisted breeding programs in apple and closely related crops.
Knowledge of GC content of a genome and codon usage is useful when devising PCR-based strategies for mapping and gene isolation, as well as for hybridization studies by microarray. The GC content in the third base position of the full-length cDNA sampled (52% GC) is higher than the overall GC ratio of 44% from the sequences of the NR sequences. This indicates some pressure to a more balanced GC ratio in coding regions compared with UTRs. Similar GC ratios in coding regions are found in grape (51%) and pear (Pyrus communis; 52%). Overall, the codon usage of apple shares many similarities with that of other dicots represented in the codon usage database (Nakamura et al., 2000). Apple codon usage differs markedly from Arabidopsis for 12 amino acids. Further comparisons with grape, pear, peach (Prunus persica), loblolly pine (Pinus taeda), poplar, tomato, citrus, potato (Solanum tuberosum), and tobacco (Nicotiana tabacum) showed that apple codon preference is most similar to that of grape and pear, differing from grape only in its preference for His (CAC), Leu (TTG), and Ser (TCT) and to pear in its preference for an additional three codons, Arg (AGG), Val (GTG), and the stop codon (TGA). The codon usage is correlated with trinucleotide repeat class frequency (R2 = 0.75). These are also correlated in Arabidopsis (Zhang et al., 2004), arguing that codon usage is selecting repeat classes because most trinucleotide repeats are found in the coding region (Morgante et al., 2002). CpG suppression is also evident in apple with a XCG:XCC ratio of 0.65, similar to that of tomato (0.58). This modest level of suppression of the CpG dinucleotides differs markedly from that of nearly no suppression in Arabidopsis (0.92) to the high level found in grape (0.35). This may well reflect different levels of methylation in the coding sequences used by different species of plants.
Predictive bioinformatics methods were employed to suggest the function of encoded proteins predicted from the apple NR sequences. Analyses included BLASTx, comparison with the MIPS-based role classification of Arabidopsis, and matches to the Inter-Pro protein family. Overall, many gene families commonly found in plant genomes are represented in the apple sequence dataset. Only approximately 5% of the apple NR sequences did not have a match in the Arabidopsis genome. Of the apple NR sequences, the most frequently represented class of genes were the protein kinases, followed by Leu-rich repeat (LRR) proteins and RNA-binding proteins. The ability to rapidly predict gene function using bioinformatic methods will speed efforts to identify genes likely to be involved in certain economically important traits. For example, the LRR class of protein (IPR001611) contains 321 NR sequences from apple, including genes involved in disease resistance, RNases involved in self-incompatibility, and many other cellular processes where protein-protein interactions are important. Included within the LRR class (IPR001611) are the NBS-LRR type of resistance genes, of which there are 59 NR sequences within the apple sequence dataset (e-20). In another Inter-Pro class (IPR007090) of plant-specific LRRs, 47 NR sequences were found in the apple sequence dataset. Disease-resistance gene candidates will also be common in other Inter-Pro classes; for example, the NB-ARC (IPR002182) and TIR (IPR000157) domain classes (63 and 56 NR sequences, respectively) are likely to consist largely of genes involved with disease resistance. In addition, the pathogenesis-related transcriptional factor and ERF (IPR001471) and protein kinase (IPR000719) classes (63 and 564 NR sequences, respectively) are also likely to contain a subset of genes that play important roles in plant defense. Other functional classes of proteins, such as many putative transcription factors, could be identified in our database. We compared the frequency of the most common transcription factor families with similar data available from the fully sequenced plants Arabidopsis (Riechmann et al., 2000) and rice (Goff et al., 2002), and found the rankings between these three species to be quite similar. The MADS-box family, ranked 17th in our analysis, was the only striking omission in apple from the top 10 in both Arabidopsis and rice.
The biosynthesis and maintenance of flavor and health compounds in fruit are important agronomic traits in apple. Presumably, such compounds have evolved as attractants and potential rewards for seed dispersers. Apple fruit produce more than 200 volatile flavor compounds, including alcohols, aldehydes, esters, ketones, and sesquiterpenes (Dimick and Hoskin, 1983). Apple also contains health-promoting phytochemicals, including complex sugars, acids, and vitamins, as well as secondary metabolites, such as polyphenolics and triterpenes (Boyer and Liu, 2004; Ma et al., 2005). Gene families potentially involved in the biosynthesis of flavor and health-related secondary metabolites are well represented within the apple NR sequences. We have shown this in two ways. First, enzymes involved in the particular biosynthetic pathways that produce secondary metabolites associated with flavor and health traits are well represented in the apple EST dataset (Figs. 4–7). Second, the classes of enzyme often found in these secondary metabolite pathways are well represented, including the cytochrome P450s (IPR001128, 159 in apple) and transferase classes (IPR003480, 62 in apple; IPR004159, 59 in apple; Table VI), for example. One hypothesis to explain how fruit have evolved the ability to produce these compounds is by duplicating these biosynthetic genes (Ohno, 1970), with the duplicate being co-opted by a biosynthetic pathway that operates in fruit. In addition, new regulatory genes would also be required to coordinate the expression of these new genes and potentially entire new biosynthetic pathways (Grotewold, 2005). We have tested the hypothesis that biosynthetic genes and regulatory genes have duplicated within the lineage, leading to apple more often for genes expressed in fruit tissues compared with genes not expressed in fruit tissues. Because our sample of apple genes is not complete, we can only infer duplications relative to a plant for which the full complement of genes is known (e.g. Arabidopsis). We find evidence for more frequent duplications of genes expressed in fruit tissues than those that are not expressed in fruit. An explanation for these results may be that we have sampled genes preferentially from fruit libraries. However, less than one-half the apple ESTs are derived from fruit libraries. Another possible explanation is that there has been gene loss in the lineages leading to the Arabidopsis genes. Given these limitations, however, it will be interesting in the future to determine just how many members of these gene families are involved in compound biosynthesis and whether gene family expansion has played an important role in allowing apple fruit to produce these compounds (Schwab, 2003) and how different biosynthetic genes have been recruited to make new secondary compounds (Ober, 2005).
There are two extreme types of gene duplication that could have given rise to the phylogenetic pattern we observe: either whole-genome duplication or multiple local duplication events. A palaeopolyploidy event has been predicted in the origin of the Maloideae (Lespinasse et al., 2000) that may well have provided an opportunity to expand the set of secondary metabolite genes and their regulators. In Arabidopsis, however, Maere et al. (2005) observed that regulatory proteins, such a transcription factors and signal transduction proteins that duplicated by palaeopolyploidy, tended to be retained, whereas these genes appear to be more rapidly lost when derived by segmental duplication. They estimate that these whole-genome duplication events are responsible for about 90% of the transcription factor in higher plants. Indeed, the rate of transcription factor evolution is thought to be higher in plants than in animals (Shiu et al., 2005). This contrasts with proteins involved in secondary metabolism or abiotic stress, where those derived by small segmental duplications tended to be retained, whereas those derived by whole-genome duplications were more rapidly lost (Maere et al., 2005). If this pattern of evolution is conserved in apple, we would expect many of the genes in secondary metabolism to be derived from segmental duplication events and therefore to be clustered.
In summary, we present an extensive set of ESTs representing what we predict is approximately one-half of the expressed genes from apple. The dataset contains SSR and SNP markers that will be useful for breeding, as well as many genes that can be tested directly for their roles in various crop traits. This gene set is also forming the basis for a microarray for apple that is being used in experiments to further identify genes encoding biosynthetic enzymes and their regulators.
MATERIALS AND METHODS
Library Construction and EST Sequencing
Tissues were collected from apples at HortResearch sites in Auckland and Havelock North in New Zealand over two seasons (see Table I for details of tissues, cultivar, and treatments). Total RNA was extracted from apple tissues by the method of either Lopez-Gomez and Gomez-Lim (1992) or Chang et al. (1993). Messenger RNA was isolated from total RNA by passage through oligo(dT)-cellulose columns (Amersham Biosciences), and either phage (Zap-cDNA synthesis kit and Zap-cDNA gigapack III gold cloning kit; Stratagene) or plasmid cDNA libraries (Superscript system for cDNA synthesis and cloning; Invitrogen) were constructed. Normalized libraries were produced essentially as described in normalization method 4 from Bonaldo et al. (1996), with the following modifications. Single-stranded DNA (ssDNA) was prepared from an apple (cv Pinkie) leaf library by production of M13 phage from the library and isolation of phage DNA, as described by Sambrook et al. (1989), and ssDNA and double-stranded DNA was selected using QiaexII resin, according to the manufacturer's instructions. ssDNA was hybridized with PCR-amplified driver DNA before isolation of rare ssDNA using hydroxyapatite chromatography. Rare ssDNA was made double stranded and transformed into DH10B using electroporation, as described by Bonaldo et al. (1996). Plasmids from the phage cDNA libraries were mass excised, according to the manufacturer's recommendations (Stratagene). Plasmid extractions were then undertaken on individual bacterial colonies of either the phage-derived or the plasmid-derived cDNA libraries and the corresponding cDNA inserts sequenced predominantly from the 5′ end. Big Dye Terminator sequencing reactions were resolved on ABI377, ABI3100, or ABI3700 sequencers, according to the manufacturer's instructions (Applied Biosystems).
For determination of the complete sequence of cDNA clones, M13R and M13F or T3 and T7 primers were used for 5′ confirmatory resequencing and 3′ end sequencing. In situations where EST clones had long poly(A) tails (generally >40 nucleotides) and therefore failed to yield good-quality sequence with standard sequencing primers, an anchored T24VN primer was used. Resulting sequences were edited manually and assembled using Sequencher software, version 4.0.5 (GeneCodes). Sequencing progress for each cDNA library was assessed manually for clone length and sequence quality. Decisions were made on the depth a library was sequenced to based on the levels of predicted sequence redundancy. This resulted in libraries made from meristematic tissues being sequenced to greater depths than libraries made from other tissues (see Table I).
Bioinformatics
EST sequences were automatically trimmed of vector, adapter, and low-quality sequence regions, and uploaded to a relational database. Automatic annotation was performed using the HortResearch BioPipe sequence annotation pipeline (a cluster-based annotation system written in PERL [R.N. Crowhurst, unpublished data]) and utilizing a relational database (MySQL; http://www.mysql.com). The EST clustering phase was performed using The Institute for Genomic Research (TIGR) gene indices clustering tools (http://www.tigr.org/tdb/tgi/software). The representation of protein families, domains, and functional sites within the apple NR sequences was determined using Inter-ProScan. The proteome for Arabidopsis (Arabidopsis thaliana) was obtained from The Arabidopsis Information Resource (TAIR; http://Arabidopsis.org; Garcia-Hernandez et al., 2002), and comparisons to proteins from Arabidopsis using BLASTx (Altschul et al., 1990) were used to identify apple NR sequences with similarity to Arabidopsis proteins. These apple NR sequences were then categorized into 21 functional categories based on functional annotations available for Arabidopsis proteins following MIPS (http://mips.gsf.de) FunCat schema (Ruepp et al., 2004). Apple sequences encoding enzymes involved in secondary metabolite biosynthetic pathways were identified by BLASTx (e-05 cutoff) using the Protein Information Resource (PIR) NREF database (Wu et al., 2003).
Detection of SSRs was undertaken using the PERL program within BioPipe that identified tandem repetition of sequence words in target sequences. SSRs were characterized by repeat type (di-, tri-, or tetranucleotide repeat units), repeat length, and position. For the purpose of reporting the frequency of repeat classes, different di- and trinucleotide sequences were combined by type; for example, AG repeats also encompassed repeats identified as GA and their complementary sequences CT or TC repeats. The repeat motifs combined are described in detail in Table II.
Prediction of SNPs and insertion/deletions and sequencing errors was performed using PERL scripts that parsed the output of contig sequences generated by the CAP3 (Huang and Madan, 1999) sequence assembly program running as part of the TIGR gene indices clustering tools.
Codon usage tables were derived from sequences of cDNAs encoding predicted full-length proteins. Clones were predicted to be full length only if they started with an ATG codon at a similar position to that of other plant genes (or have an in-frame stop codon upstream of the putative ATG) and end with an in-frame stop codon at a position equivalent to that of other plant genes. Codon usage was calculated from sequences using the CUSP program implemented within EMBOSS (Rice et al., 2000).
Members of gene families from Arabidopsis were extracted from GenBank and compared with predicted full-length family members from apple. Alignments and trees were constructed using ClustalX (version 1.81) using the default settings (Thompson et al., 1997). TreeView (version 1.6.6) was used to display resulting trees (Page, 1996).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CN848772 to CN851520, CN851527 to CN852114, CN854524 to CN860109, CN860111 to CN861528, CN861730 to CN862087, CN862091 to CN865258, CN865263 to CN870966, CN870969 to CN875894, CN875896 to CN881602, CN881608 to CN881609, CN881619 to CN884429, CN884434 to CN886998, CN887004 to CN890357, CN890361 to CN890409, CN890413 to CN896142, CN896144 to CN900284, CN900286 to CN901293, CN901299 to CN906863, CN906869 to CN907638, CN907715 to CN914192, CN914230 to CN914912, CN916097 to CN920835, CN920840 to CN925026, CN925028 to CN925934, CN925939 to CN929310, CN929396 to CN932721, CN932727 to CN933610, CN933676 to CN937515, CN937517 to CN943462, CN943466 to CN949201, CN949206 to CN949208, CN949216 to CN949629, CV126090 to CV126104, CV126106 to CV126115, DR033885 to DR033893, EB105831 to EB157590, and EB175250 to EB178034.
Acknowledgments
We thank Robert Simpson, Dave Greenwood, Maysoon Rasam, Matt Templeton, and Ross Atkinson for their work on gene annotation systems; David Chagne for advice on SNPs; Colm Carraher and Tim Holmes for graphics; and Ian Ferguson and Richard Forster for support. The ESTs reported in this article were sequenced at Genesis Research and Development Corporation, Auckland, New Zealand.
This work was supported by the Foundation for Research, Science, and Technology (grant no. C06X0207), and the Horticultural and Food Research Institute of New Zealand Limited.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard D. Newcomb (rnewcomb@hortresearch.co.nz).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076208.
References
- Adams MD, Soares MB, Kerlavage AR, Fields C, Venter JC (1993) Rapid cDNA sequencing (expressed sequence tags) from a directionally cloned human infant brain cDNA library. Nat Genet 4: 373–380 [DOI] [PubMed] [Google Scholar]
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Pharmacol Sci 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Keizer LCP, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AMML, De Vos RCH, van der Voet H, et al (2000) Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A, O'Connell AP (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–820 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9: 208–218 [Google Scholar]
- Bengtsson M, Backman A-C, Liblikas I, Ramirez MI, Borg-Karlson A-K, Ansebo L, Andreson P, Lofqvist J, Witzgall P (2001) Plant odor analysis of apple: antennal response of codling moth females to apple volatiles during phonological development. J Agric Food Chem 49: 3736–3741 [DOI] [PubMed] [Google Scholar]
- Ben-Yehudah G, Korchinsky R, Redel G, Ovadya R, Oren-Shamir M, Cohen Y (2005) Colour accumulation patterns and the anthocyanin biosynthetic pathway in ‘Red Delicious’ apple variants. J Hortic Sci Biotechnol 80: 187–192 [Google Scholar]
- Bieleski RL (1982) Sugar alcohols. In FA Loewus, W Tanner, eds, Plant Carbohydrates I. Intracellular Carbohydrates. Encyclopaedia of Plant Physiology New Series, Vol 13A. Springer, New York, pp 158–192
- Bonaldo MF, Lennon G, Soares MB (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res 6: 791–806 [DOI] [PubMed] [Google Scholar]
- Boyer J, Liu RH (2004) Apple phytochemicals and their health benefits. Nutr J 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challice JS (1974) Rosaceae chemotaxonomy and the origin of the Pomidae. Bot J Linn Soc 69: 239–259 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D (2004) Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol 136: 2641–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhou R, Reidel EJ, Sharkey TD, Dandekar AM (2005) Antisense inhibition of sorbitol synthesis leads to up-regulation of starch synthesis without altering CO2 assimilation in apple leaves. Planta 220: 767–776 [DOI] [PubMed] [Google Scholar]
- Dandekar AM, Teo G, Defilippi BG, Uratsu SL, Passey AJ, Kader AA, Stow JR, Colgan RJ, James DJ (2004) Effect of down-regulation of ethylene biosynthesis on fruit flavour complex in apple fruit. Transgenic Res 13: 373–384 [DOI] [PubMed] [Google Scholar]
- Defilippi BG, Dandekar AM, Kader AA (2005. a) Relationship of ethylene biosynthesis to volatile production, related enzymes, and precursor availability in apple peel and flesh tissues. J Agric Food Chem 53: 3133–3141 [DOI] [PubMed] [Google Scholar]
- Defilippi BG, Kader AA, Dandekar AM (2005. b) Apple aroma: alcohol acyltransferase, a rate limiting step for ester biosynthesis, is regulated by ethylene. Plant Sci 168: 1199–1210 [Google Scholar]
- Dimick PS, Hoskin JC (1983) Review of apple flavor—state of the art. Crit Rev Food Sci Nutr 4: 387–409 [DOI] [PubMed] [Google Scholar]
- Epplen JT, Kyas A, Maueler W (1996) Genomic simple repetitive DNAs are targets for differential binding of nuclear proteins. FEBS Lett 389: 92–95 [DOI] [PubMed] [Google Scholar]
- Fei ZJ, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ (2004) Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J 40: 47–59 [DOI] [PubMed] [Google Scholar]
- Fellman JK, Miller TW, Mattinson DS, Mattheis JP (2000) Factors that influence biosynthesis of volatile flavor compound in apple fruits. HortScience 35: 1026–1033 [Google Scholar]
- Ferree DC, Carlson RF (1987) Apple rootstocks. In RC Rom, RF Carlson, eds, Rootstocks for Fruit Crops. John Wiley & Sons, New York, pp 107–144
- Finnegan EJ, Genger RK, Peacock WJ, Dennis ES (1998) DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 223–247 [DOI] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Berardini TZ, Chen G, Crist D, Doyle A, Huala E, Knee E, Lambrecht M, Miller N, Mueller LA, et al (2002) TAIR: a resource for integrated Arabidopsis data. Funct Integr Genomics 2: 239–253 [DOI] [PubMed] [Google Scholar]
- Giovannoni J (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Goes da Silva F, Iandolino A, Al-Kayal F, Bohlmann MC, Cushman MA, Lim H, Ergul A, Figueroa R, Kabuloglu EK, Osborne C, et al (2005) Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol 139: 574–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Grimplet J, Romieu C, Audergon J-M, Marty I, Albagnac G, Lambert P, Bouchet J-P, Terrier N (2005) Transcriptomic study of apricot fruit (Prunus armeniaca) ripening among 13,006 expressed sequence tags. Physiol Plant 125: 281–292 [Google Scholar]
- Grotewold E (2005) Plant metabolic diversity: a regulatory perspective. Trends Plant Sci 10: 57–62 [DOI] [PubMed] [Google Scholar]
- Guilford PS, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus X domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94: 249–254 [Google Scholar]
- Harker FR, Gunson FA, Jaeger SR (2003) The case for fruit quality: an interpretive review of consumer attitudes, and preferences for apples. Postharvest Biol Technol 28: 333–347 [Google Scholar]
- Hellens RP, Allan A, Friel E, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias AR, Kindlund E, Tammi M, Wadelius C (2004) Some microsatellites may act as novel polymorphic cis-regulatory elements through transcription factor binding. Gene 341: 149–165 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Sakai H, Finnegan EJ, Cao X, Meyerowitz EM (2000) Ectopic hypermethylation of flower-specific genes in Arabidopsis. Curr Biol 10: 179–186 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Curry E (2000) Lovastatin inhibits α-farnesene synthesis without affecting ethylene production during fruit ripening in ‘Golden Supreme’ apples. J Am Soc Hortic Sci 215: 105–110 [Google Scholar]
- Kim S-H, Lee J-R, Hong S-T, Yoo Y-K, An G, Kim S-R (2003) Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci 165: 403–413 [Google Scholar]
- Knee M (1993) Pomme fruits. In GB Seymour, JE Taylor, GA Tucker, eds, Biochemistry of Fruit Ripening. Chapman and Hall, London, pp 325–346
- Lespinasse D, Grivet L, Troispoux V, Rodier-Goud M, Pinard F, Seguin M (2000) Identification of QTLs involved in the resistance to South American leaf blight (Microcyclus ulei) in the rubber tree. Theor Appl Genet 100: 975–984 [Google Scholar]
- Loescher WH, Marlo GC, Kennedy RA (1982) Sorbitol metabolism and source-sink interconversions in developing apple leaves. Plant Physiol 70: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomez R, Gomez-Lim MA (1992) A method for extraction of intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 27: 440–442 [Google Scholar]
- Ma CM, Cai SQ, Cui JR, Wang RQ, Tu PF, Masao H, Mohsen D (2005) The cytotoxic activity of ursolic acid derivatives. Eur J Med Chem 40: 582–589 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie TK, Hunt M, Barnett LE (2005) Cultivar and growing region determine the antioxidant polyphenolic concentration and composition of apples grown in New Zealand. J Agric Food Chem 53: 3065–3070 [DOI] [PubMed] [Google Scholar]
- McKeon T, Yang SF (1987) Biosynthesis and metabolism of ethylene. In PJ Davies, ed, Plant Hormones and Their Role in Plant Growth and Development. Martinus Nijhoff, Boston, pp 94–112
- Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with non repetitive DNA in plant genomes. Nat Genet 30: 194–200 [DOI] [PubMed] [Google Scholar]
- Moser C, Segala C, Fontana P, Salakhudtinov I, Gatto P, Pindo M, Zyprian E, Toepfer R, Grando MS, Velasco R (2005) Comparative analysis of expressed sequence tags from different organs of Vitis vinifera L. Funct Integr Genomics 5: 208–217 [DOI] [PubMed] [Google Scholar]
- Moyle R, Fairbairn DJ, Ripi J, Crowe M, Botella JR (2005) Developing pineapple fruit has a small transcriptome dominated by metallothionine. J Exp Bot 56: 101–112 [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res 31: 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Gojobori T, Ikemura T (2000) Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negm FB, Loescher WH (1981) Characterization of aldose 6-phosphate reductase (alditol 6-phosphate: NADP 1-oxidoreductase) from apple leaves. Plant Physiol 67: 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober D (2005) Seeing double: gene duplication and diversification in plant secondary metabolism. Trends Plant Sci 10: 444–449 [DOI] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer-Verlag, New York
- Page RDM (1996) Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pechous SW, Watkins CB, Whitaker BD (2005) Expression of α-farnesene synthase gene AFS1 in relation to levels of α-farnesene and conjugated trienols in peel tissue of scald-susceptible ‘Law Rome’ and scald-resistant ‘Irared’ apple fruit. Postharvest Biol Technol 35: 125–132 [Google Scholar]
- Pechous SW, Whitaker BD (2004) Cloning and functional expression of an (E,E)-α-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219: 84–94 [DOI] [PubMed] [Google Scholar]
- Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5: 94–100 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin GLR, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rowan DD, Allen JM, Fielder S, Hunt MB (1999) Biosynthesis of straight-chain ester volatiles in Red Delicious and Granny Smith apples using deuterium-labeled precursors. J Agric Food Chem 47: 2553–2562 [DOI] [PubMed] [Google Scholar]
- Rowan DD, Lane HP, Allen JM, Fielder S, Hunt MB (1996) Biosynthesis of 2-methylbutyl, 2-methyl-2-butenyl, and 2-methylbutanoate esters in red delicious and Granny Smith apples using deuterium-labeled substrates. J Agric Food Chem 44: 3276–3285 [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sangwan I, O'Brian MR (2002) Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol 129: 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab W (2003) Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62: 837–849 [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Shih M-C, Li W-H (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souleyre EJF, Greenwood DR, Friel EN, Karunairetnam S, Newcomb RD (2005) An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavour. FEBS J 272: 3123–3144 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Tanksley MG (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a larger expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari J, Kobae Y, Yamaki S, Yamada K, Toyofuku K, Tabuchi T, Shiratake K (2004) Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant Cell Physiol 45: 1031–1041 [DOI] [PubMed] [Google Scholar]
- Wolfe K, Wu XZ, Liu RH (2003) Antioxidant activity of apple peels. J Agric Food Chem 51: 609–614 [DOI] [PubMed] [Google Scholar]
- Wu CH, Yeh L-SL, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, Suzek BE, et al (2003) The Protein Information Resource. Nucleic Acids Res 31: 345–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahia EM (1994) Apple flavour. Hortic Rev (Am Soc Hortic Sci) 16: 197–234 [Google Scholar]
- Young H, Gilbert JM, Murray SH, Ball RD (1996) Causal effects of aroma compounds on Royal Gala apple flavours. J Sci Food Agric 71: 329–336 [Google Scholar]
- Young JC, Chu CLG, Lu X, Zhu H (2004) Ester variability in apple varieties as determined by solid-phase microextraction and gas chromatography mass spectrometry. J Agric Food Chem 52: 8086–8093 [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–878 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yuan D, Yu S, Li Z, Cao Y, Miao Z, Qian H, Tang K (2004) Preference of simple sequence repeats in coding and non-coding regions of Arabidopsis thaliana. Bioinformatics 20: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Zhou R, Cheng L, Wayne R (2003) Purification and characterization of sorbitol-6-phosphate phosphatase from apple leaves. Plant Sci 165: 227–232 [Google Scholar]
- Zohary D, Hopf M (2000) Domestication of Plants in the Old World, Ed 3. Oxford University Press, New York