Abstract
Nonribosomal peptide synthetases (NRPSs) and polyketide synthases are large, multidomain enzymes that biosynthesize a number of pharmaceutically important natural products. The recognition of biosynthetic intermediates, displayed via covalent attachment to carrier proteins, by catalytic domains is critical for NRPS and polyketide synthase function. We report the use of combinatorial mutagenesis coupled with in vivo selection for the production of the Escherichia coli NRPS product enterobactin to map the surface of the aryl carrier protein (ArCP) domain of EntB that interacts with the downstream elongation module EntF. Two libraries spanning the predicted helix 2 and loop 2/helix 3 of EntB-ArCP were generated by shotgun alanine scanning and selected for their ability to support enterobactin production. From the surviving pools, we identified several hydrophobic residues (M249, F264, and A268) that were highly conserved. These residues cluster near the phosphopantetheinylated serine in a structural model, and two of these positions are in the predicted helix 3 region. Subsequent in vitro studies are consistent with the hypothesis that these residues form a surface on EntB required for interaction with EntF. These results suggest that helix 3 is a major recognition element in EntB-ArCP and demonstrate the utility of selection-based approaches for studying NRPS biosynthesis.
Keywords: nonribosomal peptide synthetase, siderophore
A number of medicinally useful natural products are biosynthesized in their cognate producer organisms by nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) mutidomain enzymes (1). Examples of NRPS- and PKS-derived compounds include the antibiotics erythromycin (PKS) and vancomycin (NRPS) (2, 3), the antitumor agents epothilone and bleomycin (both from hybrid PKS/NRPS systems) (4, 5), and the immunosuppressant rapamycin (hybrid PKS/NRPS) (6). At various points during the biosynthesis of these molecules, substrates for catalytic operations are presented as thioesters tethered to carrier proteins through a 4′-phosphopantetheine cofactor. These carrier proteins, peptidyl carrier proteins (PCPs) in NRPSs and acyl carrier proteins (ACPs) in PKSs, are homologous to the ACPs that are involved in production of fatty acids from primary metabolism (7). ACPs and PCPs are small (≈80–100 residues) and contain a central serine residue that is the site of attachment for the phosphopantetheine arm (1, 7). Carrier proteins thus provide the protein scaffolding for display of the growing acyl chains in each module.
Critical to the function of NRPSs and PKSs are the interactions between domains (8). Because the biosynthetic intermediates are covalently tethered to carrier proteins, studying the determinants of carrier-protein recognition by other synthetase domains remains an important goal. In a typical cycle of NRPS elongation (that is, formation of an amide bond between two amino acids), a PCP must be recognized by the adenylation domain, which acylates the phosphopantetheine with an amino acyl group and then by the downstream condensation domain, which catalyzes formation of the amide bond (9). In addition, carrier proteins that are located on terminal modules of polypeptide chains must mediate protein–protein interactions to channel substrate intermediates to the correct downstream module (8, 10–12). We and others have reported the characterization of short, nonconserved linker regions that mediate, to a certain extent, communication between modules on separate proteins (10–12). However, subsequent studies suggest that these linkers may not be the only determinants for recognition of carrier proteins (13, 14).
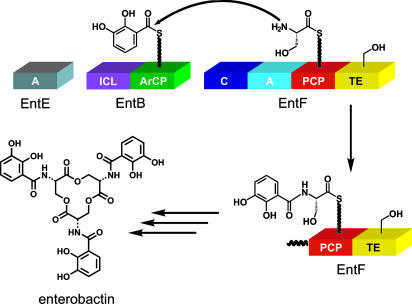
The four-component synthetase that produces enterobactin, an iron-chelating siderophore from Escherichia coli generated in response to iron deficiency, serves as an excellent model for studying the enzymology of NRPS systems (Fig. 1) (15, 16). Enterobactin is a trilactone produced from three molecules of 2,3-dihydroxybenzoate (DHB), derived from diversion of the shikimate pathway, and three molecules of serine. The biosynthesis of enterobactin follows typical NRPS logic (reviewed extensively in ref. 15). EntE, a free-standing adenylation domain, acylates the phosphopantetheine prosthetic group of EntB, a dual-function isochorismate lyase-aryl carrier protein (ArCP) didomain protein, with DHB (17, 18). EntF contains a single NRPS module that activates serine and condenses it with DHB (presented on EntB) to form DHB-Ser (16, 19–21). Three such elongation cycles are followed by macrolactonization by the thioesterase domain of EntF, resulting in release of enterobactin Fig. 1 shows the scheme for the condensation of DHB and serine catalyzed by EntF.
Fig. 1.
Schematic for the enterobactin synthetase and the condensation step of enterobactin biosynthesis.
Here, we describe a selection-based approach to revealing recognition determinants of EntB-ArCP by the downstream enterobactin synthetase elongation module EntF. The ArCP domain of EntB must participate in three protein–protein interactions during enterobactin biosynthesis: the first with EntD upon phosphopantetheinylation; the second with EntE upon acylation with DHB; and the third with EntF for condensation with serine. We used a combinatorial mutagenesis technique, shotgun alanine scanning (22–24), to identify crucial surface residues in EntB-ArCP that are required for the survival of E. coli on iron-depleted media. We then performed biochemical studies with EntB-ArCP mutants in vitro to determine whether the conserved residues from the selection were involved in interactions with various synthetase components. We identified several hydrophobic residues that constitute an interaction face on EntB-ArCP for EntF; in a structural homology model of EntB-ArCP, this face is located proximal to the phosphopantetheinylated serine. These results provide insights into recognition determinants in NRPSs.
Results
Structural Homology Model for the ArCP Domain of EntB.
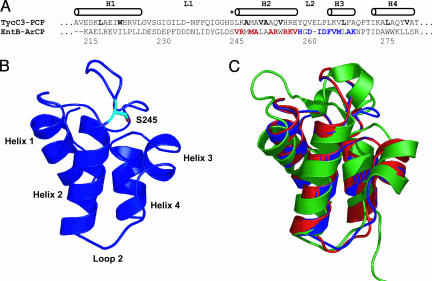
The prototypic PCP structure from the tyrocidine synthetase (TycC3-PCP) served as the template for an EntB-ArCP-homology model (25). An alignment with TycC3-PCP (Fig. 2A) revealed predicted core residues in EntB-ArCP, assuming high structural homology between these two carrier proteins. The solution structures for several carrier proteins from primary and secondary metabolism have been previously reported, all with well conserved folds (25–29). The sequence alignment of EntB-ArCP and TycC3-PCP was submitted to swiss-model to produce the EntB-ArCP homology model shown in Fig. 2B. Comparison of this EntB-ArCP homology model with the structure of TycC3-PCP and another carrier protein from the frenolicin type II PKS system (26) (Fig. 2C) reveals good agreement with both the template for the homology model (TycC3) and an unrelated ACP structure (FrnN). The three structures bear particular resemblance to one another in the helix 2/helix 3 regions. Although some inaccuracies may exist in the predicted structure of EntB-ArCP, it provided a rough model on which we based our library design. As is observed with other carrier proteins, the predicted EntB-ArCP structure is a four-helix bundle. The phosphopantetheinylated serine lies at the N terminus of the long helix 2. A short, one-turn helix (helix 3) buttresses the phosphopantetheinylated serine and is perpendicular to helices 1, 2, and 4. A long loop connects helices 1 and 2.
Fig. 2.
Structural model of EntB-ArCP and regions of randomization. (A) Sequence alignment of TycC3-PCP and EntB-ArCP. Core residues in TycC3-PCP are shown in bold. The helical regions in the solution structure are indicated (25). The phosphopantetheinylated serine is indicated with an asterisk. The residues on EntB that were subjected to shotgun alanine scanning are indicated in red (the H2 library) and blue (L2H3 library). The residue numbering is for full-length EntB. (B) EntB-ArCP structural homology model based on optimized sequence alignment with TycC3-PCP. (C) The overlay of this structural homology model with TycC3-PCP (red), and FrnN (green) from frenolicin biosynthesis shows good structural conservation among carrier proteins and validates the EntB-ArCP homology model.
Design of Libraries That Scan Selected Residues on the Surface of EntB.
Mutational and crystallographic studies have shown that helix 2 of some carrier proteins is involved in protein–protein interactions (29–31). For example, the cocrystal structure of the phosphopantetheinyl transferase from primary metabolism in Bacillus subtilis ACPS with ACP revealed multiple interactions between the phosphopantetheinylation enzyme and helix 2 of ACP (29). Subsequent studies by Marahiel and coworkers (30) demonstrated that mutating helix 2 residues of TycC3-PCP, normally not accepted as a substrate for ACPS, such that they match those of ACP is sufficient to enable the TycC3-PCP mutant to be recognized by ACPS. In addition, Khosla and coworkers (31) have shown that a residue on helix 2 of ACPs from type II PKS systems determines specificity for interaction with initiation or elongation ketosynthase domains. Structural studies with the ACP from the frenolicin PKS as well as other ACPs from primary metabolism have collectively suggested that the helix 3 region of certain carrier proteins is able to adopt multiple conformations (26, 28). These data suggest that helix 3 might be another important element for protein–protein interactions.
Based on the above analysis, we designed libraries H2 and L2H3 (Fig. 2A) to vary residues in helix 2 and loop 2/helix 3, respectively, of EntB-ArCP. All amino acids in these regions predicted to be surface-exposed were subjected to shotgun alanine scanning randomization, in which side chains were allowed to vary among WT, Ala, and occasionally a third or fourth residue, as required by the result of partial codon variation. The surface-exposed alanines in these regions (A250, A253, and A268) were allowed to vary among WT (Ala), Lys, Glu, and Thr. Gly 260 and the predicted core residues (M248, L251, A252, W255, and L267) of EntB-ArCP were not altered.
Library Production and Format of the Selection.
The E. coli K12-derived strain entB::kanR contains a chromosomal replacement of entB with a kanamycin-resistance gene [obtained from H. Mori (Osaka University, Japan)]. When this entB knockout strain was complemented with a pET22b-based plasmid containing the entB gene from E. coli CFT073, the cells were able to grow quickly on minimal media made iron-deficient by addition of the metal chelator 2,2′-dipyridyl (19). However, when the knockout strain was transformed with empty vector, no growth was observed on iron-deficient media after 3 days (see Fig. 5, which is published as supporting information on the PNAS web site). We hypothesized that entB complementation in this format could be used to rapidly process libraries of many EntB variants for their ability to support growth on iron-deficient media.
Libraries H2 and L2H3 were generated by using standard methods (see Materials and Methods) and introduced into entB::kanR cells by electroporation. The resulting transformants were plated onto iron-deficient media for selection of enterobactin producers. Surviving clones of varying colony diameters were observed after ≈40 h of growth at 37°C. The largest colonies were isolated and sequenced. The theoretical diversities of these libraries, 6.5 × 104 for H2 and 3.3 × 104 for L2H3, were adequately represented among our total 5 × 105 and 3 × 104 clones, respectively.
Conservation from the Surviving Pool.
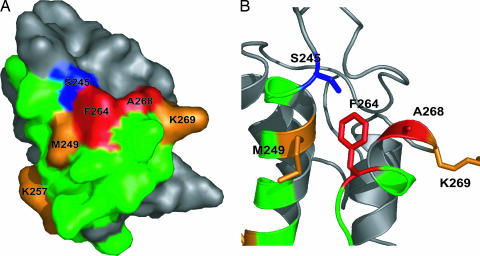
More than 100 nonredundant clones from the surviving pool of each library were sequenced. A graphical representation of the homology model, color-coded for degree of conservation among surviving clones, is shown in Fig. 3A. These sequence data are also represented numerically in Table 1 as the ratio of the number of times WT was observed at a position to the number of times a particular mutation was observed among the survivors (WT/mut). Two positions in the predicted helix 3 region were particularly intolerant to substitution, F264 and A268 (shown in red in Fig. 3). In the case of F264, only 1 surviving clone of 113 was found to contain Ala at this position (WT/mut = 95.0 in Table 1), although a few contained the third and fourth alternatives serine and valine (Table 5, which is published as supporting information on the PNAS web site). None of the surviving pool contained either Lys or Glu in place of Ala at position 268, and only one clone contained Thr at this position. Therefore, the WT/mut ratio for Lys and Glu at position 268 represents only an estimated lower limit.
Fig. 3.
Conservation of residues from library selections. (A) Surface representation of the EntB-ArCP homology model with residues color-coded for the observed degree of conservation from the surviving pools after selection. Red indicates high WT/mut ratios (>50), orange indicates intermediate (6 <WT/mut <50), and green indicates low conservation (<6). The phosphopantetheinylated serine is shown in blue. The ratios shown here and numerically in Table 1 are the results from sequencing of >100 clones from each surviving pool. (B) Ribbon diagram representation of A with side chains for the hotspot near S245 shown as stick.
Table 1.
WT/mut ratios for surviving clones of H2 and L2H3 libraries
| Helix 2 |
Loop 2/helix 3 |
||
|---|---|---|---|
| Residue | WT/mut* | Residue | WT/mut |
| V246 | 2.9 (A) | H259 | 1.9 (A) |
| R247 | 1.0 (A) | D261 | 1.3 (A) |
| M249 | 7.7 (A) | I262 | 5.2 (A) |
| A250 | 0.5 (K); 0.8 (E) | D263 | 5.3 (A) |
| A253 | 1.7 (K); 3.9 (E) | F264 | 95.0 (A) |
| R254 | 2.1 (A) | V265 | 1.7 (A) |
| R256 | 4.2 (A) | M266 | 1.2 (A) |
| K257 | 9.6 (A) | A268 | >112 (K); >112 (E) |
| V258 | 4.7 (A) | K269 | 12.0 (A) |
*For each position, a WT/mut ratio is shown where the mutant (indicated in parentheses) is alanine, in most cases. For the positions that contain WT Ala, the WT/mut ratios are shown for both lysine and glutamate. For a complete listing of WT/mut ratios, including third and fourth possible side chains, where applicable; see Table 5. Highly conserved residues, depicted in orange and red in Fig. 3, are shown in bold.
The strong preference for WT side-chain identity at F264 and A268 contrasts markedly with the majority of the positions in the randomized regions. Most positions displayed only moderate preference for WT (WT/mut ratios of 1–6, shown in green in Fig. 3). Arg 247 of helix 2, for example, exhibits no preference for WT or Ala (WT/mut = 1.0). Interestingly, position 250 showed a modest preference for Lys and Glu over the WT Ala (WT/mut <1). Neither A250 nor A253 displayed drastically different WT/mut ratios for Lys or Glu, indicating that the charged state of the side chain is not conserved at these positions. Several other positions had intermediate preference for the WT residue (colored in orange in Fig. 3). The charged residues K257 and K269 both preferred WT over Ala by >9-fold. In addition, M249 (which lies directly adjacent the phosphopantetheinylated serine on helix 2) was also highly conserved and is predicted to pack against the highly conserved F264 (Fig. 3B).
Overall, the sequence data indicate that in vivo enterobactin production does not require conservation at the majority of the surface residues within EntB-ArCP helix 2 and loop 2/helix 3. The two highly immutable residues F264 and A268 are clustered together on the homology model, and both are proximal to the phosphopantetheinylated serine. These positions are flanked by residues of intermediate importance, M249 and K269. This type of pattern (a few very important surface-exposed hydrophobic residues surrounded by residues of intermediate importance) is typical of protein–protein interfaces. For example, Cunningham and Wells (32) have demonstrated that, in the protein interaction interface of human growth hormone with its receptor, the majority of the binding energy can be attributed to only a small number of residues that are buried upon binding. Our observations suggest that selected residues in the helix 3 region, along with surrounding residues in helix 2 and helix 3, constitute a hot spot for protein–protein interaction in enterobactin biosynthesis.
Biochemical Characterization of Point Mutants.
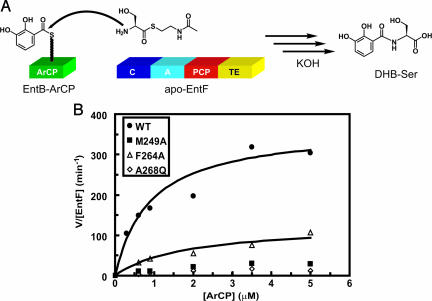
To determine whether the conservation shown in Fig. 3 was, indeed, because of interaction with other enterobactin synthetase components, EntD, -E, and -F, we cloned and expressed several EntB-ArCP point mutants at positions found to be conserved in the above in vivo studies: M249A, F264A, A268Q, and K269A. We have shown that the isochorismate lyase domain of EntB is not required for in vitro reconstitution of enterobactin synthetase activity if DHB is provided as a substrate (16). Therefore, we performed our biochemical experiments with the excised ArCP domains. The WT EntB-ArCP and all four mutants were purified as C-terminal His6-tagged proteins in good yield. We found that the presence of a C-terminal His6 tag on EntB did not significantly affect enterobactin production in an in vivo growth experiment (data not shown). The other enterobactin synthetase components EntD, -E, and -F were expressed and purified as reported in refs. 16, 18, and 33.
We performed enterobactin synthetase reconstitution assays (16) in which EntE, holo-EntB-ArCP (WT or mutants), and holo-EntF are incubated with DHB, serine, and ATP. In these assays, three of the four mutants above exhibited a decreased rate of enterobactin production relative to WT (Table 2). The most dramatic effect was observed for the mutation A268Q, which resulted in a 90-fold reduction in rate of enterobactin production. This observation is consistent with the in vivo selection results in which A268 of EntB was found to be nearly immutable in the L2H3 library. Two of the other mutations examined, M249A and F264A, also showed significant effects on enterobactin production under these conditions (9-fold and 4-fold slower, respectively). Surprisingly, the K269A mutant behaves similarly to WT in this assay. This discrepancy between in vivo and in vitro behavior may be a result of other interactions not examined in the reconstitution assay (34) or may arise from a possible role of K269 in the solubility and proper folding of EntB under selection conditions. Nonetheless, these data suggest that M249A, F264A, and A268Q are deficient in their ability to recognize the other enterobactin synthetase components (EntE or EntF). Because we used the broad-specificity enzyme B. subtilis Sfp to phosphopantetheinylate the carrier proteins in this assay (35), the interaction between EntD and each of the mutants is not measured here. We found that each of the mutants could be recognized and posttranslationally modified efficiently by Sfp (data not shown). Thus, the deficiency of the mutants for enterobactin production was not due to their inability to be phosphopantetheinylated.
Table 2.
Initial rates for enterobactin production in vitro
| EntB-ArCP | kobs, min−1 | kobs,WT/kobs,mut |
|---|---|---|
| WT | 36 ± 8 | — |
| M249A | 4 ± 1 | 9 |
| F264A | 9 ± 2 | 4 |
| A268Q | 0.4 ± 0.3 | 90 |
| K269A | 42 ± 5 | 0.9 |
To determine which of these interactions (EntE or EntF) was affected by the mutations M249A, F264A, and A268Q, we examined kinetic parameters for each of the EntB-ArCP mutants in two previously reported assays: condensation by EntF (20) and salicylation by EntE (17). In the EntF condensation assay (Fig. 4), a soluble serine N-acetylcysteamine thioester (Ser-S-NAC) substrate was used as a mimic for the Ser-S-EntF(PCP) intermediate. Upon basic workup, the condensation product DHB-Ser is obtained. Fig. 4 and Table 3 show the results of kinetic analysis for the condensation assay at 5 mM Ser-S-NAC for WT, M249A, F264A, and A268Q. Mutation of all three positions resulted in significantly decreased efficiency of condensation catalyzed by EntF. The mutation F264A had effects in both kcat and Km. We were not able to obtain saturation kinetics with M249A and A268Q because of issues with protein solubility >5 μM, but estimated catalytic efficiencies (kcat/Km) were reduced at least 68-fold relative to WT. These results suggest that M249, F264, and A268 are critical for interactions between EntB-ArCP and EntF and, therefore, constitute an interaction surface on EntB for EntF.
Fig. 4.
Condensation assay. (A) Schematic of the Ser-S-NAC condensation assay. A soluble thioester substrate acts as a mimic for natural PCP-bound intermediate. Upon basic work-up, the DHB-Ser condensation product is obtained. (B) Kinetics of condensation catalyzed by EntF by using WT EntB-ArCP or mutants M249A, F264A, and A268Q. Full parameters are listed in Table 3.
Table 3.
Kinetic parameters for condensation of DHB and serine by EntF
| EntB-ArCP | kcat, min−1 | Km, μM | kcat/Km, min−1 μM−1 |
|---|---|---|---|
| WT | 370 ± 20 | 0.9 ± 0.1 | 410 |
| M249A | ND* | ND* | ≈6 |
| F264A | 130 ± 20 | 1.8 ± 0.6 | 72 |
| A268Q | ND* | ND* | ≈2 |
*Saturation was not observed at concentrations of ArCP up to 5 μM; therefore, full kinetic data could not be obtained.
We used an EntE acylation assay that measures incorporation of radiolabeled salicylate, a substrate surrogate, onto EntB-ArCP to determine effects of the mutations on interactions of EntB with EntE (17). Table 4 shows kinetic parameters for WT EntB-ArCP and for the M249A and A268Q mutants (because F264A was only moderately deficient in the enterobactin production assay, we did not examine its salicylation kinetics). The WT parameters were in good agreement with published values (17) and were not significantly different from the kinetic parameters of either mutant. These results suggest that neither M249A nor A268Q are deficient in their ability to recognize EntE upon acylation and, therefore, that the inability of M249A and A268Q mutants to interact with EntF does not simply arise from a gross inability of these EntB mutants to fold properly.
Table 4.
Kinetic parameters for salicylation by EntE
| EntB-ArCP | kcat, min−1 | Km, μM |
|---|---|---|
| WT | 140 ± 20 | 0.5 ± 0.2 |
| M249A | 120 ± 20 | 1.1 ± 0.3 |
| A268Q | 140 ± 20 | 0.7 ± 0.4 |
Although our in vitro enterobactin production assays used the broad substrate specificity phosphopantetheinylation enzyme Sfp from B. subtilis, the natural carrier protein components of enterobactin synthetase are posttranslationally modified by the dedicated phosphopantetheinyltransferase EntD (33). We therefore examined the degree to which each of the ArCPs could recognize and be phosphopantetheinylated by EntD (see Fig. 6, which is published as supporting information on the PNAS web site). We found that F264A was accepted as efficiently as WT by EntD, and A268Q was accepted at an ≈2-fold lower rate. These results suggest that F264 is not involved in recognition by EntD and that A268 may be involved to a small degree. Surprisingly, M249A showed an enhanced rate of phosphopantetheinylation by EntD relative to WT. These data might account for the observation that the mutation M249A was tolerated to a higher degree than F264A in the in vivo selection, despite the fact that M249A is far more impaired in its ability to recognize EntF in vitro. It is possible that the deficiency of M249A to recognize EntF is offset partially by the enhanced rate of phosphopantetheinylation of M249A by EntD in vivo, which allows the enterobactin synthetase to produce enterobactin more efficiently with M249A than with F264A.
Discussion
Carrier proteins, comprising 80–100 residues that fold autonomously into the three- or four-helix structures shown in Fig. 2C, are key way stations in each module of every enzymatic assembly line that produces fatty acids, polyketides, and nonribosomal peptides, including the iron-chelating siderophores, such as enterobactin (1, 7, 15). The posttranslational priming with phosphopantetheine equips each carrier protein with this 20-Å-long prosthetic group and its terminal thiol that is the site of covalent tethering of the growing acyl chain. In many assembly lines, carrier proteins are embedded within multidomainal subunits, in cis, with catalytic domains that carry out chemical transformations on acyl chains tethered to the carrier proteins. In others, including the two-module enterobactin and several other siderophore synthetases, a free-standing carrier protein exists and must be recognized in trans by partner proteins. Whether in cis or in trans, carrier proteins in all assembly lines must present the tethered acyl groups to partner proteins with enzymatic activity in a spatially and temporally ordered fashion. We have deemed that evaluation of the recognition features of the free-standing EntB ArCP by its partner enzymes EntE (adenylation), EntD (phosphopantetheinylation), and EntF (condensation of DHB onto the Ser-S-PCP) could be most readily assessed as a first step to deciphering which face of the carrier protein scaffold was being recognized by partner enzymes.
The enterobactin synthetase is advantageous compared with other NRPS systems, because its function is essential for growth of E. coli in iron-limited medium and, so, offers the opportunity for selection for enterobactin synthetase components that are functional (19). In this study, we have used this feature to map the protein-interaction surface on EntB for EntF. The EntB residues M249, F264, and A268 constitute an interface for interaction with EntF; these three residues are predicted to cluster near the phosphopantetheinylated serine in our structural homology model based on TycC3-PCP. Both F264 and A268 are located on the predicted helix 3 of EntB-ArCP, suggesting that helix 3 may be an important recognition element in carrier proteins. In NMR studies with the ACP from frenolicin biosynthesis, Li et al. (26) found that the helix 3 region likely adopted multiple conformations but was able to assign only the major conformer. Nonetheless, the observed flexibility in this region may indicate that it is involved in protein–protein interactions in this system too. Interestingly, in the type II polyketide synthases of frenolicin and R1128, the position aligning with M249 of EntB-ArCP (helix 2) was found by Tang et al. (31) to mediate specificity for interaction. In these systems, initiation and elongation of a poly-β-ketone chain are carried out by two separate ketosynthase/ACP pairs. The initiation ketosynthase, a homologue of KSIII from fatty acid biosynthesis, is able to recognize the initiation ACP but not the elongation ACP. Mutations in this position in the elongation ACP, such that the residue matched that of the initiation ACP, resulted in ability of the initiation KSIII to recognize the elongation ACP. These observations suggest that the positions we have identified as important for protein–protein interactions with EntB-ArCP may also play functional roles in other systems.
At the protein–protein junctions of longer NRPS and PKS assembly lines, peptide-linker regions have been described that mediate transfer of biosynthetic intermediates to the appropriate downstream module (10–12). In the three-protein synthase that produces the aglycone scaffold of erythromycin, 6-deoxyethronolide B, Khosla and coworkers (10) have demonstrated that appending these short linker regions onto noncognate donor carrier proteins allows for recognition by downstream elongation modules. In the hybrid PKS/NRPS interface of the first two modules of epothilone biosynthesis, our laboratory has shown that removal of these linker regions disrupts the channeling of thiotemplated substrate intermediates to the downstream module (11, 36). In addition, similar communication-mediating domains have been discovered in the NRPS that produces tyrocidine (12). In many cases where noncognate interfaces have been engineered by appending these linkers to carrier proteins, the thiotemplated substrate is not processed by the downstream elongation module with the same efficiency as substrates presented on the natural carrier protein (13, 14). Our results with EntB, where much of the interface between a donor carrier protein (EntB-ArCP) and the downstream elongation module (EntF) lies in residues proximal to the phosphopantetheinylated serine, suggest that other recognition elements may be involved in such interfaces.
Although the EntB-ArCP mutants we studied were found to be significantly deficient in their ability to interact with EntF, M249A and A268Q could still be efficiently acylated by the free-standing adenylation domain EntE. In addition, all three mutants served as good substrates for the phosphopantetheinylation enzymes Sfp and EntD. These data suggest that there may be separate interaction faces for each of the proteins with which EntB-ArCP must interact to produce enterobactin. The implication that individual domains have distinct interaction faces on carrier proteins would suggest that reprogramming of NRPS or PKS machinery for production of noncognate natural product derivatives in high efficiency should be possible by appropriate engineering of carrier protein interactions. Furthermore, these results reinforce the idea that carrier proteins are information-rich scaffolds, in accord with results that show differentiation of carrier proteins by cognate vs. noncognate partner enzymes during assembly-line function (13, 14, 37). As precedent, it appears that the eukaryotic protein ubiquitin, about the same size as carrier protein domains, is also an information-rich scaffold, with at least two surfaces that can be recognized and differentiated by ubiquitin-binding proteins to direct the many distinct outcomes initiated by ubiquitylation of proteins (38, 39). It remains to be established whether all condensation domain–PCP domain interactions will use the equivalent surface residues on their partner carrier protein domains, in cis or in trans, but this hypothesis forms a testable set to evaluate carrier-protein domains as information-rich scaffolds.
Materials and Methods
Production of an EntB-ArCP Structural Homology Model.
The ArCP domain of EntB (residues M188–K285) was aligned with TycC3-PCP [Protein Database (PDB) ID 1DNY; an average structure was produced from the ensemble of NMR structures] by using the clustalw algorithm. The alignment was optimized for structural considerations then submitted to swiss-model (http://swissmodel.expasy.org//SWISS-MODEL.html) for calculation of a homology model. All structural figures were generated by using pymol (DeLano Scientific, http://pymol.sourceforge.net).
Library Production and Screening.
The entB gene from E. coli CFT073 (American Type Culture Collection 700928) was amplified from genomic DNA and cloned into pET22b (Novagen) to produce the plasmid pJRL16. For each library, an inactive template based on pJRL16 was generated that contained two sequential TAA stop codons followed by a unique restriction site (SacI for H2 and EcoRI for L2/H3) in the region of entB to be randomized. These inactive templates were used for full plasmid replication using the primers 5′-GAC TAC GGT CTG GAT TCG GYT SST ATG RYG RMA CTG GCG RMA SST TGG SST RMA GYT CAT GGT GAT ATC GAC TTT GTC-3′ H2, and 5′-GCG CGC TGG CGC AAA GTG SMT GGT GMT RYT GMT KYT GYT RYG CTG RMA RMA AAT CCG ACC ATC GAC GCC TGG TGG-3′ for L2H3, respectively (hybridization sequences indicated in italics, sites of randomization indicated in bold; K = G/T, M = A/C, R = A/G, S = G/C, Y = C/T). Plasmid replication using these library primers resulted in replacement of the stop codons with the desired regions of randomization. Double digestion with DpnI and SacI (H2) or EcoRI (L2/H3) resulted in destruction of the template. The purified library DNA product was then transformed into electrocompetent entB::kanR cells and plated onto minimal media made iron deficient by the addition of 100 μM 2,2′-dipyridyl (19). In a typical selection, 104 to 105 transformants were plated onto 241- × 241-mm plates of iron-deficient media and grown over two nights at 37°C. The largest colonies were picked, restreaked onto iron-deficient media, and sequenced. Sequencing was performed at the Dana Farber Cancer Institute Molecular Biology Core Facilities (Boston).
Site-Directed Mutagenesis, Protein Expression, and Purification.
The DNA-encoding site-directed mutants of EntB-ArCP were generated by splice overlap extension (40) and cloned into pET22b. EntB-ArCP (WT and mutants) were expressed as C-terminal His6-tagged proteins as previously described. EntD, EntE, EntF, and Sfp were purified as described (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site) (16, 19, 35).
Biochemical Assays.
The enterobactin reconstitution assay was performed by using a protocol modified from ref. 16 under the following conditions: 75 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 mM TCEP, 500 μM DHB, 1 mM serine, 10 mM ATP, 300 nM EntE, 450 nM EntB-ArCP (WT or mutants), and 300 nM EntF. Reaction progress was monitored by HPLC with water/acetonitrile/trifluoroacetic acid mobile phases. Assays for condensation by EntF, salicylation by EntE, and phosphopantetheinylation by EntD and Sfp were conducted as described in refs. 17, 20, and 35 and in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Hirotada Mori for the entB::kanR strain and Alexander Koglin, Volker Doetsch, and Kevin Phillips for helpful discussions. This research was supported by National Institutes of Health Grants AI042738, GM020011, and GM047467 (to C.T.W.) and GM065400 (to D.R.L.) and by the Howard Hughes Medical Institute (D.R.L.). J.R.L. acknowledges a postdoctoral fellowship from the Helen Hay Whitney Foundation. M.A.F. is funded by a predoctoral fellowship from the Hertz Foundation.
Abbreviations
- ACP
acyl carrier protein
- ArCP
aryl carrier protein
- DHB
2,3-dihydroxybenzoate
- NRPS
nonribosomal peptide synthetase
- PCP
peptidyl carrier protein
- PKS
polyketide synthase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Cane D. E., Walsh C. T., Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 2.Pieper R., Luo G., Cane D. E., Khosla C. Nature. 1995;378:263–266. doi: 10.1038/378263a0. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard B. K., Walsh C. T. Angew. Chem. Int. Ed. Engl. 2003;42:730–765. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- 4.Tang L., Shah S., Chung L., Carney J., Katz L., Khosla C., Julien B. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 5.Shen B., Du L., Sanchez C., Edwards D. J., Chen M., Murrell J. M. J. Nat. Prod. 2002;65:422–431. doi: 10.1021/np010550q. [DOI] [PubMed] [Google Scholar]
- 6.Aparicio J. F., Molnar I., Schwecke T., Konig A., Haydock S. F., Khaw L. E., Staunton J., Leadlay P. F. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 7.Walsh C. T., Gehring A. M., Weinreb P. H., Quadri L. E. N., Flugel R. S. Curr. Opin. Chem. Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji S. Y., Wu N., Khosla C. Biochemistry. 2001;40:2317–2325. doi: 10.1021/bi002462v. [DOI] [PubMed] [Google Scholar]
- 9.Belshaw P. J., Walsh C. T., Stachelhaus T. Science. 1999;284:486–489. doi: 10.1126/science.284.5413.486. [DOI] [PubMed] [Google Scholar]
- 10.Gockhale R. S., Tsuji S. Y., Cane D. E., Khosla C. Science. 1999;284:482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 11.O'Connor S. E., Walsh C. T., Liu F. Angew. Chim. Int. Ed. Engl. 2003;42:3917–3921. doi: 10.1002/anie.200352077. [DOI] [PubMed] [Google Scholar]
- 12.Hahn M., Stachelhaus T. Proc. Natl. Acad. Sci. USA. 2004;101:15585–15590. doi: 10.1073/pnas.0404932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu N., Tsuji S. Y., Cane D. E., Khosla C. J. Am. Chem. Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 14.Wu N., Cane D. E., Khosla C. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 15.Crosa J. H., Walsh C. T. Microbiol. Mol. Biol. Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring A. M., Mori I., Walsh C. T. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 17.Gehring A. M., Bradley K. A., Walsh C. T. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 18.Rusnak F., Faraci W. S., Walsh C. T. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 19.Roche E. D., Walsh C. T. Biochemistry. 2003;42:1334–1344. doi: 10.1021/bi026867m. [DOI] [PubMed] [Google Scholar]
- 20.Ehmann D. E., Trauger J. W., Stachelhaus T., Walsh C. T. Chem. Biol. 2000;7:765–772. doi: 10.1016/s1074-5521(00)00022-3. [DOI] [PubMed] [Google Scholar]
- 21.Ehmann D. E., Shaw-Reid C. A., Losey H. C., Walsh C. T. Proc. Natl. Acad. Sci. USA. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss G. A., Watanabe C. K., Zhong A., Goddard A., Sidhu S. S. Proc. Natl. Acad. Sci. USA. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison K. L., Weiss G. A. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 24.Gregoret L. M., Sauer R. T. Proc. Natl. Acad. Sci. USA. 1993;90:4246–4250. doi: 10.1073/pnas.90.9.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber T., Baumgartner R., Renner C., Marahiel M. A., Holak T. A. Structure (London) 2000;8:407–418. doi: 10.1016/s0969-2126(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 26.Li Q., Khosla C., Puglisi J. D., Liu C. W. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 27.Crump M. P., Crosby J., Dempsey C. E., Parkinson J. A., Murray M., Hopwood D. A., Simpson T. J. Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y., Prestegard J. H. Biochemistry. 1989;28:8792–8797. doi: 10.1021/bi00448a017. [DOI] [PubMed] [Google Scholar]
- 29.Parris K. D., Lin L., Tam A., Mathew R., Hixon J., Stahl M., Fritz C. C., Seehra J., Somers W. S. Structure (London) 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 30.Mofid M. R., Finking R., Marahiel M. A. J. Biol. Chem. 2002;277:17023–17031. doi: 10.1074/jbc.M200120200. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y., Lee T. S., Kobayashi S., Khosla C. Biochemistry. 2003;42:6588–6595. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham B. C., Wells J. A. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 33.Lambalot R. H., Gerhing A. M., Flugel R. S., Zuber P., LaCelle M., Marahiel M. A., Reid R., Khosla C., Walsh C. T. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 34.Hantash F. M., Earhart C. F. J. Bacteriol. 2000;182:1768–1773. doi: 10.1128/jb.182.6.1768-1773.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 36.Liu F., Garneau S., Walsh C. T. Chem. Biol. 2004;11:1533–1542. doi: 10.1016/j.chembiol.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Kim C. Y., Alekseyev V. Y., Chen A. Y., Tang Y., Cane D. E., Khosla C. Biochemistry. 2004;43:13892–13898. doi: 10.1021/bi048418n. [DOI] [PubMed] [Google Scholar]
- 38.Aguilar R. C., Wendland B. Curr. Opin. Cell Biol. 2003;15:184–190. doi: 10.1016/s0955-0674(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 39.Walsh C. T. Posttranslational Modification of Proteins: Expanding Nature's Inventory. Englewood, CO: Roberts; 2005. pp. 243–279. [Google Scholar]
- 40.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.