Abstract
In this paper, we report an in vivo model for the chimerins, a family of Rac GTPase-activating proteins (Rac-GAPs) that are uniquely regulated by the lipid second messenger diacylglycerol and have been implicated in the control of actin dynamics, migration, and proliferation. We cloned the zebrafish homologue of mammalian α2-chimerin (chn1) and determined that it possesses Rac-GAP activity and a C1 domain with phorbol ester/diacylglycerol-binding capability. chn1 morpholino knockdown embryos exhibit severe abnormalities, including the development of round somites, lack of yolk extension, and a kinked posterior notochord. These zebrafish morphants show Rac hyperactivation and progress faster through epiboly, leading to tailbud-stage embryos that have a narrow axis and an enlarged tailbud with expanded bmp4 and shh expression. Phenotypic rescue was achieved by mRNA microinjection of chn1 or an active chimerin Rac-GAP domain into the yolk syncytial layer but not by a chn1 mutant deficient in Rac-GAP activity, suggesting that the lack of chn1 Rac-GAP activity in the yolk syncytial layer was causative of the misbalance in morphogenetic movements. Our results reveal a crucial role for chn1 in early development and implicate Rac as a key regulator of morphogenetic movements during zebrafish epiboly.
The Rho family of small GTPases, which include Rho, Cdc42, and Rac, are key regulators of cell morphology, movement, and mitogenesis. These small GTPases cycle between active GTP-bound and inactive GDP-bound states. Activation is controlled by guanine exchange factors that promote GTP loading. On the other hand, functional inactivation is regulated by the action of GAPs (GTPase-activating proteins), molecules that stimulate the intrinsic GTP hydrolyzing activity of the small G proteins (1). Numerous Rac-GAPs have been identified, which have the ability to down-modulate Rac-mediated signaling. Chimerins, a family of Rac-GAPs, have emerged as important modulators of Rac function in various cellular models. Four mammalian chimerins with differential tissue distribution have been identified (α1-, α2-, β1-, and β2-chimerins), which are spliced variants of the CHN1 (α) and CHN2 (β) chimerin genes. A unique property of mammalian chimerins is the presence of a C1 domain upstream from the Rac-GAP domain that is highly homologous to those in PKC isozymes. This C1 confers responsiveness for the lipid second messenger diacylglycerol (DAG) and phorbol esters. Thus, mammalian chimerins represent a previously undescribed example of Rac down-modulators regulated by this lipid second messenger. Biochemical and structural analysis reveal that lipid binding to the chimerin C1 domain leads to allosteric activation of the Rac-GAP, therefore triggering a mechanism that limits Rac activation in response to various stimuli (2–5).
Although little is known about the cellular regulation and functional properties of chimerins, emerging evidence suggests that they are highly expressed in brain and several peripheral tissues, and that they play important roles in the control of actin dynamics, cell cycle progression, and neuritogenesis (6–8). β2-chimerin inhibits actin cytoskeleton reorganization and migration in response to growth factors and regulates cell proliferation by modulating Rac-mediated control of cyclin D1 expression and Rb phosphorylation. Because chimerins have been found to be down-regulated in various types of cancers (7, 9), a tumor suppressor role for these Rac-GAPs has been postulated.
The lack of in vivo models and limited information available in lower organisms has represented a serious limitation in our understanding of the functional properties of chimerin Rac-GAPs. In this paper, we report the identification of the product of the zebrafish α-chimerin gene (chn1) and its functional characterization. To gain further understanding on the role of chimerins in vivo, we used morpholino antisense oligonucleotides to knock down chn1 in zebrafish. Morphant embryos show enhanced Rac activity and display a characteristic phenotype that includes the development of round somites, lack of yolk extension, and a kinked posterior notochord. A thorough analysis revealed that chn1 controls morphogenetic movements during epiboly. Thus, our studies establish a relevant role for chimerin Rac-GAPs in early development.
Results
Cloning of chn1 and Expression During Development.
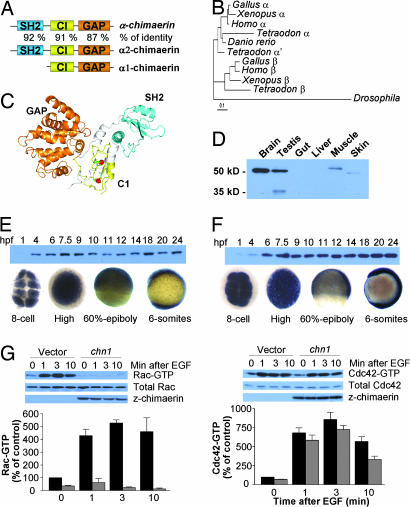
We cloned the zebrafish α-chimerin homologue from zebrafish brain cDNA by using a PCR approach. The protein, which we named Chn1 (GenBank accession no. AY684586), has an overall 86% amino acid sequence identity with human α2-chimerin and 74% with human β2-chimerin. Like the mammalian chimerin isoforms, chn1 contains three well defined domains: a N-terminal SH2 domain, a central C1 domain, and a C-terminal Rac-GAP domain. The SH2, C1, and GAP modules are highly conserved, having 92%, 91% and 87% identity, respectively, to those domains in human α2-α2-chimerin (Fig. 1A). Sequences available in public databases show that the chimerin gene is duplicated early in the vertebrate lineage. All available ESTs belong to the same gene, suggesting the possibility that a single chimerin that belongs to the α-chimerin subfamily is present in the zebrafish genome; however, the presence of another chimerin gene(s) cannot be completely ruled out (Fig. 1B). In silico modeling of chn1 to the recently solved structure of human β2-chimerin revealed a similar overall topology (Fig. 1C). Previous studies on lipid regulation of chimerins and structural information revealed that mammalian chimerins are subject to allosteric activation by DAG, an effect that involves the collapse of their “closed” or inactive conformation. Such conformation is kept by strong intramolecular contacts among the different domains (5). Residues involved in intradomain contact are highly conserved in chn1 (Fig. 5, which is published as supporting information on the PNAS web site), suggesting a similar autoinhibitory regulation than mammalian chimerins.
Fig. 1.
α-chimerin is a Rac-GAP that is zygotycally and maternally expressed in zebrafish. (A) Structure of chn1 and identity to α-chimerins. (B) Phylogenetic analysis of chimerin genes. (C) Molecular modeling of chn1. Cyan, SH2 domain; yellow, C1 domain; orange, GAP domain. (D) Western blot showing the expression of chn1 in different adult zebrafish organs. (E and F) Expression of chn1 and z-Rac in the zebrafish embryo at different stages by Western blot (Upper) and in situ hybridization (Lower). (G) Determination of Rac-GTP and Cdc42-GTP levels in COS-1 cells transfected with either pcDNA3-HA-chn1 (black bars) or pcDNA3-HA (empty vector; gray bars). Forty-eight hours after transfection, cells were serum starved (18 h) and then stimulated with EGF (100 ng/ml) for the times indicated in the figure. Rac-GTP and Cdc42-GTP levels were determined by using a pull-down assay, as described in Materials and Methods. Representative Western blots are included. Densitometric analysis of Rac-GTP and Cdc42-GTP levels normalized to total levels in each case is presented. Data are expressed percentage relative to Rac-GTP or Cdc42-GTP levels before EGF stimulation and represent the mean ± SE of five independent assays.
Using a monoclonal anti-chimerin antibody, we detected strong immunoreactivity in brain extracts from adult zebrafish at the expected molecular size (50 kDa). Immunoreactivity was also detected in testis and muscle. An ≈35-kDa band also was found in testis extracts (Fig. 1D), which presumably corresponds to a splice variant similar to that described in mammalian testis (10).
chn1 Is Maternally and Zygotically Expressed in the Zebrafish Embryo.
A temporal analysis of expression revealed that the chn1 transcript is maternally expressed, because it can be detected in eight-cell stage embryos and it is widely distributed during the cleavage, blastula, and gastrula periods (Fig. 1E). chn1 protein begins to be detected at 4 h postfertilization (hpf) (sphere stage). During the segmentation period, the expression of chn1 becomes restricted to neural tissue, and by the pharyngula period, the protein is highly expressed in brain. At the larval stages, chn1 is present in brain and in liver, gut, pancreas, and pharyngeal structures (Fig. 6, which is published as supporting information on the PNAS web site). Expression of chn1 during early development coincides with that of its target, zebrafish Rac1 (rac1). rac1 was cloned from brain cDNA by PCR with specific primers designed from partial EST sequences (accession no. AY682791), and we found that it is 99% identical to human and Xenopus Rac1. In situ hybridization analysis showed that rac1 mRNA is also maternally expressed and widely distributed during cleavage, blastula, and gastrula periods. rac1 was detected in embryonic extracts by Western blot from 4 hpf (Fig. 1F).
chn1 Is a Rac-Specific GAP and a Phorbol Ester Receptor.
Based on sequence and structural analysis, we predicted that chn1 has Rac-GAP activity. COS-1 cells were transfected with an expression vector encoding chn1 (pcDNA3-HA-chn1) or empty vector, and Rac-GTP levels in serum-growing cells were determined after 48 h by using a p21-binding domain (PBD) pull-down assay. Reduced Rac-GTP levels were observed in cells expressing chn1 relative to vector-transfected cells (Fig. 1G). We have recently found that mammalian chimerins inhibit receptor-induced stimulation of Rac activity (7). Likewise, expression of chn1 in COS-1 cells attenuates the elevations in Rac-GTP levels induced upon EGF receptor stimulation. On the other hand, chn1 did not affect elevations in Cdc42 induced by EGF. These results suggest that chn1 is a Rac-specific GAP.
Mammalian chimerins are high-affinity receptors for the lipid second messenger DAG and DAG mimetics such as the phorbol esters (11). The C1 domain in chn1 possesses all of the structural requirements for DAG responsiveness (Fig. 5). Indeed, recombinant chn1 generated in Escherichia coli binds [3H]PDBu (phorbol 12, 13-dibutyrate) in in vitro radioligand assays. Scatchard plot analysis revealed a Kd of 1.9 ± 0.6 nM (n = 3) (Fig. 7A, which is published as supporting information on the PNAS web site), which is in the same range as mammalian chimerins and other C1-domain containing proteins such as PKCs. chn1 is also responsive to phorbol esters in cellular models, because it can be translocated to membranes upon PMA (phorbol 12-myrystate 13-acetate) treatment (Fig. 7B). More importantly, PMA activates chn1 Rac-GAP activity (Fig. 7C), suggesting that chn1 represents a distinct class of Rac-GAP regulated by lipid binding to its C1 domain. Although the implications of this regulation are beyond the scope of this paper, the striking similarities of chn1 with its mammalian homologues α2- and β2-chimerin provide us with a great opportunity to explore the functional relevance of chimerins in the developing embryo.
chn1 Is Essential During Early Development.
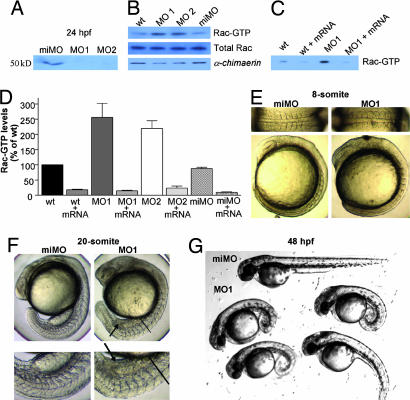
To assess the role of chn1 during zebrafish embryogenesis, we blocked mRNA translation of chn1 by using two different antisense morpholino oligonucleotides that target the 5′ UTR (MO1 and MO2) (12). A mismatched morpholino (miMO) of MO1 with five base exchanges was used as a control. After microinjection of either MO1 or MO2 at the one-cell stage, significant reductions in chn1 protein levels were observed in embryos both at 50% epiboly (≈50% reduction) and at 24 hpf (>90% reduction) (Fig. 2A and B). Interestingly, significant elevations in Rac-GTP levels were observed in whole embryos that had been injected with either MO1 or MO2 but not in those that received miMO (Fig. 2 B–D). Thus, chn1 plays a significant role as a physiological modulator of Rac activity in zebrafish. On the other hand, injection of chn1 mRNA (100 pg) into one-cell-stage embryos led to a significant decrease in Rac-GTP levels in control embryos. Moreover, coinjection of chn1 mRNA with either MO1 or MO2 embryos reverses the elevations in Rac-GTP levels caused by the morpholinos. Taken together, these results indicate that chn1 acts as a Rac-GAP in vivo.
Fig. 2.
Phenotypic changes during zebrafish development caused by morpholino-induced depletion of chn1. Injection of morpholinos against chn1 (MO1 or MO2) or a miMO was carried out at a one-cell stage. (A) Western blot in embryos injected with the different morpholinos against chn1 at 24 hpf by using an anti-chimerin antibody (4). (B) Rac-GTP levels in whole embryos at 50% epiboly. Rac-GTP levels were determined by using the PBD pull-down assay. Similar results were observed in three independent experiments. (C) Effect of chn1 mRNA injection on Rac-GTP levels in embryos injected with MO1. Rac-GTP levels were determined in whole embryos at 50% epiboly. A representative experiment is shown. Similar results were observed in at least three independent experiments. (D) Densitometric analysis of Rac-GTP levels in morpholino-injected embryos at 50% epiboly. Rac-GTP levels were determined by using the PBD pull-down assay and normalized to total Rac levels. Data are expressed as percentage relative to noninjected embryos (wt) and represent the mean ± SE of at least three independent assays. (E) Somite changes in MO1 embryos at 13 hpf (eight-somite). Upper, detailed dorsal views; Lower, lateral views. (F) Embryos at 19 hpf (20-somite). Upper, lateral views; Lower, detailed lateral views. Arrows indicate the accumulation of cells in lieu of a yolk extension. Lines indicate the starting point of the notochord defect. (G) Representative embryos at 48 hpf injected with either miMO or MO1.
Zebrafish morphants injected with either MO1 or MO2 show a peculiar phenotype with considerable morphogenic defects. At the eight-somite stage, these embryos display a narrower mediolateral extent of the axis with rounded somites as compared with the chevron-shaped somites observed in miMO-injected embryos (Fig. 2E). At the 20-somite stage, the majority of MO1-injected embryos lack or have a reduced yolk extension and have a kinked notochord and rounded somites (Fig. 2F). Some of these features, such as the narrow axis and rounded somites, resemble the ventralized mutants ogo and din (13, 14). However, unlike these mutants, chn1 morphants showed no signs of ventralization, as revealed by the expression pattern of marker genes (fkd3, gsc, dlx3, gata2, bmp4, eve, and shh) at different stages in MO1-injected embryos (Fig. 3D and E; see also Fig. 8, which is published as supporting information on the PNAS web site) and the lack of tail fin duplications (data not shown). At 48 hpf, most MO-injected embryos show considerable developmental abnormalities, which include a curled body shape and the lack of yolk extension (Fig. 2G). The majority of these embryos die before day 5. Indistinguishable phenotypes were observed for MO1- and MO2-injected embryos, and no differences were observed between miMO-injected and WT embryos (data not shown).
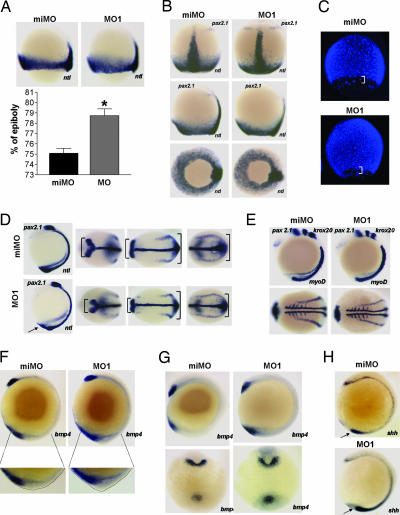
Fig. 3.
Morpholino depletion of chn1 leads to a faster epiboly progression and changes in anteroposterior cell distribution. Injection of MO1 or a miMO was carried out at a one-cell stage. (A) ntl expression and percentage of epiboly in embryos injected with either miMO or MO1. Embryos were staged and kept at 28°C until WT siblings reached 75% epiboly. Histogram shows quantification of percentage of epiboly (n = 10; ∗, P < 0.001). (B) ntl and pax2.1 expression in embryos injected with either miMO or MO1. Embryos were staged and kept at 28°C until WT siblings reached 90% epiboly. Top, dorsal; Middle, lateral; Bottom, vegetal views. (C) DAPI stain of embryos injected with either miMO or MO1. Embryos were staged and kept at 28°C until WT siblings reached 90% epiboly. Brackets, YSL nuclei. (D) ntl and pax2.1 expression in three-somite embryos. Images, from left to right, are lateral, dorsal anterior, dorsal medial, and dorsal posterior views. Arrows indicate enlargement of the tb. Brackets indicate the narrowing of midbrain–hindbrain boundary in MO1-injected embryos. (E) Lack of changes in patterning, as shown with pax2.1, myoD, and krox20 expression in six-somite embryos. (F) Lateral views of bmp4 expression in one-somite embryos and details of the outlined tb morphologies. MO1 embryos show an enlargement of the tb. (G) Lateral and ventral views of bmp4 expression in six-somite embryos showing expansion of the tb expression of bmp4. (H) Expansion of shh expression dorsal to the tb in five-somite embryos.
Morphological Analysis of chn1 Knockdown Embryos.
The striking phenotypic changes observed upon morpholino depletion of chn1 prompted us to pursue a more detailed morphogenetic analysis. Initial experiments in which we performed time-lapse microscopy of developing embryos revealed that although MO1- and miMO-injected embryos appeared essentially identical before epiboly, those with chn1 depletion showed a slight but reproducible faster progression through epiboly of the blastoderm, the enveloping layer (EVL), and the yolk syncytial layer (YSL) from its onset until it reached 90–95% (data not shown). This effect could be measured by in situ hybridization in embryos fixed at 75% epiboly by using the mesoderm marker ntl (Fig. 3A). A quantitative analysis revealed a higher percentage of epiboly completed in MO1-injected embryos relative to miMo-injected age-matched embryos. Additionally, expression of pax2.1 in the mid-hindbrain boundary initiates concurrently in miMO and MO1 siblings, although MO1-injected embryos showed an advanced epiboly stage (Fig. 3B). DAPI and phalloidin staining of aged-matched embryos confirmed the faster epiboly progression in the blastoderm, EVL, and YSL of chn1-depleted embryos (Fig. 3C; see also Fig. 9, which is published as supporting information on the PNAS web site). Onset of somitogenesis was normal in MO1-injected embryos. Expression of pax2.1 in the midbrain hindbrain boundary and ntl at the three-somite stage revealed a narrower axis in MO1-injected embryos compared with controls (Fig. 3D; see also Fig. 10A, which is published as supporting information on the PNAS web site). On the other hand, no significant differences were observed in the length of the notochord (Fig. 10A). Staining of krox20 in rhombomeres 3 and 5, myoD in the somites and paraxial mesoderm, and pax2.1 at the six-somite stage, confirmed the presence of a narrower axis in MO1 morphants and reveal that there were no major changes in dorsoventral patterning (Fig. 3E). A distinct feature of MO1-injected embryos is the presence of a large tailbud (tb) (Fig. 3 D and F). Although there are no described mutants with faster epiboly progression, there is a recent report on mutants with delayed epiboly, poky (pky) and slow (sow). Slowing down epiboly results in delayed extension. Indeed, these mutants show a thicker axis with broader pax2.1 and ntl expression in the mid-hindbrain boundary and in the axial mesoderm, respectively (15). Other mutants, such as volcano and half baked, show slower blastoderm epiboly due to uncoupling of the different layers (16, 17).
We hypothesized that altering the rate of epiboly could lead to changes in the distribution of cells along the anteroposterior axis. To further investigate this issue we measured the expression of bmp4, a gene that is expressed in the prechordal plate (pcp) and the ventral tb (18). At the tb stage, chn1 morphant embryos show normal expression of bmp4 in the pcp but an expansion of the expression in the ventral tb (Fig. 3F). Moreover, an abnormal accumulation of cells was observed in the tb of MO1-injected embryos (Fig. 3F). This difference in expression also was observed in six-somite stage embryos (Fig. 3G). At the six-somite stage, the expression of the dorsal mesoderm marker shh also was expanded at the posterior end of MO1-injected embryos (Fig. 3H). However, the pattern of expression of bmp4 and shh was not affected at shield and 75% epiboly stages (Fig. 8 D and E), suggesting that the expansion of the expression of these genes in the tb could be due to an increase in the number of cells that reach the posterior end rather than to a priori changes in cell fates. The improper formation of the yolk extension may be secondary to the accumulation of cells in the tb, although we cannot rule out that it may relate to the depletion of chn1 at late stages. This accumulation of cells may also be a primary event that drives subsequent notochord defects and the curly shaped tail of the chn1-depleted embryos.
chn1 Acts in the YSL to Regulate Epiboly.
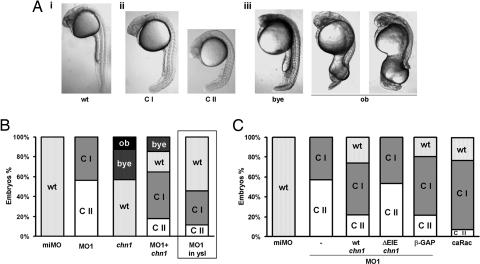
MO1-injected embryos were placed into different classes based on the severity of the phenotype at 24 hpf (Fig. 4A). Class II embryos (≈60% of the MO1-injected embryos) show no yolk extension and a kinked notochord. Class I embryos (≈40%) show a milder phenotype (reduced yolk extension without appreciable changes in the notochord). To confirm the specificity of the morpholino effect, we attempted a phenotypic rescue by microinjection of chn1 mRNA. This mRNA lacked the 5′ UTR to prevent morpholino interference. Injection of chn1 mRNA (200 pg) into MO1-injected embryos causes a significant rescue, with only 18% class II embryos, 48% having the mild phenotype, and 22% WT. Interestingly, we observed that a fraction (15%) of the embryos display bigger yolk extensions.
Fig. 4.
Phenotypic rescue of MO1-injected embryos by chn1 mRNA injection. (A) The phenotypic classes of chn1-depleted and -overexpressing embryos: i, WT embryo at 24 hpf; ii, MO1-injected embryos were divided into class I (CI; thin yolk extension) and class II (CII; thin or no yolk extension and kinked notochord); iii, bigger yolk extension (bye) and open back (ob) phenotypes observed upon injection of chn1 mRNA. (B) Percentage of embryos in each class after injecting miMO, MO1, chn1 mRNA (200 pg), or MO1 together with chn1 mRNA (200 pg) at the one-cell stage. The right column shows the injection of MO1 in the YSL at high stage. Graph shows the average of at least three independent experiments with 30–50 embryos per group. (C) Effect of microinjection of wt-chn1, ΔEIE-chn1, β-GAP domain, or caRac mRNA into the YSL. Embryos were injected with either miMO or MO1 at the one-cell stage. At high stage, MO1-injected embryos were injected with either chn1, ΔEIE-chn1, or β-GAP domain mRNA, and WT siblings were injected with caRac mRNA (50 pg). Graph shows the percentage of embryos in each of the groups, which are the average of three independent experiments with 30–50 embryos per group.
In the next series of experiments, we analyzed the effect of injecting chn1 mRNA (200 ng) into one-cell stage WT embryos. Remarkably, 30% of the embryos displayed bigger yolk extensions and 13% exhibited an “open back” phenotype characteristic of slower epiboly mutants (Fig. 4B; ref. 15).
It has been proposed that the YSL is a driving force in epiboly (19), an event that is dynamically regulated by microtubules and actin (20, 21). Because Rac plays a crucial role in the regulation of the actin cytoskeleton (1, 22), and mammalian chimerins inhibit actin cytoskeleton reorganization mediated by Rac (6), we decided to deplete chn1 in the YSL by injecting MO1 into the syncytium at high stage. Remarkably, this depletion led to a significant number of class I and class II embryos, although the phenotype was milder than when injected in one-cell stage embryos (Fig. 4B). Furthermore, when chn1 mRNA (50 pg) was injected in the YSL of embryos previously injected with MO1, there was a partial rescue of the yolk extension and the notochord phenotype, because 22% of the embryos show no phenotype, 62% were class I, and only 16% were class II (Fig. 4C). In addition, the expansion of bmp4 in the tb also was rescued by chn1 mRNA injection (Fig. 10B). These experiments confirm the specificity of the morpholino knockdown and suggest that the YSL is a major site of action of chn1 for epiboly regulation. Thus, the lack of chn1 activity in the YSL is causative of a misbalance in morphogenetic movements.
Finally, to determine whether the phenotypic changes caused by chn1 depletion relate to Rac hyperactivation, we investigated the role of the Rac-GAP domain by using rescue experiments. A chn1 mutant was generated in which essential residues in the GAP domain have been deleted (ΔEIE, positions 291–293). A similar mutant in mammalian chimerins is GAP-deficient, because it is incapable of hydrolyzing GTP from Rac1 (4, 23). To confirm that ΔEIE-chn1 was Rac-GAP-deficient, we assessed its activity by using Rac-GTP pull-down assays in COS-1 cells and found indeed that it was unable to reduce Rac-GTP levels (data not shown). Injection of ΔEIE-chn1 mRNA (50 pg) into the YSL of MO1 embryos was unable to rescue the phenotype, therefore confirming that the Rac-GAP activity of chn1 was required for the rescue effect (Fig. 4C). To further strengthen these results, we also injected mRNA encoding for the Rac-GAP domain of mammalian β2-chimerin (β-GAP) into the YSL (4, 6, 7), and, in this case, we observed a significant rescue of the MO1 effect. Furthermore, the MO1 phenotype could be partially mimicked by expression of caRac in the YSL of WT embryos. Injection of 50 pg of caRac mRNA lead to 70% class I, 7% class II, and 23% of the embryos showing no changes in the yolk extension and notochord (Fig. 4C). These results not only suggest that the phenotypic changes caused by depletion of chn1 involve the up-regulation Rac activity in the YSL but also support the concept that chn1 has a critical role in the control of early events in zebrafish embryogenesis.
Conclusion
Considerable attention has been directed toward studying Rac in mammalian cells as an effector of cell-shape changes, migration, mitogenesis, and malignant transformation. However, little is known on the role of Rac and other Rho GTPases in early development. Our studies have provided evidence that dysregulation of Rac activity by depletion of a specific Rac-GAP leads to significant morphogenetic changes in the zebrafish embryo. Studies using dominant-negative and constitutively active forms of Rac and Rho suggested a role for these GTPases in the control of cell motility during Xenopus gastrulation. Disruption of gastrulation occurs through the regulation of convergence and extension movements by controlling actin cytoskeleton dynamics (24). Also in Xenopus, coactivation of Rac and Rho by Wnt/Frizzled signaling is required for cell movement during vertebrate gastrulation (25). More recently, studies in zebrafish based on dominant-negative and gain-of-function approaches have suggested a role for Rac in dorsal migration of lateral cells during gastrulation downstream of hyaluronan (HA), an extracellular polysaccharide implicated in migration (26). Has2, an enzyme that catalyzes the synthesis of HA, is required for dorsoventral morphogenesis but not for dorsoventral patterning in zebrafish, an effect that can be mimicked by constitutively active Rac1 and blocked by dominant-negative Rac1. Indeed, these results have been the only reported genetic evidence for a mechanism controlling dorsal convergence without affecting extension. Has2 and constitutively active Rac promote lamellipodia formation in dorsal convergence in zebrafish embryos (26). In our studies, gain of Rac function by specific depletion of chn1 resulted in changes in cell distribution along the anteroposterior axis. These changes are probably due to a dysbalance in cell movements that results in fewer cells reaching the axis with the consequent accumulation of cells in the tb. Altogether, these results support a key role for Rac in the regulation and coordination of cellular movements during the zebrafish gastrulation. Rac must play an important role in the regulation of epiboly, as suggested by the rescue of the MO1 morphant phenotype by injecting chn1 mRNA into the YSL. Epiboly may involve an extensive actin remodeling, because disruption of actin-based structures with cytochalasin B leads to the arrest of epiboly (21). Thus, we hypothesize that the phenotypic changes caused by chn1 depletion are a consequence of Rac signaling dysregulation of actin dynamics in the YSL during epiboly.
Gain-of-function studies in various mammalian systems in culture by using overexpression approaches have established an important role for chimerins in actin dynamics, migration, and cell cycle control, as well as in neuritogenesis (6–8). However, the lack of animal models so far represents a serious limitation in our understanding of the function and regulation of chimerins in living organisms. The fact that morpholino depletion of the single chimerin gene in zebrafish results in a significant gain of function of Rac and profound phenotypic changes attests to a prominent physiological role for this Rac-GAP in zebrafish.
Chimerins represent the only example of DAG-regulated GAPs. In mammalian cellular models, we have observed that in response to receptors that generate DAG via phospholipase C activation (such as the EGF receptor), chimerins relocalize to the plasma membrane, where they inactivate Rac (unpublished observations). The role of DAG and the nature of the cues that generate this lipid second messenger in early stages of development in vertebrates are essentially unknown. The fact that chn1 GAP activity is regulated by DAG mimetics suggests the possibility of a lipid-regulated mechanism that modulates Rac activity in early development.
Materials and Methods
Cloning of α-chimerin.
The 1,766-bp fragment containing the coding region of chn1 (accession no. AY684586) was amplified by PCR from adult zebrafish brain cDNA by using the primers 5′-CACTTCATGCTGGGCTGC and 5′-TCCAGAGTCTGAGGAACAGTGC. The PCR fragment was subcloned into pCRII (Invitrogen).
Phylogenetic Analysis.
Protein sequences were retrieved from GenBank (accession nos. for chimerin homologues: human α, NP_001813; human β, U28926; Xenopus α, AAH56112; Xenopus β, AAH73525; chicken α, NP001012970; chicken β, XP_425997; Tetraodon α, CAG09973; Tetraodon α′, CAF98187; Tetraodon β, CAG12004; Drosophila, AF46047). Sequence alignments were done by using clustalx (27). Poorly aligned regions were excluded from analysis. phyml (http://atgc.lirmm.fr/phyml) was used to construct a maximum likelihood tree by using a WAG substitution model (28). Site-to-site rate variation was modeled on a gamma distribution with four categories of substitution rates and invariable sites, with the alpha parameter and proportion of invariable sites estimated from the data.
Western Blot Analysis.
Western blots were done as described in ref. 4 by using the following antibodies: anti-chimerin (1:1,000; ref. 4), anti-HA (1:3,000; Babco, Richmond, CA), anti-Rac (1:3,000; Upstate Biotechnology, Lake Placid, NY), and anti-Cdc42 (1:3,000; Upstate Biotechnology). Immunoreactive bands were visualized by using chemiluminescence. Densitometry analysis was performed by using imagej (National Institutes of Health, Bethesda).
In Situ Hybridization.
Whole mount in situ hybridization was carried out as described in ref. 15. The chn1 probe was generated by digestion from pCRII-chn1 by using MluI and transcribed with T7 RNA polymerase. Other probes used were ntl, pax2.1, krox20, myoD, shh (15), and bmp4 (29).
Cell Culture.
COS-1 cells were cultured as described in ref. 11. Transfections were carried out in six-well plates by using fugene6 (Roche Molecular Biochemicals), according to the manufacturer’s protocol. pcDNA3-HA-chn1 was produced by subcloning the chn1 coding region into BamHI and EcoRI sites in pcDNA3-HA (kind gift of Margaret M. Chou, University of Pennsylvania).
Determination of Rac-GTP and Cdc42-GTP Levels.
A pull-down assay was used to isolate Rac-GTP or Cdc42-GTP by binding to the PBD of PAK1, as described in ref. 4. For determination of Rac-GTP levels in whole embryos, 40 embryos at 50% epiboly were lysed by passing them through a needle in 1 ml of GLPB buffer (20 mM Tris·HCl, pH 7.5/1 mM DTT/5 mM MgCl2/150 mM NaCl/0.5% Nonidet P-40/5 mM β-glycerophosphate/protease inhibitor mixture from Sigma-Aldrich containing 8 μg of GST-PBD.
Site-Directed Mutagenesis.
Deletion of amino acids 291–293 from chn1 (mutant ΔEIE-chn1) was generated by PCR as described in ref. 30 by using the following oligonucleotide: 5′-GGTGGACATGTGCATACGAGCACGAGGGTTGCAGTC.
mRNA Synthesis, Morpholino Oligonucleotides, and Microinjection.
miMOs were obtained from Gene Tools (Philomath, OR). Sequences were as follows: MO1, 5′-GCCATTGCAGACAGTGATTCAGCCG; MO2, 5′-AAGACAAATAAACAATGACCCGCCT; miMO (MO1 with five nucleotide mismatches, indicated in lowercase), 5′-GCgATTgCAGACAcTGATTgAGgCG′. Lyophilized morpholinos were dissolved in water, diluted in Danieau buffer (12), and then injected (3 ng) into one-cell stage embryos. Volume of injection was in all cases 1 nl. Coding regions of chn1, the β-GAP (amino acids 290–466 from β2-chimerin) and human constitutively active Rac1 were amplified with primers containing BamHI and EcoRI restriction sites and cloned into pCS2+. Capped mRNA was in vitro transcribed from pCS2+ plasmids linearized with NotI by using SP6 mMessage mMachine kits (Ambion, Austin, TX) and injected into embryos at the one-cell stage (50–200 pg). For the YSL injections, 50 pg of RNA were injected into the syncytiun of a random subset of MO1-injected embryos at the high stage.
Photography.
Images where captured on Prog/Res/3012 (Kontron, Zurich) and Cool Snap by using openlab (Improvision, Lexington, MA) and processed by using photoshop (Adobe Systems, San Jose, CA).
Supporting Information.
For more information, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Dr. Peter M. Blumberg and Nancy A. Lewin (National Institutes of Health) for their help with [3H]PDBu-binding assays, Dr. Michael Pack (University of Pennsylvania) for help in the early phases of this study, and Dr. Claudio Slamovits (University of British Columbia, Vancouver) for help with the phylogenetic analysis. This work was supported by National Institutes of Health Grants CA74197 (to M.G.K.) and ES11248 (to M.C.M.).
Abbreviations
- DAG
diacylglycerol
- HA
hyaluronan
- hpf
hours postfertilization
- miMO
mismatched morpholino
- PBD
p21-binding domain
- tb
tailbud
- YSL
yolk syncytial layer.
Footnotes
References
- 1.Etienne-Manneville S., Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 2.Caloca M. J., Garcia-Bermejo M. L., Blumberg P. M., Lewin N. E., Kremmer E., Mischak H., Wang S., Nacro K., Bienfait B., Marquez V. E., Kazanietz M. G. Proc. Natl. Acad. Sci. USA. 1999;96:11854–11859. doi: 10.1073/pnas.96.21.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caloca M. J., Wang H., Delemos A., Wang S., Kazanietz M. G. J. Biol. Chem. 2001;276:18303–18312. doi: 10.1074/jbc.M011368200. [DOI] [PubMed] [Google Scholar]
- 4.Caloca M. J., Wang H., Kazanietz M. G. Biochem. J. 2003;375:313–321. doi: 10.1042/BJ20030727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canagarajah B., Leskow F. C., Ho J. Y., Mischak H., Saidi L. F., Kazanietz M. G., Hurley J. H. Cell. 2004;119:407–418. doi: 10.1016/j.cell.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Menna P. L., Skilton G., Leskow F. C., Alonso D. F., Gomez D. E., Kazanietz M. G. Cancer Res. 2003;63:2284–2291. [PubMed] [Google Scholar]
- 7.Yang C., Liu Y., Leskow F. C., Weaver V. M., Kazanietz M. G. J. Biol. Chem. 2005;280:24363–24370. doi: 10.1074/jbc.M411629200. [DOI] [PubMed] [Google Scholar]
- 8.Hall C., Michael G. J., Cann N., Ferrari G., Teo M., Jacobs T., Monfries C., Lim L. J. Neurosci. 2001;21:5191–5202. doi: 10.1523/JNEUROSCI.21-14-05191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan S., Miller D. W., Barnett G. H., Hahn J. F., Williams B. R. Cancer Res. 1995;55:3456–3461. [PubMed] [Google Scholar]
- 10.Hall C., Monfries C., Smith P., Lim H. H., Kozma R., Ahmed S., Vanniasingham V., Leung T., Lim L. J. Mol. Biol. 1990;211:11–16. doi: 10.1016/0022-2836(90)90006-8. [DOI] [PubMed] [Google Scholar]
- 11.Caloca M. J., Fernandez N., Lewin N. E., Ching D., Modali R., Blumberg P. M., Kazanietz M. G. J. Biol. Chem. 1997;272:26488–26496. doi: 10.1074/jbc.272.42.26488. [DOI] [PubMed] [Google Scholar]
- 12.Nasevicius A., Ekker S. C. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 13.Solnica-Krezel L., Stemple D. L., Mountcastle-Shah E., Rangini Z., Neuhauss S. C., Malicki J., Schier A. F., Stainier D. Y., Zwartkruis F., Abdelilah S., Driever W. Development (Cambridge, U.K.) 1996;123:67–80. doi: 10.1242/dev.123.1.67. [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt M., Pelegri F., Mullins M. C., Kane D. A., van Eeden F. J., Granato M., Brand M., Furutani-Seiki M., Haffter P., Heisenberg C. P., et al. Development (Cambridge, U.K.) 1996;123:95–102. doi: 10.1242/dev.123.1.95. [DOI] [PubMed] [Google Scholar]
- 15.Wagner D. S., Dosch R., Mintzer K. A., Wiemelt A. P., Mullins M. C. Dev. Cell. 2004;6:781–790. doi: 10.1016/j.devcel.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Kane D. A., McFarland K. N., Warga R. M. Development (Cambridge, U.K.) 2005;132:1105–1116. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- 17.Kane D. A., Maischein H. M., Brand M., van Eeden F. J., Furutani-Seiki M., Granato M., Haffter P., Hammerschmidt M., Heisenberg C. P., Jiang Y. J., et al. Development (Cambridge, U.K.) 1996;123:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- 18.Sepich D. S., Myers D. C., Short R., Topczewski J., Marlow F., Solnica-Krezel L. Genesis. 2000;27:159–173. doi: 10.1002/1526-968x(200008)27:4<159::aid-gene50>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 19.Betchaku T., Trinkaus J. P. J. Exp. Zool. 1978;206:381–426. doi: 10.1002/jez.1402060310. [DOI] [PubMed] [Google Scholar]
- 20.Strahle U., Jesuthasan S. Development (Cambridge, U.K.) 1993;119:909–919. doi: 10.1242/dev.119.3.909. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J. C., Miller A. L., Webb S. E. Dev. Dyn. 2004;231:313–323. doi: 10.1002/dvdy.20144. [DOI] [PubMed] [Google Scholar]
- 22.Hall A. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed S., Lee J., Wen L. P., Zhao Z., Ho J., Best A., Kozma R., Lim L. J. Biol. Chem. 1994;269:17642–17648. [PubMed] [Google Scholar]
- 24.Tahinci E., Symes K. Dev. Biol. 2003;259:318–335. doi: 10.1016/s0012-1606(03)00206-9. [DOI] [PubMed] [Google Scholar]
- 25.Habas R., Dawid I. B., He X. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakkers J., Kramer C., Pothof J., Quaedvlieg N. E., Spaink H. P., Hammerschmidt M. Development (Cambridge, U.K.) 2004;131:525–537. doi: 10.1242/dev.00954. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S., Gascuel O. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 29.Connors S. A., Trout J., Ekker M., Mullins M. C. Development (Cambridge, U.K.) 1999;126:3119–3130. doi: 10.1242/dev.126.14.3119. [DOI] [PubMed] [Google Scholar]
- 30.Ito W., Ishiguro H., Kurosawa Y. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.