Abstract
Wilson disease protein (ATP7B) is a copper-transporting P1B-type ATPase that regulates copper homeostasis and biosynthesis of copper-containing enzymes in human tissues. Inactivation of ATP7B or related ATP7A leads to severe neurodegenerative disorders, whereas their overexpression contributes to cancer cell resistance to chemotherapeutics. Copper-transporting ATPases differ from other P-type ATPases in their topology and the sequence of their nucleotide-binding domain (N-domain). To gain insight into the structural basis of ATP7B function, we have solved the structure of the ATP7B N-domain in the presence of ATP by using heteronuclear multidimensional NMR spectroscopy. The N-domain consists of a six-stranded β-sheet with two adjacent α-helical hairpins and, unexpectedly, shows higher similarity to the bacterial K+-transporting ATPase KdpB than to the mammalian Ca2+-ATPase or Na+,K+-ATPase. The common core structure of P-type ATPases is retained in the 3D fold of the N-domain; however, the nucleotide coordination environment of ATP7B within this fold is different. The residues H1069, G1099, G1101, I1102, G1149, and N1150 conserved in the P1B-ATPase subfamily contribute to ATP binding. Analysis of the frequent disease mutation H1069Q demonstrates that this mutation does not significantly affect the structure of the N-domain but prevents tight binding of ATP. The structure of the N-domain accounts for the disruptive effects of >30 known Wilson disease mutations. The unique features of the N-domain provide a structural basis for the development of specific inhibitors and regulators of ATP7B.
Keywords: ATP7A, ATP7B, copper, P-type ATPase, NMR structure
Copper is an essential cofactor of key metabolic enzymes involved in respiration, neurotransmitter biosynthesis, radical detoxification, and other essential physiological processes (1). In humans, genetic abnormalities of copper metabolism result in severe metabolic disorders, Menkes disease and Wilson disease (2–5). The disease symptoms, which include neurological and developmental problems and connective tissue and liver pathology, are caused by malfunction of copper-transporting ATPases ATP7A or ATP7B (Menkes disease protein and Wilson disease protein, respectively). The ATP7A and ATP7B are homologous membrane proteins, which use the energy of ATP hydrolysis to maintain intracellular copper concentration and to deliver copper to the secretory pathway, an essential step in the biosynthesis of copper-dependent enzymes (6). Interestingly, overexpression of ATP7A and ATP7B in tumor cells correlates with the increased resistance of these cells to platinum-based chemotherapeutic drugs (7–9).
The structural and biochemical basis of copper transport is poorly understood. ATP7A and ATP7B belong to the P1B-subtype of the large family of P-type ATPases, which differs significantly from the better characterized P2-ATPases, such as Ca2+-ATPase or Na+,K+-ATPase (10). The central step in the catalytic cycle of all P-type ATPases, including ATP7A and ATP7B, is the formation of a transient acyl-phosphate intermediate upon ATP hydrolysis (11, 12). This reaction takes place in the ATP-binding domain, which is composed of two subdomains, the phosphorylation domain (P-domain) and the nucleotide-binding domain (N-domain) (13, 14). The residues involved in the transfer of the γ-phosphate from ATP are located in the P-domain and are invariant in all P-type ATPases. By contrast, there is little sequence similarity between the N-domains of the P1B-ATPases and those of other P-type ATPases. There are also differences in the selectivity and affinity of the P1B N-domain for the nucleotides (15). The mechanistic significance of these differences is unknown.
The structure of the ATP7B N-domain determined in this study revealed a distinct nucleotide-coordination environment in the P1B-ATPases within the context of a structural fold that is common to all P-type pumps. The structure provides a basis for the future design of specific inhibitors of ATP7B and predicts consequences of >30 Wilson disease mutations found in this domain (16–18).
Results
(i) The N-Domain of ATP7B Is a Well Folded Protein with a Large Flexible Loop Ala-1114-Thr-1143.
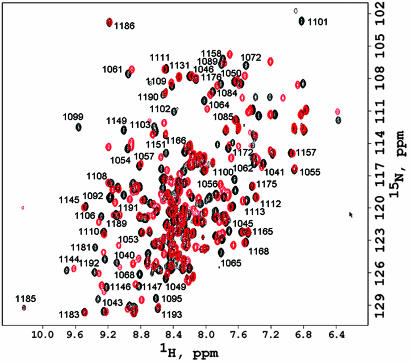
The N-domain of ATP7B is a 17-kDa protein, which specifically binds adenine-based nucleotides (15). The N-domain is highly soluble and retains its monomeric form at the concentrations up to 30 mg/ml. Nevertheless, attempts to crystallize this protein have been unsuccessful (our data). At the same time, 2D 1H,15N-chemical shift correlation spectra of the N-domain in both the apo- and the nucleotide-bound form showed very good chemical shift dispersion (Fig. 1). The number of signals in the spectrum was consistent with the size and amino acid composition of the protein.
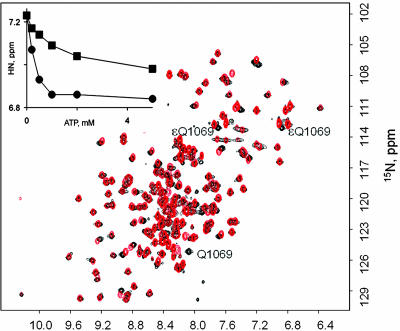
Fig. 1.
1H,15N-HSQC spectra of the ATP7B N-domain recorded in the presence (black) and absence (red) of 5 mM ATP. Some of the backbone amide assignments made in the presence of ATP are shown. Residue numbers correspond to the full-length ATP7B.
Although the isolated N-domain has no ATPase activity, ATP markedly increases stability of the protein. Samples containing ATP showed no changes in the fingerprint spectra for several weeks, whereas without ATP, the protein began to precipitate after 24–36 h. Therefore, we chose to solve the structure of the N-domain in the ATP-bound form. All of the accessible backbone chemical shifts and ≈95% of the side-chain chemical shifts have been assigned. The rms deviation of the NMR bundle from the mean structure calculated for the backbone atoms of residues 1,038–1,113 and 1,143–1,196 was 0.9 Å, indicating that the backbone conformation of the N-domain is well defined. Statistics for the structure calculation are shown in the Table 1, which is published as supporting information on the PNAS web site.
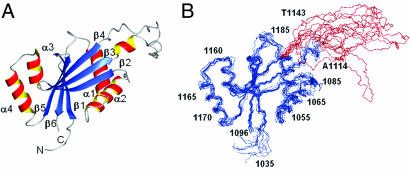
The N-domain structure consists of a central six-stranded anti-parallel β-sheet and two α-helical hairpins, one on each side of the β-sheet (Fig. 2A). The overall topology of the domain is βααβββααββ. The structure is fairly compact with the noticeable exception of the Ala-1114-Thr-1143 region, which is largely unfolded and does not seem to form contacts with the structured regions (Fig. 2B). This region corresponds exactly to the sequence insert, which is found in mammalian copper-ATPases, but absent in bacteria or lower eukaryotes (Fig. 7, which is published as supporting information on the PNAS web site). Therefore, it is possible that the Ala-1114-Thr-1143 region may be involved in regulatory functions unique to the mammalian ATPases (see Discussion).
Fig. 2.
Structure of the N-domain in the presence of ATP. (A) Ribbon diagram of the structure with the α-helices shown in red-yellow and β-sheet in blue. (B) The backbone traces of the 10 lowest energy structures of the N-domain illustrating folded regions (blue) and the flexible loop Ala-1114-Thr-1143 (red).
(ii) The P1B-ATPases Share a Common Core N-Domain Structure with Other P-Type ATPases.
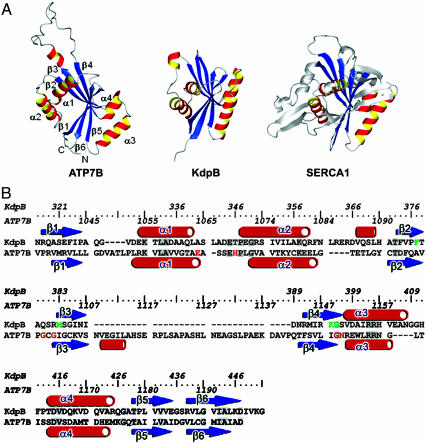
The N-domains of the P1B-ATPases display little sequence homology to other proteins, including members of the P-type ATPase family. Comparison of the ATP7B N-domain fold with the known 3D protein structures by using dali (www.ebi.ac.uk/dali) did not initially detect any similarity with either Ca2+-ATPase or Na+,K+-ATPase. The only significant structural match was the N-domain of KdpB, a potassium-transporting ATPase from Escherichia coli (19) (Fig. 3A, see stereo view of the overlaid structures in Fig. 8, which is published as supporting information on the PNAS web site). Given the low sequence identity (11.5%) between these two proteins (Fig. 3B), the similarity of their 3D folds was particularly striking and pointed to the importance of this fold for the overall molecular architecture of the P-type pumps. The unexpectedly higher similarity of the ATP7B N-domain to bacterial, rather than mammalian, P-type ATPases, likely reflects an early evolutionary divergence of the copper-transporting ATPases from other P-type pumps (20, 21).
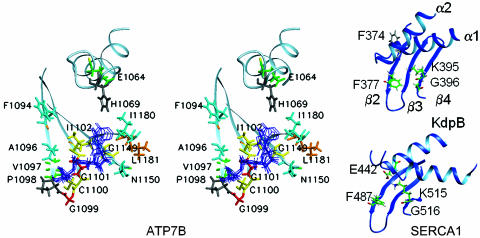
Fig. 3.
Comparison of the N-domains of ATP7B (P1B-ATPase), KdpB (P1A-ATPase), and SERCA1 Ca2+-ATPase (P2-ATPase). (A) The structures of KdpB (1SVJ) and SERCA1 (1T5S) N-domains were aligned to the ATP7B structure by using dali. rms deviation for the α-carbons of the aligned regions was 3.6 Å (ATP7B–KdpB alignment) and 4.2 Å (ATP7B–SERCA1 alignment). The common core is shown in red-yellow (α-helices) and blue (β-sheet). (B) Alignment of the secondary structure elements for ATP7B and KdpB. The residues predicted to be involved in ATP binding are shown in red (ATP7B) or green (KdpB). The identical residues are highlighted in gray.
Previously, the N-domain structure of KdpB was proposed to represent the minimal nucleotide-binding core of the P-type ATPase (19). The structural similarity of the ATP7B N-domain to KdpB prompted us to use dali to perform a pairwise structural alignment of the N-domains of ATP7B and Ca2+-ATPase. The regions that aligned corresponded exactly to the proposed N-domain core (Fig. 3A). Thus, all classes of P-type ATPases appear to share the same core nucleotide-binding structure despite their primary sequence dissimilarities.
(iii) ATP7B Has a Distinct Nucleotide-Binding Environment.
The amino acid residues involved in nucleotide coordination in several P-type ATPases have been identified (22–26); these residues are not conserved in ATP7B and other P1B-type pumps. Furthermore, overlay of KdpB and ATP7B structures revealed that in ATP7B the positions equivalent to F377 and K395 of KdpB, which belong to the proposed rudimentary ATP binding motif (19), are occupied by A1096 and I1148, respectively (Fig. 3B). These observations and the characteristic nucleotide-binding properties of the ATP7B N-domain (15) suggested that ATP7B and the homologous P1B-ATPases have a distinct coordination environment for nucleotides (15, 27). To identify the region of the N-domain involved in ATP coordination, we performed detailed sequence analysis of the P1B-ATPases, measured residue-specific secondary chemical shifts induced by binding of ATP, ADP, or AMP, and attempted to detect ATP-protein nuclear Overhauser effects (NOEs) directly.
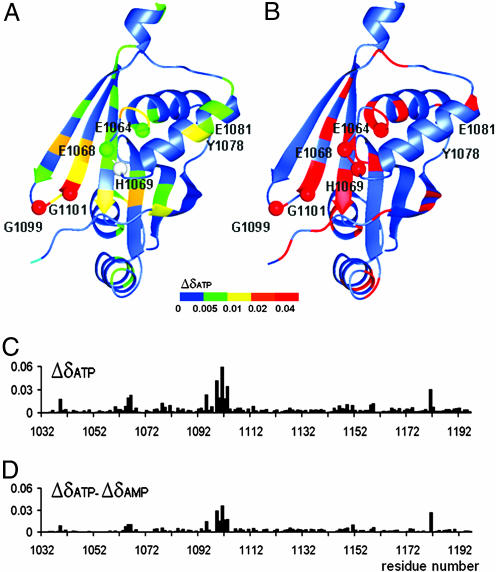
Multiple sequence alignment of the N-domains of P1B-type ATPases reveals several highly conserved residues, four of which are invariant. In ATP7B, the invariant residues are E1064, H1069, G1099, and G1101 (Fig. 7). Significantly, in the 3D structure of ATP7B, these residues are in close spatial proximity to each other, suggesting the location of the ATP binding site (Fig. 4A). This location is consistent with the chemical shift perturbations observed in the 1H,15N- HSQC spectra of the N-domain upon additions of ATP.
Fig. 4.
Residues affected by nucleotide binding and the location of Wilson disease-causing mutations. (A) The structure of the ATP7B N-domain with a color map of backbone amide chemical shift changes induced by ATP binding. The secondary chemical shifts for each residue are expressed as Δδ = (((δHNNUC − δHNfree)/δHNNUC)2 + ((δNNUC − δNfree)/δNNUC)2)1/2, where δHNNUC and δNNUC are 1H and 15N chemical shifts of the backbone amide group in the presence of the nucleotide, and δHNfree and δNfree are chemical shifts measured without nucleotide present. Data for H1069 (gray) is not available. The residues invariant in the P1B-ATPases are shown as spheres. The disordered loop A1114-T1143 is not shown. (B) The location of disease causing mutations is shown in red. (C) Chemical shift changes (Δδ, defined above) at each amino acid residue upon binding of ATP. (D) Chemical shift changes at each residue observed on binding ATP minus the corresponding chemical shift changes on binding AMP.
ATP induces significant changes in the chemical shifts of ≈40 N-domain residues (Fig. 1; see also Table 2, which is published as supporting information on the PNAS web site). The segment 1,098–1,103 in the β2-β3 loop and the lower part of the β3-strand was the most affected (Fig. 4). All residues in this segment showed considerable changes of the backbone amide chemical shifts, especially the invariant G1099 and G1101. Signal overlap prevented the determination of chemical shift perturbations for the invariant H1069. However, a chemical shift change was detected for the adjacent residue E1068 and for the Q1069 in the H1069Q mutant protein (see below). Significant changes also were observed in the segment 1,060–1,065, which includes the invariant residue E1064. Thus, at least three invariant amino acid residues are among those most affected by ATP, suggesting that these residues contribute to the nucleotide binding site. In the other regions, the nonconserved L1181 was notably affected by ATP addition, also suggesting proximity to the binding site. Interestingly, the residues affected by ATP and the amino acids in their immediate vicinity represent frequent sites of Wilson disease mutations (Fig. 4B), which further emphasizes their functional significance.
To characterize the nucleotide coordination environment in more detail, we performed NOESY experiments and compared the secondary chemical shifts induced by ATP, ADP, and AMP. The NOEs between 1H1′ of the ribose moiety and the 1Hαs of G1149 and 1Hα, 1Hβ, and 1HN of N1150 were detected in a 13C-NOESY experiment with the N-domain and 13C-labeled ATP. Both G1149 and N1150 are conserved in the P1B-ATPases (Fig. 7). The backbone amide of G1149 displays ATP-dependent chemical shift change (Fig. 4C), consistent with its close proximity to ATP. Several NOESY experiments using different isotope-labeling patterns and isotope-filtering schemes were recorded, but very few additional NOEs between the ATP and the protein were observed. The scarcity of detectable protein-ATP NOEs could be due to the intermediate exchange regime, where substantial signal broadening may prevent observation of NOE peaks. The ATP dissociation constant for the N-domain is ≈70 μM and is in the range (10−4 to 10−5 M) where the intermediate exchange conditions are often observed for protein-ligand systems.
The residues in the vicinity of the ATP phosphate tail were identified by comparing the backbone amide chemical shift changes induced by ATP, ADP, and AMP. All three nucleotides bind to the N-domain with similar affinity (15), suggesting that the differences in the nucleotide-induced secondary chemical shifts should be attributed to the difference in the number and the location of the phosphate groups. Binding of ATP and ADP produced very similar shifts in the signals (Fig. 9, which is published as supporting information on the PNAS web site). This similarity indicates that the γ-phosphate of ATP is not in close contact with any N-domain residue and does not contribute to the binding of nucleotides. In contrast, the effect of AMP differed from that of ATP or ADP (Fig. 9). Although the residues in the 1,060–1,065 region showed comparable chemical shifts changes upon ATP or AMP binding, the 1,098–1,103 region displayed greater chemical shift changes in the presence of ATP/ADP, the changes being especially large for G1099 and G1101 (Fig. 4D and Table 2). Thus, these invariant Gly residues are likely to be in the immediate vicinity of the α- and β-phosphate groups of the bound ATP.
(iv) The Disease Mutation H1069Q Does Not Significantly Affect the Structure of the N-Domain but Disrupts Tight Binding of ATP.
Many Wilson disease-causing mutations are located in the N-domain. Among them, H1069Q is the most frequent mutation, found in ≈40% of patients (3, 16, 28). Previous attempts to understand the consequences of this mutation for ATP7B structure produced conflicting results. Circular dichroism measurements, limited proteolysis, and functional assays using the full-length ATP7B or purified ATP7B N-domain suggested that the H1069Q mutation did not significantly affect the ATP7B structure but markedly diminished the ability of this protein to bind and hydrolyze ATP (15, 29). In contrast, the mislocalization of the ATP7B H1069Q mutant in cells and tissues suggested that this mutation may affect protein folding (30, 31). To address this issue, we prepared a15N-labeled N-domain containing the H1069Q substitution and recorded its 1H,15N-HSQC spectrum.
The fingerprint spectra of the WT and mutant N-domain (Fig. 5) in the absence of ATP were nearly identical, indicating that the structural changes induced by H1069Q substitution are minimal. Addition of ATP to the H1069Q mutant induced changes in the 1H,15N-HSQC spectrum with many of the same residues affected (data not shown), indicating that the ability to bind ATP was not abolished. Furthermore, the ATP titration curves for G1101 and Q1069 located in different parts of the binding site were very similar, suggesting that the geometry of ATP binding was unaltered. However, the affinity for ATP, measured from the backbone amide chemical shift titration of either G1101 or Q1069, was markedly reduced (apparent Ka = 1.0 ± 0.2 mM) (Fig. 5 Inset). The reduced affinity of the H1069Q mutant for ATP is likely due to introduced steric hindrance or disrupted energetically important interaction between the adenine moiety and the imidazole group.
Fig. 5.
Characterization of the H1069Q mutant. Overlay of the fingerprint spectra of the WT ATP7B N-domain (red) and the H1069Q mutant (black) are shown. Tentative Q1069 assignment is shown. (Inset) Chemical shift of the G1101 1HN signal as a function of added ATP in the wild type (•) and H1069Q mutant (■) of the ATP7B N-domain.
Discussion
Mechanistic Implications of the N-Domain Structure.
The P-type ATPases transport various metal ions via a common mechanism involving formation of the phosphoenzyme intermediate. The present work shows that the nucleotide-binding fold is conserved in the P-type ATPase family despite the lack of sequence conservation. The structure, consisting of a central six-stranded β-sheet and two pairs of α-helices, has been proposed to represent the core module of the P-type ATPase N-domain (19). Our results indicate that this fold is also conserved in the P1B-ATPase structure and, therefore, represents a truly universal structural motif of the P-type ATPases. It seems particularly important that the fold of the domain is preserved, even though most of the residues involved in ATP binding are not conserved. Rather than simply providing a scaffold for ATP coordination in the nucleotide-binding center, the structure of the N-domain must have evolved to fit more fundamental aspects of the P-type ATPase architecture and molecular mechanism.
The structure of the ATP7B N-domain is very similar to that of KdpB with two notable exceptions. First, the orientation of the α-helical hairpins with respect to the central β-sheet is different (Fig. 3A). This difference may simply represent a structural dissimilarity of two proteins with little sequence homology. Alternatively, the change in the position of the hairpins of the ATP7B N-domain compared with KdpB could be due to the presence of ATP in the ATP7B structure (the reported structure of the KdpB N-domain is in the apo form).
The second distinct feature of the ATP7B N-domain is a large and flexible loop Ala-1114–Thr-1143. Substitution of this insert by an unrelated sequence had no effect on the ATP-binding activity of the N-domain (data not shown). Chemical shifts of the residues in this region were not significantly affected by ATP binding. Thus, this flexible region in ATP7B seems to participate in the functions unrelated to ATP binding and specific to the mammalian copper transporters. Such functions may include interactions with the other domains or binding of the components of intracellular trafficking machinery that regulate the intracellular localization of ATP7B.
Organization of the ATP Binding Site in P1B-ATPases Compared with P2-ATPases.
Previous mutagenesis studies on P1B-ATPases pointed to the important role of the invariant His and the GxG motif (H1069 and G1099 × G1101 in ATP7B) in ATP-binding (15, 32–34). Our data support the role of these residues in ATP coordination. The chemical shifts of G1099 and G1101 are among the most changed in the presence of ATP, and the H1069Q substitution dramatically decreases ATP binding affinity without any apparent effect on the fold of the apoprotein. Interestingly, neither the key His nor the GxG motif was found in the nucleotide-binding site of other P-type ATPases. Thus, despite common fold and the same general location of the ATP-binding site (Fig. 10A, which is published as supporting information on the PNAS web site), the residues that form the nucleotide environment in P1B-ATPases appear to differ from those in other P-type pumps.
The crystal structures of SERCA (a P2-type ATPase), with a bound ATP analogue (25, 26), show that the N-domain residue F487 is involved in a stacking interaction with the adenine moiety of the nucleotide and that the side chain of another N-domain residue R560 interacts with the β-phosphate of ATP. In KdpB, residue F377 homologous to SERCA F487, participates in adenine binding through a π–π interaction, and the positively charged ε-amino-group of K395 forms a cation–π interaction with the adenine ring (19). Structure alignments (Figs. 3B and 10B) show no lysine residue in the putative ATP binding site of ATP7B that could interact with ATP in a similar fashion. A phenylalanine (F1094) is present close to the ATP-binding site; however, its location corresponds to that of noncoordinating F374 in KdpB, rather than of F377 (Fig. 3B and 6A). Also, in the other P1B-ATPases, this position can be occupied by residues as diverse as Ser, Gln, and Val (Fig. 7). Therefore, it seems unlikely that F1094 plays a central role in ATP coordination.
The ATP-dependent secondary chemical shift pattern, the NOEs between ATP and the protein, and the analysis of H1069Q mutant indicate that H1069, G1099, G1101, G1149, and N1150 are located in the immediate vicinity of bound ATP in the N-domain of ATP7B. ATP docking performed by simulated annealing based on the distance restraints derived from these data provided more detailed view of the binding site (Fig. 6A). In the ensemble of the lowest energy structures, adenine is positioned close to the H1069 imidazole group, but the available data are insufficient to determine the nature of interaction between adenine and H1069. The distance between H1069 and the adenine ring of ATP and their relative orientation suggest a possibility of a hydrogen bond between the imidazole and the adenine amino group rather than a stacking interaction between the rings. The adenine moiety is surrounded by the hydrophobic side chains of I1102 and I1180 (Fig. 6A). Consistent with its role in positioning the adenine ring, I1102 shows large ATP-dependent secondary shift and is conserved in P1B-ATPases (Fig. 7), with some ATPases having a valine in this position. The I1102T substitution is a disease-causing mutation, which further points to the functional significance of this residue. The proximity of another isoleucine, I1180, to the bound ATP is reflected in the large chemical shift change observed for the backbone amide of the neighboring L1181 (Fig. 4C). The backbone amide of L1181 is closer to bound ATP than that of I1180. However, the side chain of L1181 is located on the opposite side of the β-sheet, away from the ATP binding center, whereas the side chain of I1180 is facing the adenine moiety (Fig. 6A).
Fig. 6.
Organization of the nucleotide-binding site in the N-domains of ATP7B, SERCA1, and KdpB. (A) Stereo view of the ensemble of the ATP molecules docked into the ATP-binding site of ATP7B. The residues in proximity to ATP are colored according to the magnitude of the ATP-dependent secondary chemical shifts as in Fig. 4A. Data are not available for H1069 (shown in gray). (B) Fragments of the KdpB and SERCA1 N-domain structures with the residues involved in ATP binding.
The sugar moiety is in the immediate vicinity of G1149 and N1150. Comparison of the nucleotide-binding pockets of the P-type ATPases illustrates that G1149 in ATP7B, G396 in KdpB, and G516 in SERCA appear to occupy equivalent positions in the structure (Fig. 6B) and may contribute to the formation of the ribose-binding pocket. In ATP7B, the close proximity of G1149 and N1150 to the ribose is directly supported by the NOE data, whereas in the KdpB N-domain, no NOEs were detected between the protein and the ribose moiety (19). This observation may explain a 10-fold higher affinity of ATP for the ATP7B N-domain as compared to KdpB.
In SERCA, the α-phosphate is stabilized by R489 and the β-phosphate by R560, whereas in ATP7B, the large ATP-dependent secondary chemical shifts clearly demonstrate proximity of the phosphate tail to G1099 and G1101 (Fig. 6A). Similar binding affinities of ATP, ADP, and AMP suggest that the β- and γ-phosphates are not involved in energetically important interactions with the N-domain residues. The difference observed between the spectra recorded with ATP and AMP may reflect a balance of additional attractive and repulsive electrostatic interactions caused by the β-phosphate proximity without major net effect on the binding energy. It should be noted that Mg2+ does not noticeably affect ATP binding to the isolated N-domain (15), but Mg2+ is required for ATPase activity of the native enzyme, presumably because of its role in the coordination of the ATP phosphate groups. The ATP phosphate tail, thus, may adopt a different conformation in the full-length protein in the presence of Mg2+.
The question about the possible role of the invariant E1064 remains open. The current model shows no contacts between this residue and ATP. The side-chain carboxyl of E1064 is located within 5Å from the imidazole ring of H1069 and resembles the location of E442 in SERCA (Fig. 6B), which is involved in proper positioning of K515 for interaction with adenine. Mutations of E1064 or the equivalent residue in ZntA drastically decrease affinity for ATP (15, 33). Altogether, these observations suggest that E1064 may be involved in stabilizing a possible interaction between H1069 and the adenine moiety of ATP.
ATP-Dependent Conformational Changes in the N-Domain of ATP7B.
In SERCA, ATP binding induces large-scale movement of the entire N-domain relative to the P-domain. This results in a “closed” protein conformation in which the ATP molecule bridges the gap between the domains, allowing the γ-phosphate of ATP to reach the phosphorylation site (25, 26). In SERCA or KdpB, ATP binding does not significantly alter the structure of the N-domain (19), and fairly minor ATP-dependent conformational changes were observed in the N-domain of the Na+,K+-ATPase (35).
In ATP7B, at least 20% of the N-domain residues show significant backbone amide chemical changes when ATP binds. The number and location of the affected residues taken together with large magnitude of many secondary shifts point to conformational changes in the protein in addition to the direct effect of the bound nucleotide. Indeed, some of the affected residues in the ATP7B N-domain, for example, 1,079–1,081 and 1,064–1,066, cannot be in direct contact with ATP (Fig. 4A). The pattern of ATP-induced chemical shift changes suggests that the conformational change may involve movement of the two α-helices comprising residues 1,053–1,065 (α1) and 1,072–1,083 (α2). A possible effect of such rearrangement may be to bring the side chain of H1069 into a contact with the adenine moiety and to lock the ATP molecule in the binding pocket. In the full-length ATP7B, another important consequence of the hairpin reorientation could be facilitation of the interdomain interactions, which are critical for the catalytic cycle of the P-type ATPases.
The Effects of Wilson Disease Causing Mutations.
There are ≈40 missense mutations in the N-domain that result in Wilson disease phenotype (18). The N-domain structure provides an explanation for the effect of most of these mutations, which appear to fall into two distinct categories. The first group includes 16 residues in the regions 1,061–1,069, 1,099–1,106, and 1,146–1,153, all located in the immediate vicinity of the ATP-binding site (Fig. 4B). Substitutions of these residues are likely to directly affect the nucleotide binding, as demonstrated for the H1069Q mutant. Residues 1,083, 1,088, and 1,089 are located very close to the residues 1,061–1,069. Thus, mutations at positions 1,083–1,089 may inhibit ATP binding indirectly by disrupting the proper configuration of the ATP-binding site.
The second group of mutations affects residues in the 1,033–1,038 segment, which is a part of the hinge region connecting the N- and P-domains, and residues in the segment 1,168–1,176 (helix α3), which in the structure of the full-length protein would face the P-domain. These mutations may affect interactions between the N- and P-domain and interfere with proper domain closure. The few remaining mutations may have diverse effects, either disrupting proper folding of the protein, for example, C1104F (15), or interfering with the regulatory interactions.
In summary, the structure of a P1B-ATPase N-domain presented here proves the universal nature of the 3D structural motif involved in the ATP binding in all P-type ATPases. It reveals the distinct ATP-binding local environment in the P1B-ATPases and provides the basis for further investigation of the mechanism of these remarkable enzymes.
Materials and Methods
Preparation of the Isotopically Labeled N-Domain.
Plasmid pTYB12-NABD (15) was used for expression of the ATP7B N-domain. The 13C and 15N labeling was done essentially as described in ref. 36.
Purification of 15N- and 15N-,13C-labeled N-domain was performed as reported for nonlabeled protein (15). The efficiency of isotopic labeling (≈95%) was determined by mass spectrometry. To verify the lack of ATP hydrolysis during NMR experiments the N-domain (0.5 mM) was incubated in 50 mM phosphate buffer in the presence of 1 mM ATP for 18 h at room temperature. The EnzCheck Phosphate kit (Invitrogen) was used to monitor the formation of inorganic phosphate, which was found to be negligible.
For NMR experiments, protein samples were dialyzed against 50 mM NaHPO4, pH 6.0/5 mM DTT/50 μM NaN3 buffer and concentrated to 0.5–1 mM. Gel filtration on Superose 12 HR 10/30 column (Amersham Pharmacia) confirmed that the protein was monomeric under these conditions.
NMR Experiments and Structure Calculation.
The backbone chemical shift assignments in the presence of 5 mM ATP were performed by using a set of 3D NMR experiments including HNCO, HNCA, HNCACB, HN(CO)CACB, and HN(CA)CO as input to the autoassign (37) and mapper (38) software. The side-chain assignments were derived primarily from 3D H(CCO)NH, HCCH-COSY, and HCCH-TOCSY data. The assignments have been deposited at BioMagResBank (accession no. 6914). Most of the backbone amide assignments were easily transferable to the apoform of the protein. The HN(CO)CACB, HNCA, and HNCACB experiments were used to verify the backbone amide assignments in the apoform. The secondary structure elements were determined from secondary chemical shifts, proton amide exchange rates, and direct detection of hydrogen bonds in a long-range HNCO-type experiment (39). Backbone angles were predicted from chemical shifts by the semiempirical method implemented in the talos program (40), and the 3D structure was determined from distance constraints obtained from 3D 15N- and 13C-edited NOESY and 4D 13C,15N-edited NOESY data. The cyana (41) package was used to assign NOEs and to calculate the structure. cns (42) was used to dock the ATP on the basis of distance constraints derived from protein-ATP NOEs detected in 13C-edited NOESY with U-13C-ATP (Cambridge Isotope Laboratories, Cambridge, MA) and ATP-induced secondary chemical shifts. The structures were analyzed by using molmol (43). The atomic coordinates have been deposited at the Protein Data Bank (ID code 2ARF).
Supplementary Material
Acknowledgments
O.D. thanks Dr. Bob Fillingame for encouraging this foray to the frontiers of the ATPase world. We thank Jack Kaplan and Maria Schumacher for helpful suggestions on the manuscript preparation, Dr. Haupt and Dr. Rosenzweig for sharing with us their unpublished data, and Dr. Ralle at the Oregon Health & Science University Metal Iron Core for performing mass spectroscopy measurements. This study used the National Magnetic Resonance Facility at Madison, supported by funds from the National Institutes of Health (NIH), the University of Wisconsin, the National Science Foundation, and the Department of Agriculture. This work was funded by NIH Grants P01 GM 067166-01 and R01 DK071865 (to S.L.).
Abbreviations
- N-domain
nucleotide-binding domain
- NOE
nuclear Overhauser effect
- P-domain
phosphorylation domain.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 6914). The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2ARF).
References
- 1.Puig S., Thiele D. J. Curr. Opin. Chem. Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 2.Daniel K. G., Harbach R. H., Guida W. C., Dou Q. P. Front. Biosci. 2004;9:2652–2662. doi: 10.2741/1424. [DOI] [PubMed] [Google Scholar]
- 3.Ferenci P. Aliment. Pharmacol. Ther. 2004;19:157–165. doi: 10.1046/j.1365-2036.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- 4.Gitlin J. D. Gastroenterology. 2003;125:1868–1877. doi: 10.1053/j.gastro.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Shim H., Harris Z. L. J. Nutr. 2003;133:1527S–1531S. doi: 10.1093/jn/133.5.1527S. [DOI] [PubMed] [Google Scholar]
- 6.Lutsenko S., Petris M. J. J. Membr. Biol. 2003;191:1–12. doi: 10.1007/s00232-002-1040-6. [DOI] [PubMed] [Google Scholar]
- 7.Katano K., Kondo A., Safaei R., Holzer A., Samimi G., Mishima M., Kuo Y. M., Rochdi M., Howell S. B. Cancer Res. 2002;62:6559–6565. [PubMed] [Google Scholar]
- 8.Safaei R., Howell S. B. Crit. Rev. Oncol. Hematol. 2005;53:13–23. doi: 10.1016/j.critrevonc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Samimi G., Katano K., Holzer A. K., Safaei R., Howell S. B. Mol. Pharmacol. 2004;66:25–32. doi: 10.1124/mol.66.1.25. [DOI] [PubMed] [Google Scholar]
- 10.Tsivkovskii R., Purnat T., Lutsenko S. In: Handbook of ATPases. Futai M., Kaplan J. H., Wada Y., editors. Weinheim, Germany: Wiley; 2003. pp. 99–158. [Google Scholar]
- 11.Voskoboinik I., Mar J., Strausak D., Camakaris J. J. Biol. Chem. 2001;276:28620–28627. doi: 10.1074/jbc.M103532200. [DOI] [PubMed] [Google Scholar]
- 12.Tsivkovskii R., Eisses J. F., Kaplan J. H., Lutsenko S. J. Biol. Chem. 2002;277:976–983. doi: 10.1074/jbc.M109368200. [DOI] [PubMed] [Google Scholar]
- 13.Kuhlbrandt W. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 14.Toyoshima C., Nakasako M., Nomura H., Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 15.Morgan C. T., Tsivkovskii R., Kosinsky Y. A., Efremov R. G., Lutsenko S. J. Biol. Chem. 2004;279:36363–36371. doi: 10.1074/jbc.M404553200. [DOI] [PubMed] [Google Scholar]
- 16.Riordan S. M., Williams R. J. Hepatol. 2001;34:165–171. doi: 10.1016/s0168-8278(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 17.Hsi G., Cox D. W. Hum. Genet. 2004;114:165–172. doi: 10.1007/s00439-003-1045-y. [DOI] [PubMed] [Google Scholar]
- 18.Cox D. W., Moore S. D. J. Bioenerg. Biomembr. 2002;34:333–338. doi: 10.1023/a:1021293818125. [DOI] [PubMed] [Google Scholar]
- 19.Haupt M., Bramkamp M., Coles M., Altendorf K., Kessler H. J. Mol. Biol. 2004;342:1547–1558. doi: 10.1016/j.jmb.2004.07.060. [DOI] [PubMed] [Google Scholar]
- 20.Axelsen K. B., Palmgren M. G. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmgren M. G., Axelsen K. B. Biochim. Biophys. Acta. 1998;1365:37–45. doi: 10.1016/s0005-2728(98)00041-3. [DOI] [PubMed] [Google Scholar]
- 22.Morsomme P., Slayman C. W., Goffeau A. Biochim. Biophys. Acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 23.Clausen J. D., McIntosh D. B., Vilsen B., Woolley D. G., Andersen J. P. J. Biol. Chem. 2003;278:20245–20258. doi: 10.1074/jbc.M301122200. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen P. L., Hakansson K. O., Karlish S. J. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- 25.Toyoshima C., Mizutani T. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 26.Sorensen T. L., Moller J. V., Nissen P. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 27.Efremov R. G., Kosinsky Y. A., Nolde D. E., Tsivkovskii R., Arseniev A. S., Lutsenko S. Biochem. J. 2004;382:293–305. doi: 10.1042/BJ20040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caca K., Ferenci P., Kuhn H. J., Polli C., Willgerodt H., Kunath B., Hermann W., Mossner J., Berr F. J. Hepatol. 2001;35:575–581. doi: 10.1016/s0168-8278(01)00219-7. [DOI] [PubMed] [Google Scholar]
- 29.Tsivkovskii R., Efremov R. G., Lutsenko S. J. Biol. Chem. 2003;278:13302–13308. doi: 10.1074/jbc.M300034200. [DOI] [PubMed] [Google Scholar]
- 30.Huster D., Hoppert M., Lutsenko S., Zinke J., Lehmann C., Mossner J., Berr F., Caca K. Gastroenterology. 2003;124:335–345. doi: 10.1053/gast.2003.50066. [DOI] [PubMed] [Google Scholar]
- 31.Payne A. S., Kelly E. J., Gitlin J. D. Proc. Natl. Acad. Sci. USA. 1998;95:10854–10859. doi: 10.1073/pnas.95.18.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okkeri J., Bencomo E., Pietila M., Haltia T. Eur. J. Biochem. 2002;269:1579–1586. doi: 10.1046/j.1432-1033.2002.02810.x. [DOI] [PubMed] [Google Scholar]
- 33.Okkeri J., Laakkonen L., Haltia T. Biochem. J. 2004;377:95–105. doi: 10.1042/BJ20030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bissig K. D., Wunderli-Ye H., Duda P. W., Solioz M. Biochem. J. 2001;357:217–223. doi: 10.1042/0264-6021:3570217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilge M., Siegal G., Vuister G. W., Guntert P., Gloor S. M., Abrahams J. P. Nat. Struct. Biol. 2003;10:468–474. doi: 10.1038/nsb924. [DOI] [PubMed] [Google Scholar]
- 36.Marley J., Lu M., Bracken C. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 37.Moseley H. N., Monleon D., Montelione G. T. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 38.Guntert P., Salzmann M., Braun D., Wuthrich K. J. Biomol. NMR. 2000;18:129–137. doi: 10.1023/a:1008318805889. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y. X., Jacob J., Cordier F., Wingfield P., Stahl S. J., Lee-Huang S., Torchia D., Grzesiek S., Bax A. J. Biomol. NMR. 1999;14:181–184. doi: 10.1023/a:1008346517302. [DOI] [PubMed] [Google Scholar]
- 40.Cornilescu G., Delaglio F., Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 41.Guntert P. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 42.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Koradi R., Billeter M., Wuthrich K. J. Mol. Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.