The immune response to viral infections comes in two well described flavors. First, innate immunity [for example, natural killer (NK) cells and IFN responses] nonspecifically recognizes viral infections to trigger autocrine and paracrine signals that limit viral replication. Second, adaptive immunity (for example, T cells and B cells) specifically recognizes viral infections through secreted and cell-meditated factors. However, for retroviral infections, there is a third component of viral recognition and subsequent restriction that has been called “intrinsic immunity” (1). Intrinsic immunity differs from innate and adaptive immunity in that it is cell autonomous (does not rely on secreted factors), it is present in many cells (rather than only in specialized immune cells), it does not need to be induced by viral infections, and it probably evolves with viral infections on a longer evolutionary time scale than the innate and adaptive immune responses. Trim5α is a cytoplasmic protein that forms part of the intrinsic immune system that restricts retroviral infections in primates. This protein targets the viral capsid (CA) protein in a species-specific manner, but the mechanism of how Trim5α inhibits retroviral infections has been unclear. In this issue of PNAS, Stremlau et al. (2) address this important question by showing that Trim5α from rhesus macaques inhibits HIV type 1 (HIV-1) by direct recognition of the viral capsid protein followed by accelerated “uncoating” of capsid from the incoming viral particle.
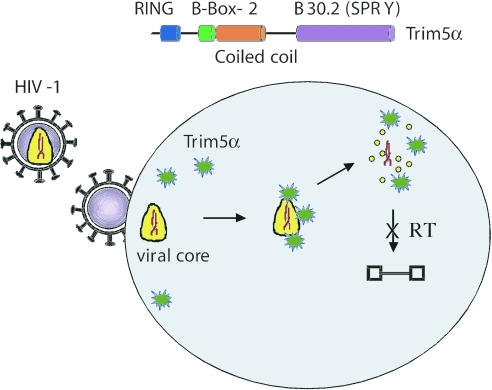
The importance of the Trim5α gene was originally discovered by the Sodroski laboratory because it protects cells derived from Old World monkeys (OWM) from infection by HIV-1 (3). This gene encodes a protein that is a member of the tripartite motif family defined by the presence of RING, B-box 2, and coiled-coil domains (4, 5). In addition, Trim5α is part of the subset of this family that also contains a C-terminal B30.2 (SPRY) domain (Fig. 1). Expression of Trim5α from OWM either in its natural context or in heterologous cells inhibits the accumulation of new viral genomes that are the product of the viral enzyme reverse transcriptase. The human version of Trim5α is not effective against HIV but does inhibit a mouse retrovirus, N-tropic murine leukemia virus (N-MLV).
Fig. 1.
Proposed mechanism of Trim5α. HIV (at left) enters a cell by membrane fusion and deposits its core (yellow) containing viral RNA (red) and other viral proteins into the cytoplasm. Trim5α (green) binds to the mutimeric CA on the viral core and leads to accelerated uncoating (free CA monomers as yellow circles). This activity prevents reverse transcription (RT) of the viral RNA into proviral DNA.
Using swaps of genes from related Trim5α-sensitive and -resistant viruses and also viral mutants, several groups identified the CA protein of retroviruses as the critical viral determinant for recognition by Trim5α (reviewed in ref. 6). Moreover, Trim5α restriction can be saturated by preinfecting cells with virus-like particles from a virus that is sensitive to the restriction but not with monomeric CA (7, 8). Thus, it was predicted that Trim5α recognized an epitope of CA that depends on the multimeric complex of CA formed only upon assembly of CA proteins into particles. Stremlau et al. (2) were able to demonstrate direct recognition (binding) of Trim5α for capsids by using a recombinant version of CA still fused to the nucleocapsid (NC) protein that was known to spontaneously form virus-like particles in vitro (9). This experiment confirms a previous report that had shown that human Trim5α could bind detergent-treated virions of N-MLV (10).
Because both virus-binding studies used Trim5α expressed in cell lysates (2, 10), it is still formally possible that there is an “adapter” protein that bridges Trim5α to viral capsids. However, this scenario is very unlikely because of the unusual genetics of restriction factors. That is, Trim5α shows strong evidence for positive selection in primates (11–13). Selection pressures that would lead to the accelerated evolution of Trim5α to recognize new viral infections that become endemic in a population are most simply explained by a direct recognition of Trim5α for new variants of CA. Indeed, the regions of Trim5α that have been identified as showing the highest amounts of positive selection in the B30.2 domain and, to a lesser extent, in the coiled-coil domain are those domains that confer specificity of restriction, and those two regions are also the two regions identified by Stremlau et al. (2) as being essential for the biochemical recognition of capsids by Trim5α.
When does Trim5α recognize the viral capsids? The stage of the viral life-cycle after entry into the cytoplasm but before completion of reverse transcription and entry into the nucleus is called uncoating, and it is thought that CA becomes dissociated from the viral particle shortly after entry. It has been proposed that this early uncoating step is essential for HIV to enter the nucleus before mitosis (14). Thus, for Trim5α to affect the virus after entry, it must affect the virus before it loses its capsid. Indeed, HIV-1 becomes resistant to restriction within minutes of infection (15). This result indicates that Trim5α acts at a very early stage of the viral life-cycle immediately after entry (Fig. 1).
Mutations in CA that both stabilize and destabilize the association of CA with the core affect reverse transcription (16). These results suggest that any cellular process that changes the kinetics of uncoating might affect reverse transcription. Thus, Stremlau et al. (2) considered three different possibilities regarding how rhesus Trim5α affects HIV uncoating: Trim5α could prevent uncoating, it could accelerate uncoating, or it could cause degradation of the capsids. This latter hypothesis was based on the E3 ubiquitin ligase activity of a shorter isoform of TRIM5, TRIM5δ, which depends on the RING domain that is present in both Trim5 isoforms (17). However, by using a sucrose gradient technique to capture intracellular CA protein and separate it into particulate (presumably still containing a higher-order structure) and soluble (disassembled) forms, they found that rhesus Trim5α does not cause an apparent proteolysis of HIV-1 capsids because bulk CA levels remain constant in the presence or absence of Trim5α (3). Rather, Trim5α causes a decrease in particle-associated CA. Significantly, they saw the same result when they examined the consequences of human Trim5α restriction of N-MLV. Because the association of CA with N-MLV is more stable than for HIV-1, they could directly follow the conversion of particle-associate N-MLV CA into soluble forms. Stremlau et al. (2) interpret these results to suggest that Trim5α executes its antiviral effects by disrupting what is usually an ordered process of uncoating and reverse transcription (Fig. 1).
How does Trim5α accelerate uncoating? The B-box and RING domains are essential for this effector (as opposed to recognition) function (18, 19). Because proteasome inhibitors had only a minor effect on the overall activity of Trim5α (2, 18), it is less likely that the ubiquitin-ligase activity of the RING domain is directly involved in degradation of a protein that holds the capsid together. Cyclophilin A, a peptidyl-prolyl isomerase, is important for the activity of rhesus Trim5α (20), and it is likely that other proteins are also involved in the process. The development of in vitro uncoating assays (16, 21) should shed light on whether Trim5α recruits factors that accelerate the usual uncoating process of HIV. One caveat to these speculations is that another form of Trim5α that is found in squirrel monkeys appears to exert its antiviral affect after, rather than before, reverse transcription (22). Thus, it is possible that multiple or alternative mechanisms of inhibition might exist.
What is the “normal” function of Trim5α? One could argue that it does not have one except as surveillance against retroviral infections because the rapid evolution of the gene (13) would mean that any normal function of the gene would also have to be evolving in parallel. Because the positive selection of Trim5α is centered on the B30.2 domain, it is possible that the other Trim5 isoforms that do not contain this domain (5) do have a normal function. However, this function is unlikely to be essential because there are humans with deleterious homozygous mutations in a conserved residue of a domain present in all of the isoforms (23).
Trim5α genes that have been cloned and tested from a variety of primates show that each one has a unique viral specificity, but in no case so far do they have substantial activity against retro-viruses that currently endemically infect that species. Because host populations evolve much more slowly than viral populations do, the recognition of a new virus will therefore always lag behind unless coincidental viral restriction already existed in the newly infected host. That is, the Trim5α that each species currently possesses protects that species from past infections (and possibly, serendipitously, future ones) but not current ones. Because nearly 8% of our genome is made of the relics of past infections in the form of extinct endogenous retroviruses (24), and because positive selection of Trim5α in primates dates back to >35 million years, some of these ancient events are presumably examples of prior episodes of Trim5α recognition and surveillance (13). Thus, our immunity to some retroviral infections endures far longer than any active immunological response. The elucidation of TRIM5 mechanism may facilitate pharmacological and genetic interventions to coax currently nonrestrictive human TRIM genes to target and restrict important pathogens like HIV-1.
Conflict of interest statement: No conflicts declared.
See companion article on page 5514.
References
- 1.Bieniasz P. D. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 2.Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D. J., Sundquist W. I., Sodroski J. Proc. Natl. Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 4.Meroni G., Diez-Roux G. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 5.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., et al. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Towers G. J. Hum. Gene Ther. 2005;16:1125–1132. doi: 10.1089/hum.2005.16.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodding M. P., Bock M., Yap M. W., Stoye J. P. J. Virol. 2005;79:10571–10577. doi: 10.1128/JVI.79.16.10571-10577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forshey B. M., Shi J., Aiken C. J. Virol. 2005;79:869–875. doi: 10.1128/JVI.79.2.869-875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganser B. K., Li S., Klishko V. Y., Finch J. T., Sundquist W. I. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 10.Sebastian S., Luban J. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H. L., Wang Y. Q., Liao C. H., Kuang Y. Q., Zheng Y. T., Su B. Gene. 2005;362:109–116. doi: 10.1016/j.gene.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 12.Song B., Gold B., O'Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer S. L., Wu L. I., Emerman M., Malik H. S. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita M., Emerman M. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Caballero D., Hatziioannou T., Zhang F., Cowan S., Bieniasz P. D. J. Virol. 2005;79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C. J. Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L., Yang L., Moitra P. K., Hashimoto K., Rallabhandi P., Kaul S., Meroni G., Jensen J. P., Weissman A. M., D'Arpa P. Exp. Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. J. Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speelmon E. C., Livingston-Rosanoff D., Li S. S., Vu Q., Bui J., Geraghty D. E., Zhao L. P., McElrath M. J. J. Virol. 2006;80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthoux L., Sebastian S., Sokolskaja E., Luban J. Proc. Natl. Acad. Sci. USA. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayan S., Young J. A. T. Proc. Natl. Acad. Sci. USA. 2004;101:7721–7726. doi: 10.1073/pnas.0401312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ylinen L. M., Keckesova Z., Wilson S. J., Ranasinghe S., Towers G. J. J. Virol. 2005;79:11580–11587. doi: 10.1128/JVI.79.18.11580-11587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawyer S. L., Wu L. I., Akey J. M., Emerman M., Malik H. S. Curr. Biol. 2006;16:95–100. doi: 10.1016/j.cub.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 24.Bannert N., Kurth R. Proc. Natl. Acad. Sci. USA. 2004;101(Suppl. 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]