Abstract
Histone arginine methylation is a posttranslational modification linked to the regulation of gene transcription. Unlike other posttranslational modifications, methylation has generally been regarded as stable, and enzymes that demethylate histone arginine residues have not been identified. However, it has recently been shown that human peptidylarginine deiminase 4 (PAD4), a Ca2+-dependent enzyme previously known to convert arginine residues to citrulline in histones, can also convert monomethylated arginine residues to citrulline both in vivo and in vitro. Citrullination of histone arginine residues by the enzyme antagonizes methylation by histone arginine methyltransferases and is thus a novel posttranslational modification that regulates the level of histone arginine methylation and gene activity. Here we present the crystal structures of a Ca2+-bound PAD4 mutant in complex with three histone N-terminal peptides, each consisting of 10 amino acid residues that include one target arginine residue for the enzyme (H3/Arg-8, H3/Arg-17, and H4/Arg-3). To each histone N-terminal peptide, the enzyme induces a β-turn-like bent conformation composed of five successive residues at the molecular surface near the active site cleft. The remaining five residues are highly disordered. The enzyme recognizes each peptide through backbone atoms of the peptide with a possible consensus recognition motif. The sequence specificity of the peptide recognized by this enzyme is thought to be fairly broad. These observations provide structural insights into target protein recognition by histone modification enzymes and illustrate how PAD4 can target multiple arginine sites in the histone N-terminal tails.
Keywords: calcium binding, histone modification, rheumatoid arthritis, protein deimination/citrullination, x-ray crystal structure
The eukaryotic genome is organized within the nucleosome core particle, which is composed of core histones (H2A, H2B, H3, and H4) and their associated 146 bp of DNA (1). A diverse array of core particle modifications, involving both histones and the DNA, regulate genomic functions. In particular, a variety of covalent modifications, including acetylation of lysines, phosphorylation of serines and threonines, methylation of lysines and arginines, and ubiquitination of lysines play a fundamental role in specific nucleosome regulations. These modifications are located on flexible and unstructured histone N- and C-terminal tails that protrude from the nucleosome core particle (2, 3).
Together with these modifications, Ca2+-dependent citrullination (or deimination) of histone arginine residues by peptidylarginine deiminase 4 (PAD4) (Fig. 1A) has come into focus as a novel posttranslational modification linked to transcriptional regulation in eukaryotes (4, 5). In fact, the enzyme has been shown to target multiple arginine sites in histones H3 (Arg-2, Arg-8, Arg-17, and Arg-26) and H4 (Arg-3), including those sites methylated by coactivator-associated arginine methyltransferase 1 (H3/Arg-17) and protein arginine methyltransferase 1 (H4/Arg-3) (6, 7) (Fig. 1A). Furthermore, transcription of the estrogen-regulated genes is activated by dimethylations of H3/Arg-17 and H4/Arg-3 but is suppressed by citrullination (6, 7). In fact, PAD4 is recruited to an estrogen-responsive gene (pS2 promoter) in MCF-7 cells with the subsequent appearance of citrullinated histones and the disengagement of RNA polymerase II (4, 5). In addition to these histone modifications, Arg-2142 of coactivator p300 is also both citrullinated by PAD4 and methylated by coactivator-associated arginine methyltransferase 1. These modifications result in a significant change of the assembly structure of the coactivator complex (8). These findings show that PAD4 participates in transcriptional regulation of gene expression by antagonizing protein arginine methylation by coactivator-associated arginine methyltransferase 1 and protein arginine methyltransferase 1.
Fig. 1.
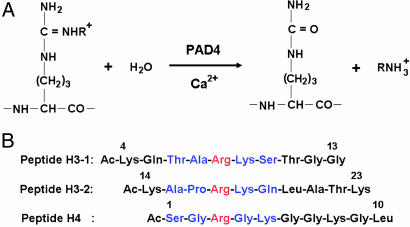
Citrullination (or deimination) of peptide substrates by PAD4. (A) Citrullination of arginine (R = H) and NG-monomethylarginine (R = CH3) residues by PAD4. Arginine residue is methylated to give NG-monomethylarginine, symmetric NG,NG-dimethylarginine, or asymmetric NG,NG-dimethylarginine by enzymes such as protein arginine methyltransferases (33). (B) Three histone N-terminal peptides, H3-1, H3-2, and H4. For each peptide, the target arginine residue is colored red and the five successive residues that form a β-turn-like bent conformation upon interaction with PAD4 are colored blue.
To date, five types of human PADs, PAD1–PAD4 and PAD6, have been characterized by cDNA cloning (9). Some of these PADs are associated with human diseases such as rheumatoid arthritis (RA) (10, 11), where autoantibodies against proteins citrullinated by PAD are found at early phases of the manifestation of the disease in the majority of RA patients. Moreover, the presence of these antibodies is very specific to RA (12). Furthermore, a significant association was reported between RA and functional variants of the gene encoding PAD4 in the Japanese population (13). PAD4 is expressed in bloodstream granulocytes and is the only type of PAD that has a nuclear localization signal and is thus localized in the cell nucleus (14).
In view of this consideration, we previously determined the crystal structures of Ca2+-free PAD4 and of a Ca2+-bound inactive mutant with and without a substrate, benzoyl-l-arginine amide (BA), and showed a Ca2+-induced activation mechanism of the enzyme (15). However, the substrate recognition by the Ca2+-bound inactive mutant for BA is likely to differ from the recognitions of physiological substrates, because BA is an artificial substrate having only one peptide bond between the benzoic acid and the arginine amide (16). To fully understand molecular recognition of natural target proteins by the enzyme, it is of great importance to use histone N-terminal tails as the substrate (4, 5, 17). In this report, we define the structural basis for the histone N-terminal recognition by determining the high-resolution crystal structures of the Ca2+-bound inactive C645A mutant in complex with the three histone N-terminal peptides shown in Fig. 1B.
Results and Discussion
The Three Histone N-Terminal Peptides Are Citrullinated by PAD4.
Arg-2, Arg-8, Arg-17, and Arg-26 in histone H3 and Arg-3 in histone H4 have been shown to be citrullinated both in vivo and in vitro by the enzyme, respectively (4, 5). However, citrullination of Arg-8, Arg-17, and Arg-3 in the chemically synthesized histone N-terminal peptides H3-1, H3-2, and H4 (Fig. 1B) has not been demonstrated. Therefore, we assessed whether these three chemically synthesized histone N-terminal peptides are citrullinated by the wild-type enzyme. Table 1summarizes PAD activities of the enzyme toward these three histone peptides. The PAD activities were 21.1, 19.6, and 46.0 for peptides H3-1, H3-2, and H4, respectively, when PAD activity of the enzyme toward BA is defined as 100. The results demonstrate that the enzyme targets the arginine residue in each histone N-terminal peptide and can convert it to a citrulline residue.
Table 1.
PAD activities toward BA and peptides H3-1, H3-2, and H4
| Peptide | PAD4 (wild type) | PAD4 (R374A) |
|---|---|---|
| BA | 100 | 5.7 |
| Peptide H3-1 | 21.1 | 0.4 |
| Peptide H3-2 | 19.6 | 0.6 |
| Peptide H4 | 46.0 | 1.0 |
PAD activity is shown relative to the wild-type enzyme (49.8 units/mg−1), which was defined as 100.
Structures of PAD4.
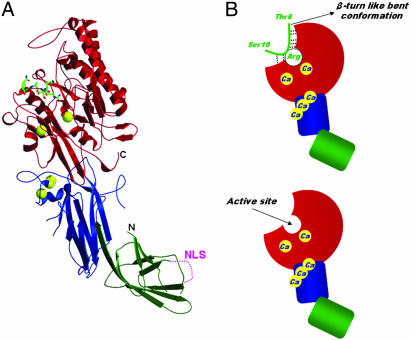
The respective Ca2+-bound PAD4 (C645A) structures in complexes with the three histone N-terminal peptides H3-1, H3-2, and H4 are similar to each other and also to the previously determined structure bound to BA (15). PAD4 has five non-EF-hand Ca2+ binding sites and adopts an elongated fold with two domains (Fig. 2A). The N-terminal domain consists of amino acid residues Met-1 to Pro-300 and is further divided into two Ig-like subdomains (subdomains 1 and 2). Subdomain 1 has nine β-strands and a nuclear localization signal (56PPAKKKST63) on the molecular surface in a loop region. Functional variants of the gene encoding the enzyme in the Japanese RA population are positioned in subdomain 1, far from the active site (which is in the C-terminal domain), and are unlikely to affect catalytic function directly. Subdomain 2 has ten β-strands, four short α-helices, and three Ca2+ ions. Residues Asn-301 to Pro-663 form the C-terminal domain, which has a structure of five ββαβ modules arranged circularly in a pseudofivefold symmetric structure called an α/β propeller (18). The active site cleft, in which each histone N-terminal peptide and two Ca2+ ions are bound (Fig. 2B), is found in the α/β propeller structure in the C-terminal domain.
Fig. 2.
Structure of Ca2+-bound PAD4 (C645A) in complex with peptide H3-1. (A) Ribbon representation of the structure. Ca2+ ions and the histone peptide are shown as yellow balls and as a green stick model, respectively. The N-terminal subdomains 1 (residues 1–118) and 2 (residues 119–300) and the C-terminal domain (residues 301–663) are colored green, blue, and red, respectively. The nuclear localization signal (NLS) region in the subdomain 1 is shown as a dotted line. (B Upper) Schematic representation of the structure shown in A. The colors of the N-terminal domain (subdomains 1 and 2) and the C-terminal domain are the same as in A. Dotted lines show hydrogen bonds that form a consensus recognition motif at the molecular surface near the active site. (B Lower) For reference, the structure of the Ca2+-bound PAD4 (C645A) is shown.
Recognition of Histone N-Terminal Peptides by PAD4.
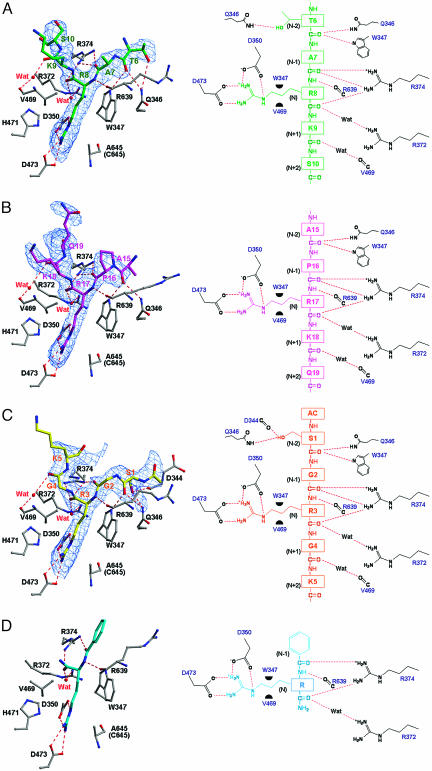
In the histone N-terminal peptides (each of which consists of ten residues), five successive residues from Thr-6H to Ser-10H (a superscripted H indicates a histone peptide residue), from Ala-15H to Gln-19H, and from acetylated Ser-1H to Lys-5H (Fig. 1B) were unambiguously assigned in Fo − Fc maps for the complexes with peptides H3-1, H3-2, and H4, respectively. However, the electron densities that correspond to the side-chain moieties of Lys-9H in peptide H3-1 and Lys-5H in peptide H4 are highly smeared (Fig. 3A and C). These five residues are positioned at (N − 2), (N − 1), N, (N + 1), and (N + 2), respectively, where “N −” and “N +” define the positions immediately before and after the target arginine residue (Fig. 3 A–C). The remaining five residues are highly disordered and flexible in the three structures. This result indicates that the peptide recognition as presented here (Figs. 2B and 3 A–C) is also likely to occur with the flexible/unstructured histone N-terminal tails that extend out from the α-helical core structures of histones H3 and H4.
Fig. 3.
Structures around the active sites of the Ca2+-bound PAD4 (C645A) in complex with peptides H3-1, H3-2, H4, and BA. (Left) Ball-and-stick representation of the structures. The protein moiety is colored gray, and the peptides, H3-1 (A), H3-2 (B), H4 (C), and BA (D), are colored green, magenta, yellow, and cyan, respectively. Superimposed are Fo − Fc electron densities of the peptides, contoured at 2σ. (Right) Schematic diagrams of the structures in Left. Dotted lines and green half-circles show hydrogen bonds and hydrophobic interactions, respectively. The structure of the complex with BA was drawn by using the refined coordinates deposited in the Protein Data Bank (accession code 1WDA).
The side chain of the target arginine residue (Arg-8H in peptide H3-1, Arg-17H in peptide H3-2, and Arg-3H in peptide H4) is recognized within the active site cleft (Fig. 3 A–C) in the same manner as that seen for the previously determined BA complex (15) (Fig. 3D): Three side-chain nitrogen atoms (Nε, Nη1, and Nη2) are hydrogen-bonded to the two catalytic residues, Asp-350 and Asp-473, positioning the guanidino group near the remaining two catalytic residues, His-471 and Cys-645. The aliphatic portion of the arginine side chain is in close hydrophobic contact with Trp-347 and Val-469. Further peptide recognition by the enzyme occurs through a series of hydrogen bonds at the molecular surface near the active site cleft (Fig. 2B).
It is noticeable that peptide recognition at the molecular surface near the active site cleft occurs commonly through backbone atoms of the peptides at the (N − 2), (N − 1), N, and (N + 1) positions (Fig. 3 A–C). Thus, Gln-346 and Trp-347 recognize the carbonyl oxygen at the (N − 2) position, Arg-374 recognizes the carbonyl oxygens at the (N − 1) and N positions, Arg-639 recognizes the amido nitrogen at the N position, and Arg-372 and Val-469 recognize the carbonyl oxygens at the N and (N + 1) positions by means of water-mediated hydrogen bonds. The only nonbackbone atoms of the peptides recognized by the enzyme are the side-chain oxygens of Thr-6H and Ser-1H at the (N − 2) positions in peptides H3-1 and H4, respectively (Fig. 3 A and C). However, these nonbackbone recognitions do not occur at the (N − 2) position in peptide H3-2, because, unlike Thr-6H and Ser-1H, Ala-15H at the (N − 2) position in peptide H3-2 has no γ-oxygen (Fig. 3B). As presented here, the interactions through the backbone atoms at the (N − 2), (N − 1), N, and (N + 1) positions are, therefore, considered to be a possible consensus recognition motif by which the enzyme recognizes histone N-terminal tails.
The peptide recognition by the enzyme is extensive when compared with that observed with the artificial substrate, BA, where only four hydrogen bonds are formed with the substrate at the (N − 1) and N positions (Fig. 3D). This observation shows that the substrate interaction in the previously determined BA complex (15) is insufficient to describe the interactions between the enzyme and its physiological substrates. Furthermore, the main-chain carbonyl oxygen at the (N − 1) position is hydrogen-bonded to the Nη1 and Nη2 atoms of Arg-374 in the complexes with the histone peptides (Fig. 3 A–C), whereas the corresponding atom in the complex with BA is hydrogen-bonded to only one of the two Nη atoms of Arg-374 (Fig. 3D). The interaction with BA is therefore the simplest substrate interaction for the enzyme.
Structures of the Histone N-Terminal Peptides.
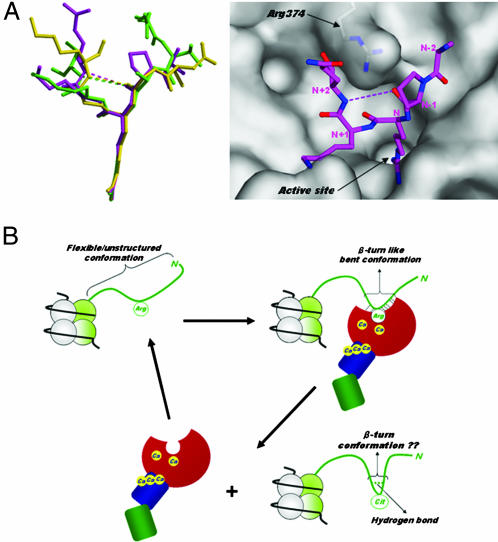
Histone N-terminal tails are known to be flexible and protrude from the nucleosome core particle (2, 3) (Fig. 4B). However, for the histone N-terminal peptides bound to PAD4, five successive residues form a bent conformation with an ordered structure (Fig. 4A). The backbone structures of these three peptides can be superimposed well with rms deviations of Cα atoms between peptides H3-1 and H3-2, between peptides H3-1 and H4, and between peptides H3-2 and H4 being 0.8 Å, 1.3 Å, and 0.7 Å, respectively. Arg-374 makes multiple hydrogen bonds with backbone carbonyl oxygens in each peptide, thus contributing to formation of the bent conformation (Figs. 3 A–C and 4B). The potential role of Arg-374 was determined experimentally by preparing PAD4 (R374A). The enzyme activity of PAD4 (R374A) toward these three histone peptides and BA was decreased to <6% that of the wild-type enzyme (Table 1), thus demonstrating that Arg-374 contributes to both substrate recognition and formation of the bent conformation.
Fig. 4.
Histone N-terminal structures. (A Left) Structural comparison of PAD4-bound forms. Peptides H3-1, H3-2, and H4 are shown as ball-and-stick representations colored green, magenta, and yellow, respectively, as in Fig. 3 A–C. (A Right) Top view of the peptide H3-2 structure shown in Left, together with a molecular surface representation near the active site cleft. The weak intrapeptide interactions between the backbone oxygen at (N − 1) position and the backbone nitrogen at (N + 2) position are shown as dotted lines. (B) Possible conformational change of the histone N-terminal tail in histone citrullination. The histone N-terminal tail protruding from the nucleosome core particle is shown as green ribbon. The structure of PAD4 is drawn in the same way as in Fig. 2B.
It is noticeable that, unlike at the (N − 2), (N − 1), N, and (N + 1) positions, there is no significant interaction with the enzyme at the (N + 2) position (Fig. 3 A–C), thereby causing poorer superimposition of the three peptide structures toward their C termini than their N termini (Fig. 4A). To compensate for the lack of the interaction at the (N + 2) position, the enzyme induces β-turn (reverse-turn)-like bent conformations to the histone peptides at the molecular surface near the active site cleft through a weak intrapeptide interaction between the backbone carbonyl oxygen at the (N − 1) position and the backbone amide nitrogen at the (N + 2) position, with interatomic distances of 4.5 Å, 3.8 Å, and 3.5 Å for peptides H3-1, H3-2, and H4, respectively (Figs. 2B and 4A). The structures of several histone modification enzymes have been determined by x-ray diffraction and NMR spectroscopy in the form of complexes with histone peptides (3), and in no case has such characteristic, local induction of substrate conformation been found. Induction of the β-turn-like bent conformation is therefore a structural insight for peptide recognition by histone modification enzymes.
Biological Implication of the Locally Induced β-Turn-Like Bent Conformation.
It is very important for target arginine recognition by PAD4 that local peptide around the target arginine residue takes a highly disordered conformation, because the enzyme recognizes a flexible/unstructured peptide at the molecular surface near the active site cleft and induces a β-turn-like bent conformation (Fig. 4B). This notion is consistent with the experimental results that the degree and rate of modification of arginine residues to citrulline residues directly correlate with the structural order of the substrate (19–21).
In addition, it has been shown that the use of cyclic citrullinated peptides in an ELISA of autoantibodies against citrullinated proteins from RA patients results in a more sensitive assay than one using linear citrullinated peptides (22). This finding suggests that the conformation of flanking regions around the citrulline residue plays an important role for epitope recognition by the antibodies. Therefore, it is of interest to know whether or not citrullination by PAD4 causes the change of the β-turn-like bent conformation into a more stable β-turn conformation through the formation of a hydrogen bond between the backbone oxygen at the (N − 1) position and the backbone nitrogen at the (N + 2) position, resulting in the release of citrullinated product from the enzyme molecule (Fig. 4B).
Sequence Specificity.
Most histone modification enzymes interact extensively with their peptide ligands and recognize a specific amino acid residue in a sequence-specific manner (3); this is not the case for PAD4. As stated above, each histone peptide is recognized by the enzyme with a possible consensus motif at the (N − 2), (N − 1), N, and (N + 1) positions through backbone atoms of the peptide (Figs. 2B and 3 A–C). This feature indicates that PAD4 can target multiple arginine sites in histone N-terminal tails without sequence specificity.
In peptides H3-1 and H4, however, Thr-6H and Ser-1H at the (N − 2) positions are recognized by the enzyme through their side-chain atoms, where Oγ atoms of Thr-6H and Ser-1H are hydrogen-bonded to Gln-346 and Gln-346/Asp-344, respectively (Fig. 3 A and C). Therefore, it may be because of the lack of these hydrogen bonds that no side-chain recognitions occur in peptide H3-2, because Ala-15H at the (N − 2) position in peptide H3-2 has no Oγ atoms (Fig. 3B). Closer inspection of the structure in the substrate binding region shows that side-chain moieties of the residues at the (N − 2) position should be small. Otherwise, they give rise to steric hindrance to the extent that the enzyme cannot recognize target arginine residues in a site-specific way. In fact, residues with small side-chain moieties occupy the (N − 2) positions of histone N-terminal arginine residues citrullinated by the enzyme (Arg-2, Arg-8, Arg-17, and Arg-26 in histone H3 and Arg-3 in histone H4) (4, 5). In contrast to the (N − 2) position, there will be no sequence specificity at the (N − 1), (N + 1), and (N + 2) positions, because the side-chain moieties of these residues are exposed to the solvent region (Fig. 3 A–C). From these results, we suggest that the enzyme recognizes five successive residues with the consensus sequence of 1ΦXRXX5, where Φ denotes amino acids with small side-chain moieties and X denotes any amino acid. In fact, the enzyme shows fairly broad sequence specificity and could target multiple arginine sites in histones such as Arg-8, Arg-17, and Arg-26 in histone H3 tail and Arg-3 in histone H4 tail both in vivo and in vitro. Furthermore, Arg-3 in the sequence 1SGRGK5 of histone H2A has recently been shown to be a target residue for the enzyme in HL-60 granulocyte cells (17). However, Arg-17, Arg-19, and Arg-23 in histone H4 tail are citrullinated by PAD4 in vitro but not in vivo (data not shown). Differences in citrullination in vitro and in vivo may be accounted for through the enzyme's having access to only a limited site of higher-order chromatin structure or through cooperation of PAD4 with an accessory factor.
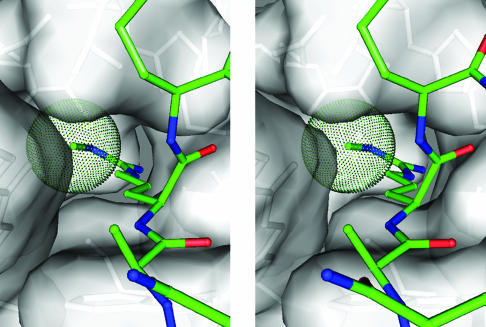
As stated above, PAD4 has been shown to target multiple arginine sites in histones H3 and H4, including those sites methylated by coactivator-associated arginine methyltransferase 1 and protein arginine methyltransferase 1 (6, 7). Therefore, we constructed a modeled structure of the active site cleft with bound monomethylated arginine to assess why PAD4 targets both methylated and unmethylated arginines. The model indicates that a NG-monomethylarginine side chain can be introduced into the cleft, with its terminal methyl group located at the bottom of the cleft (Fig. 5). The crystal structures determined in this study will be very helpful in the elucidation of detailed PAD4-substrate interactions and thereby suggest useful strategies to develop PAD-inhibiting drugs for RA.
Fig. 5.
Stereogram showing the putative active site structure with a bound NG-monomethylarginine side chain. The van der Waals representation corresponds to moieties of the protein (gray) and the methyl group of the NG-monomethylarginine side chain (green).
Materials and Methods
Synthesis of the Histone N-Terminal Peptides.
Peptides H3-1 (Ac4KQTARKSTGG13), H3-2 (Ac14KAPRKQLATK23), and H4 (Ac1SGRGKGGKGL10) were chemically synthesized by the solid-phase method (23). The N-terminal acetyl group was introduced by using acetic anhydride. All of the protecting groups and the resin were removed by anhydrous hydrogen fluoride, and the resulting peptide was extracted with 10% acetic acid, as described in ref. 24. The synthetic peptides were purified by RP-HPLC, and their identities were confirmed by MALDI-TOF MS and amino acid analysis (25, 26).
Preparation of the Crystals.
The inactive C645A mutant of PAD4 [PAD4 (C645A)] was used for the complex formation with the histone N-terminal peptides, because the wild-type enzyme was unlikely to form a stable complex with each histone peptide because of catalysis. PAD4 (C645A) was prepared as described in ref. 27. PAD4 (C645A) was overexpressed as a GST fusion protein in BL21 (DE3) cells carrying the expression plasmid. The protein was purified by glutathione-Sepharose 4B affinity chromatography and anion-exchange chromatography. Details of the purification of PAD4 (C645A) were described in ref. 27. The Ca2+-free PAD4 (C645A) thus purified was crystallized by the hanging-drop vapor-diffusion method by using a previously described protocol (27). Crystals of the Ca2+-bound PAD4 (C645A) in complex with each histone N-terminal peptide were prepared by soaking the Ca2+-free PAD4 (C645A) crystal in crystallization buffer containing 5 mM CaCl2 and each histone N-terminal peptide for 8 h at 20°C.
Data Collection and Structure Determination.
For diffraction data collection at 100 K, a cryoprotectant solution of 20% (vol/vol) ethylene glycol was included in the crystallization buffer. Diffraction data for the Ca2+-bound PAD4 (C645A) in complex with histone N-terminal peptides H3-1 and H4 were collected on BL38B1 at SPring-8 by using an ADSC Quantum 4R charge-coupled device detector, and those for the complex with peptide H3-2 were collected on NW12 at Photon Factory Advanced Ring (PF-AR) by using an ADSC Quantum 210 charge-coupled device. All of the diffraction data were indexed and scaled by using the hkl2000 program (28). Crystallographic data and data collection statistics are summarized in Table 2.
Table 2.
Crystallographic data and refinement statistics
| PAD4/H3-1 | PAD4/H3-2 | PAD4/H4 | |
|---|---|---|---|
| Crystallographic data | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, Å | 146.3 | 146.1 | 146.2 |
| b, Å | 60.8 | 60.1 | 60.6 |
| c, Å | 115.1 | 115.7 | 115.2 |
| β, ° | 124.3 | 124.3 | 124.2 |
| Resolution range, Å | 50.0–2.00 | 50.0–2.07 | 50.0–2.25 |
| Total observations | 233,822 | 222,220 | 153,298 |
| Unique reflections | 55,675 | 47,513 | 39,724 |
| Completeness, % | 97.7 (96.2) | 92.6 (63.9) | 98.3 (98.2) |
| Rmerge,* % | 5.6 (39.5) | 6.2 (33.3) | 6.1 (34.5) |
| 〈I/σ (I)〉 | 17.5 | 15.6 | 16.5 |
| Refinement statistics | |||
| Resolution, Å | 2.10 | 2.10 | 2.25 |
| Reflections used | 43,126 | 41,436 | 35,325 |
| Rwork/Rfree,† % | 20.2/24.1 | 20.2/24.6 | 19.9/24.8 |
| No. of atoms | |||
| PAD4 (C645A) | 4,952 | 4,937 | 4,943 |
| Histone peptide | 39 | 40 | 37 |
| Ca2+ ion | 5 | 5 | 5 |
| Water molecule | 224 | 191 | 154 |
| rms deviation | |||
| Bond length, Å | 0.012 | 0.012 | 0.012 |
| Bond angle, ° | 1.465 | 1.468 | 1.622 |
Values in parentheses are for the highest-resolution shell.
*Rmerge = ΣhΣi|I(h)i−〈I(h)〉|/ΣhΣiI(h)i.
†Rwork/Rfree = Σ|Fo| − |Fc|/Σ|Fo|, where Rwork and Rfree are calculated by using the working and free reflection sets, respectively. Rfree reflections (10% of the total) were held aside throughout refinement.
The initial structure of the Ca2+-bound PAD4 (C645A) in complex with each histone N-terminal peptide was derived from the atomic coordinates of the previously determined Ca2+-bound PAD4 (C645A) in complex with BA (Protein Data Bank ID code 1WDA). The structure was then refined by using the program cns (29), and manual construction was performed by using the graphic program o (30). At this stage, each peptide was identified on the Fo − Fc difference Fourier map with contour levels of >2σ. Five peaks with contour levels of >8σ on the difference Fourier map were assigned as Ca2+ ions. After rigid body refinement, the structures were further refined by simulated annealing, energy minimization, and B-individual by using the program cns and finally converged after several further cycles of refinement with the program refmac (31). The final refinement statistics are given in Table 2.
Preparation of R374A Mutant and Measurement of PAD Activity.
The R374A mutant of PAD4 [PAD4 (R374A)] was prepared with the QuikChange site-directed mutagenesis kit (Stratagene) using wild-type pGEX6P-1 PAD4 as the template, according to the manufacturer's recommendations, and confirmed by DNA sequencing. PAD4 (R374A) was expressed as a fusion protein with GST in Escherichia coli BL21 (DE3) cells and purified by using a protocol described in ref. 27.
Enzyme activity of PAD4 (R374A) was measured by using peptides H3-1, H3-2, and H4 as the substrates. The reaction mixture (50 μl) containing 0.1 M Tris·HCl (pH 8.0), 10 mM CaCl2, 5 mM DTT, and 2 mM substrate and GST-fused PAD4 (R374A) was incubated for 15 min at 37°C. The amount of citrulline derivative formed was determined by colorimetry with citrulline as the standard (32). One unit of enzyme was defined as the amount of enzyme catalyzing the formation of 1 μmol of citrulline derivative using BA as a substrate in 1 h under the assay conditions. Protein concentrations were determined by the Bradford method with BSA as the standard.
Acknowledgments
We thank Dr. M. Yamamoto, Dr. T. Kumasaka, and Dr. K. Miura for the data collection at SPring-8 and Dr. N. Matsugaki, Dr. N. Igarashi, Dr. M. Suzuki, and Prof. S. Wakatsuki for data collection at Photon Factory Advanced Ring. This work was supported by Grants-in-Aid for Young Scientists (B) (14780515 and 16770080 to H.H.) from the Japan Society of the Promotion of Science; Grants-in-Aid for Scientific Research on Priority Areas (16048226 and 17048023 to H.H. and 17054035 to T.S.); the Protein 3000 project (a national project on protein structural and functional analyses; M.S., T.S., and H.H.) of the Ministry of Education, Culture, Sports, Science and Technology; a Kaneko–Narita grant from the Protein Research Foundation (to H.H.); and a Grant-in-Aid for Research on Health Sciences Focusing on Drug Innovation (SH24402 to M.Y.) from the Japanese Health Science Foundation.
Abbreviations
- BA
benzoyl-l-arginine amide
- PAD
peptidylarginine deiminase
- RA
rheumatoid arthritis.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2DEW, 2DEX, and 2DEY).
References
- 1.Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld G., Groudine M. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 3.Khorasanizadeh S. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 4.Cuthbert G. L., Daujat S., Snowden A. W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P. D., Tempst P., Bannister A. J., et al. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Wysocka J., Sayegh J., Lee Y. H., Perlin J. R., Leonelli L., Sonbuchner L. S., McDonald C. H., Cook R. G., Dou Y., et al. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 6.Bauer U. M., Daujat S., Nielsen S. J., Nightingale K., Kouzarides T. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., et al. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y. H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. Proc. Natl. Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vossenaar E. R., Zendman A. J. W., Venrooij W. J., Pruijn G. J. M. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 10.Masson-Bessiere C., Sebbag M., Girbal-Neuhauser E., Nogueira L., Vincent C., Senshu T., Serre G. J. Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 11.van Boekel M. A., Vossenaar E. R., van den Hoogen F. H., van Venrooij W. J. Arthritis Res. 2002;4:87–93. doi: 10.1186/ar395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schellekens G. A., Visser H., de Jong B. A. W., van den Hoogen F. H. J., Hazes J. M. W., Breedveld F. C., van Venrooij W. J. Arthritis Rheum. 2000;43:155–163. doi: 10.1002/1529-0131(200001)43:1<155::AID-ANR20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A., Yamada R., Chang X., Tokuhiro S., Sawada T., Suzuki M., Nagasaki M., Nakayama-Hamada M., Kawaida R., Ono M., et al. Nat. Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 14.Nakashima K., Hagiwara T., Yamada M. J. Biol. Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 15.Arita K., Hashimoto H., Shimizu T., Nakashima K., Yamada M., Sato M. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 16.Sugawara K., Fuzisaki M. Agric. Biol. Chem. 1979;43:2407–2408. [Google Scholar]
- 17.Hagiwara T., Hidaka Y., Yamada M. Biochemistry. 2005;44:5827–5834. doi: 10.1021/bi047505c. [DOI] [PubMed] [Google Scholar]
- 18.Humm A., Fritsche E., Steinbacher S., Huber R. EMBO J. 1997;16:3373–3385. doi: 10.1093/emboj/16.12.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahara H., Okamoto H., Sugawara K. J. Biol. Chem. 1985;260:8378–8383. [PubMed] [Google Scholar]
- 20.Takahara H., Sugawara K. Agric. Biol. Chem. 1987;51:1657–1664. [Google Scholar]
- 21.Tarcsa E., Marekov L. N., Mei G., Melino G., Lee S. C., Steinert P. M. J. Biol. Chem. 1996;271:30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 22.Schellekens G. A., de Jong B. A. W., van den Hoogen F. H. J., van de Putte L. B. A., van Venrooij W. J. J. Clin. Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrifield R. B. J. Am. Chem. Soc. 1963;85:2149–2153. [Google Scholar]
- 24.Shimonishi Y., Hidaka Y., Koizumi M., Hane M., Aimoto S., Takeda T., Miwatani T., Takeda Y. FEBS Lett. 1987;215:165–170. doi: 10.1016/0014-5793(87)80134-5. [DOI] [PubMed] [Google Scholar]
- 25.Hidaka Y., Ohno M., Hemmasi B., Hill O., Forssmann W. G., Shimonishi Y. Biochemistry. 1998;37:8498–8507. doi: 10.1021/bi9731246. [DOI] [PubMed] [Google Scholar]
- 26.Hidaka Y., Shimono C., Ohno M., Okumura N., Adermann K., Forssmann W. G., Shimonishi Y. J. Biol. Chem. 2000;275:25155–25162. doi: 10.1074/jbc.M000543200. [DOI] [PubMed] [Google Scholar]
- 27.Arita K., Hashimoto H., Shimizu T., Yamada M., Sato M. Acta Crystallogr. D. 2003;59:2332–2333. doi: 10.1107/s0907444903022741. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov G. N., Vagin A. A., Dodson E. J. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima K., Hagiwara T., Ishigami A., Nagata S., Asaga H., Kuramoto M., Senshu T., Yamada M. J. Biol. Chem. 1999;274:27786–27792. doi: 10.1074/jbc.274.39.27786. [DOI] [PubMed] [Google Scholar]
- 33.Bedford M. T., Richard S. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]