Abstract
Dystrophin mechanically links the costameric cytoskeleton and sarcolemma, yet dystrophin-deficient muscle exhibits abnormalities in cell signaling, gene expression, and contractile function that are not clearly understood. We generated new antibodies specific for cytoplasmic γ-actin and confirmed that γ-actin most predominantly localized to the sarcolemma and in a faint reticular lattice within normal muscle cells. However, we observed that γ-actin levels were increased 10-fold at the sarcolemma and within the cytoplasm of striated muscle cells from dystrophin-deficient mdx mice. Transgenic overexpression of the dystrophin homologue utrophin, or functional dystrophin constructs in mdx muscle, restored γ-actin to normal levels, whereas γ-actin remained elevated in mdx muscle expressing nonfunctional dystrophin constructs. We conclude that increased cytoplasmic γ-actin in dystrophin-deficient muscle may be a compensatory response to fortify the weakened costameric lattice through recruitment of parallel mechanical linkages. However, the presence of excessive myoplasmic γ-actin may also contribute to altered cell signaling or gene expression in dystrophin-deficient muscle.
Keywords: costamere, muscular dystrophy, sarcolemma
Duchenne muscular dystrophy is caused by mutations in the gene encoding dystrophin (1). Dystrophin functions as part of a large complex of sarcolemmal proteins including dystroglycans, sarcoglycans, dystrobrevins, syntrophins, and sarcospan (2, 3). This dystrophin–glycoprotein complex is thought to link the actin-based costameric cytoskeleton with the extracellular matrix and mechanically stabilize the sarcolemma against shear stresses imposed during muscle activity (4). When dystrophin is absent the link between the costamere and sarcolemma is disrupted, resulting in compromised sarcolemma integrity (4).
Dystrophin is anchored to the sarcolemma primarily through direct interaction with β-dystroglycan (5, 6). Regarding its interaction with costameres, the amino-terminal calponin homology domain and a cluster of basic spectrin repeats within the middle rod domain of dystrophin bind directly to actin filaments (7–10). Cytoplasmic γ-actin filaments are retained in a costameric pattern on sarcolemma peeled from single myofibers of normal mouse muscle but are absent from all sarcolemma of dystrophin-deficient mdx muscle (11). Thus, dystrophin is necessary for a mechanically strong link between the sarcolemma and costameric actin filaments.
Here we peeled sarcolemma from several lines of transgenic mdx mice expressing deletion constructs of dystrophin, and we report that either actin binding domain is sufficient to retain costameric actin on peeled sarcolemma. We generated new polyclonal antibodies (pAbs) and mAbs to cytoplasmic γ-actin, and we demonstrate that γ-actin levels are elevated 10-fold in dystrophin-deficient striated muscle. We hypothesize that elevated γ-actin levels contribute to a compensatory remodeling of the dystrophin-deficient costameric cytoskeleton. Our results also provide the basis for several pathways by which increased cytoplasmic γ-actin levels may contribute to the widespread cellular perturbations observed in dystrophin-deficient muscle.
Results
Immunofluorescence Analysis of Peeled Sarcolemma.
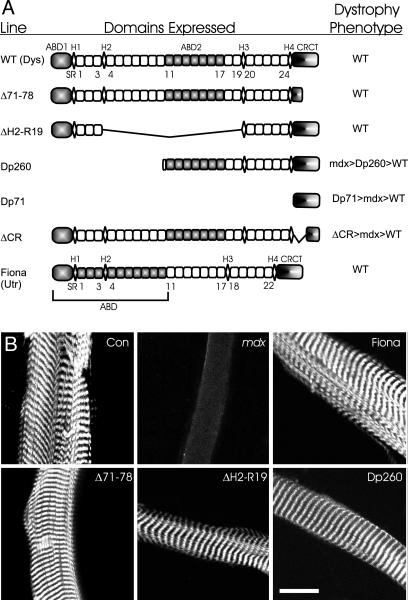
To identify the dystrophin domains necessary to anchor costameric actin to sarcolemma, we imaged phalloidin staining of peeled sarcolemma from transgenic mdx mice overexpressing dystrophin constructs deleted in different domains (Fig. 1A). As previously shown (11), actin was retained on sarcolemma peeled from control but not mdx muscle (Fig. 1B). Consistent with previous studies (12, 13), actin was retained on sarcolemma peeled from mdx muscle expressing full-length utrophin or Dp260 (Fig. 1B). Novel to this study, costameric actin was retained on all sarcolemma (n ≥ 11 sarcolemma from two different mice) from mdx mice expressing dystrophin constructs deleted for the carboxyl-terminal domain (Δ71–78) or hinge 2 through spectrin repeat 19 (ΔH2-R19). These data (Fig. 1B) indicate that either the amino-terminal domain (ΔH2-R19) or basic middle rod actin binding domain (Dp260) are sufficient to retain actin on peeled sarcolemma.
Fig. 1.
Retention of costameric actin on peeled sarcolemma from transgenic mdx mice. (A) The domain structure of dystrophin expressed in wild-type mice, the transgenic proteins expressed on the mdx background, and the phenotype of each transgenic line. The shaded regions identify the actin filament binding (ABD) or glycoprotein complex binding (CRCT) domains. H1–H4, hinge modules 1–4; SR1–SR24, spectrin repeats 1–24. (B) Confocal images of peeled sarcolemma stained with Alexa Fluor 568-phalloidin. (Scale bar: 20 μm.)
Transgenic mdx mice expressing Dp71 lacking all actin binding regions (7) and a dystrophin deleted for the cysteine-rich domain (ΔCR) important for binding to β-dystroglycan (5, 6) both exhibit a more severe phenotype than mdx (14, 15), and we hypothesized that costameric actin would not be retained because of the absence of either actin filament or β-dystroglycan binding activity. Despite numerous attempts, we were unable to isolate sarcolemma from either line because the myofibers split apart upon peeling, suggesting impaired physical coupling between myofibrils within Dp71 and ΔCR muscle cells. To circumvent this obstacle, we explored alternative methods to assess costameric actin instability.
Characterization of γ-Actin-Specific Antibodies.
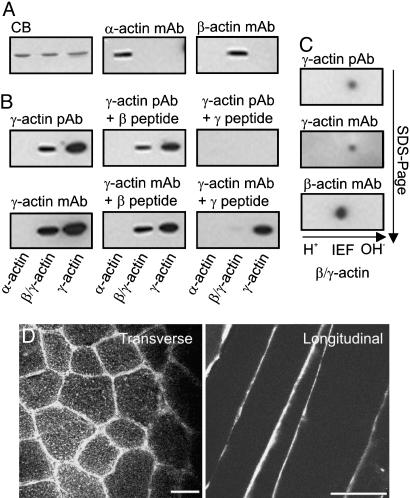
Previous studies indicated that subsarcolemmal costameric actin filaments are comprised exclusively of cytoplasmic γ-actin (11, 16). Therefore, we generated affinity-purified pAbs against the unique amino-terminal peptide of γ-actin and mAbs raised against purified γ-actin protein together with the γ-actin amino-terminal peptide. Both the pAbs and mAbs reacted strongly with purified bovine brain γ-actin, reacted less strongly to the β:γ-actin mixture from platelets, and were nonreactive with sarcomeric α-actin (Fig. 2A and B). To test specificity, synthetic peptides corresponding to the unique amino terminus of cytoplasmic γ-actin or β-actin were incubated with the antibodies before immunoblotting. The γ-actin peptide completely blocked all immunoreactivity of the pAb, whereas the β-actin peptide had a nominal effect (Fig. 2B). In contrast, the γ-actin peptide reduced, but did not eliminate, mAb reactivity to γ-actin (Fig. 2B), suggesting that the mAb recognized a distinct epitope on γ-actin. We also performed 2D electrophoresis on platelet actin to resolve β-actin from γ-actin. The γ-actin pAb and the mAb each reacted specifically with the minor and more basic protein spot, but not the more abundant and acidic spot labeled with the β-actin mAb (Fig. 2C). These results demonstrate that both the affinity-purified pAb 7577 and mAb 2–4 specifically recognize cytoplasmic γ-actin but that the two antibodies differ in their epitopes of recognition.
Fig. 2.
Characterization of γ-actin-specific antibodies. (A) Gels equally loaded with purified sarcomeric α-actin, platelet actin (a 4:1 mixture of β- and γ-isoforms), and brain γ-actin were either stained with Coomassie blue (CB) or transferred to nitrocellulose and Western-blotted with mAbs specific for α- or β-actin. (B) Western blots were stained with affinity-purified pAbs raised against the unique amino-terminal peptide sequence of cytoplasmic γ-actin or mAbs from mice immunized with both purified brain γ-actin and the amino-terminal sequence of cytoplasmic γ-actin. Where indicated, 10 μg/ml of either the amino-terminal γ-actin or β-actin peptide was preincubated with the γ-actin antibodies. (C) Purified platelet β/γ-actin was resolved by 2D electrophoresis and transferred to poly(vinylidene difluoride), and the blot was serially probed with γ-actin pAbs, γ-actin mAbs, and finally β-actin mAbs. (D) Ten-micrometer transverse and longitudinal cryosections of control tibialis anterior muscle stained with the γ-actin mAb. (Scale bar: 50 μm.)
On transverse cryosections of normal muscle, mAb 2–4 yielded a bright fluorescence signal at the periphery of each muscle cell coincident with the sarcolemma as well as a very faint honeycomb-shaped reticular lattice within the interior of each myofiber (Fig. 2D). When tested on longitudinal sections, a bright immune signal was again detected at the sarcolemma whereas internal staining was barely perceptible (Fig. 2D). No muscle-specific signal was detected in cryosections stained with peptide-specific pAb 7577 (data not shown).
γ-Actin Is Increased in Dystrophin-Deficient Skeletal Muscle.
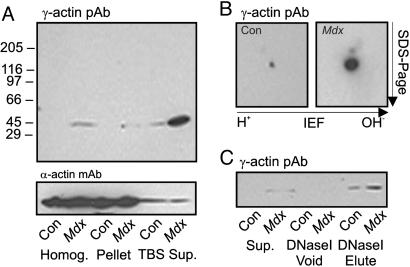
Based on our previous studies (7, 8, 11), we hypothesized that γ-actin may be more readily extracted from dystrophin-deficient muscle. The supernatants from control muscle homogenized in TBS exhibited a 42-kDa band faintly detected with γ-actin antibodies compared with an intensely stained band of identical size in mdx muscle supernatants (Fig. 3A). Although most sarcomeric α-actin was retained in the myofibrillar pellet after high-speed centrifugation, minor α-actin immunoreactivity was equal in supernatants of control and mdx skeletal muscle (Fig. 3A). Interestingly, γ-actin immunoreactivity was also detected in total homogenates from mdx muscle but not in control homogenates (Fig. 3A). Western blot analysis after 2D electrophoresis revealed γ-actin immunoreactivity in control homogenates that was dramatically elevated in mdx homogenates (Fig. 3B), suggesting that highly abundant α-sarcomeric actin obscured γ-actin detection on Western blots of control muscle resolved by 1D SDS/PAGE. Densitometric analysis after 2D electrophoresis (Fig. 3B) reported a 10-fold increase in γ-actin in mdx muscle compared with control. The 42-kDa protein stained by γ-actin antibodies in muscle high-speed supernatants was amplified by DNase I affinity chromatography while still preserving the difference in immune signal between control and mdx muscle (Fig. 3C). The results presented in Fig. 3 indicate that γ-actin protein levels are dramatically increased in dystrophin-deficient mdx muscle.
Fig. 3.
γ-Actin levels in mdx versus control muscle. (A) Western blots of control and mdx skeletal muscle homogenates and high-speed pellets or supernatants were stained with γ-actin pAb 7577 or mAb sarcomeric α-actin. (B) γ-Actin immunoreactivity in homogenates from control (Con) and mdx muscle resolved by 2D electrophoresis. (C) γ-Actin immunoreactivity in supernatants from control and mdx skeletal muscle before (Sup.) and after (Void) application to DNase I matrix and in the SDS eluate from the DNase I matrix.
Time Course and Tissue Localization of Increased γ-Actin in mdx Muscle.
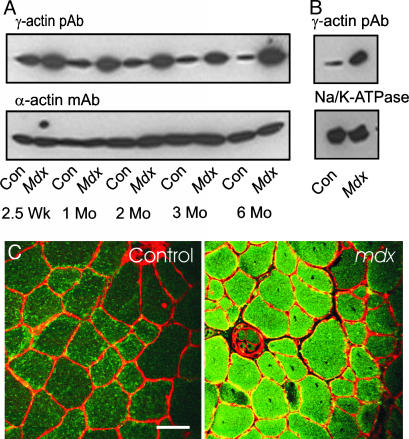
The mdx mouse experiences an acute episode of muscle cell necrosis, regeneration, and inflammation that peaks between 5 and 15 weeks of age but continues throughout adulthood at a lower rate (17). Because cytoplasmic γ-actin is highly expressed in regenerating muscle (18) and virtually all nonmuscle cells (19), the measured increase of γ-actin in mdx muscle (Fig. 3) could reflect this increased regenerative response or inflammatory cell infiltration. However, mdx mice ranging from 2.5 weeks through 6 months of age all exhibited elevated γ-actin immunoreactivity compared with age-matched controls (Fig. 4A). Confocal microscopy revealed elevated γ-actin immunoreactivity at both the periphery and within the cytoplasm of dystrophin-deficient muscle (Fig. 4C). The increased γ-actin staining intensity was observed in both peripherally nucleated and large centrally nucleated myofibers of mdx muscle (Fig. 4C). Consistent with the immunofluorescence data, γ-actin immunoreactivity was elevated in KCl-washed microsome preparations from mdx muscle (Fig. 4B). γ-Actin immunoreactivity was not detected in isolated myofibrils from control or mdx muscle by Western blotting after 2D gel electrophoresis or by immunofluorescence microscopy (data not shown). These data indicate that cytoplasmic γ-actin is markedly elevated at the sarcolemma and within the cytoplasm of dystrophin-deficient mdx muscle.
Fig. 4.
Developmental time course and localization of γ-actin in control (Con) and mdx muscle. (A) Western blots of DNase I-enriched supernatants from skeletal muscle of different age control and mdx mice were stained for γ- and α-actin. (B) Western blots of KCl-washed microsomes from control and mdx skeletal muscle stained for γ-actin and Na+/K+-ATPase immunoreactivity. (C) Transverse cryosections of control and mdx tibialis anterior muscle stained for γ-actin (green) and laminin (red). (Scale bar: 50 μm.)
Quantitation of γ-Actin in mdx Muscle and Effect of Transgenic Rescue.
The abundance of cytoplasmic γ-actin in skeletal muscle from control and mdx mice was quantified by Western blotting by using γ-actin purified from bovine brain as a standard (Fig. 6, which is published as supporting information on the PNAS web site). For control muscle, we calculated a γ-actin concentration of 0.2 ± 0.1 μM whereas the γ-actin concentration calculated for mdx muscle was 2.1 ± 1.0 μM, consistent with the 10-fold difference in immune signals measured from Western blots of whole-muscle homogenates after 2D electrophoresis (Fig. 3B). The concentration of α-actin in SDS muscle extracts was similar between control (893 ± 42 μM) and mdx (811 ± 15 μM) muscle and matched well with previously published measurements (20, 21). These data indicate that the cellular concentration of γ-actin in mdx muscle is high enough to potentially alter a variety of cellular processes affected in dystrophy.
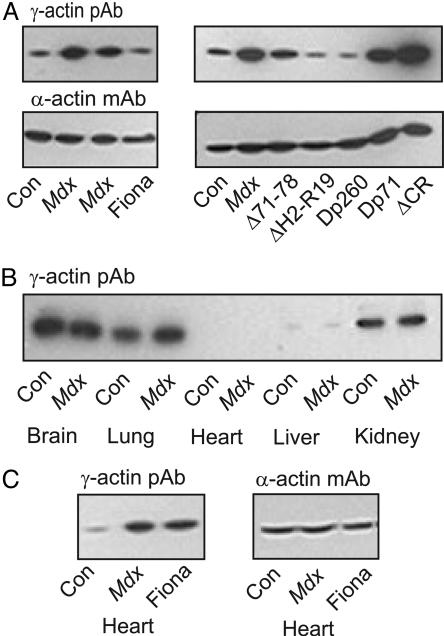
If increased γ-actin levels are an adaptive cellular response to an unstable mechanical linkage between costameric actin and the sarcolemma in dystrophin-deficient muscle, we hypothesized that restoration of the linkage should also revert γ-actin levels back to normal. As hypothesized, expression of full-length utrophin in the Fiona line of mdx mice (22) or expression of dystrophin constructs Δ71–78, ΔH2-R19, and Dp260 on the mdx background each restored γ-actin levels to those observed in control muscle (Fig. 5A). Although transgenic expression of Dp260 rescued several parameters of the mdx phenotype, the number of centrally nucleated fibers remained grossly elevated (13), which further argues against muscle regeneration as the basis for increased γ-actin levels in mdx muscle. In transgenic mdx lines expressing Dp71 (14) lacking all known actin binding regions and ΔCR (15), which is unable to form a stable linkage with β-dystroglycan, we observed elevated γ-actin levels similar to mdx muscle (Fig. 5A). Thus, there is excellent correlation between the capability of a transgene construct to restore stable coupling between costameric actin and the sarcolemma (Fig. 1) and also to restore elevated γ-actin protein levels of mdx muscle back to normal.
Fig. 5.
Tissue specificity of elevated γ-actin in mdx mice and restoration to normal levels by transgene expression. (A) Western blots of DNase I-enriched supernatants from skeletal muscle of control (Con), mdx, and transgenic mdx mice expressing utrophin or dystrophin constructs depicted in Fig. 1 were stained for γ- and α-actin. (B) γ-Actin immunoreactivity in SDS extracts from organs of control and mdx mice. (C) Western blots of DNase I-enriched supernatants of cardiac muscle from control, mdx, and Fiona transgenic mdx mice stained for γ- and α-actin.
γ-Actin Is Elevated only in Striated Muscles of mdx Mice.
To assess whether the increased γ-actin observed in mdx skeletal muscle is specific to striated muscle, we performed Western blot analysis of SDS extracts prepared from a variety of organs of control and mdx mice. Consistent with previous measurements of γ-actin mRNA (23, 24), γ-actin immunoreactivity was high in brain, lung, and kidney but barely detected in liver (Fig. 5B). In contrast to skeletal muscle, however, γ-actin protein levels were similar in normal and mdx nonmuscle tissues (Fig. 5B). γ-Actin was not detected in SDS extracts from control or mdx cardiac muscle, but γ-actin immunoreactivity was dramatically increased in DNase I-enriched supernatants from dystrophin-deficient mdx cardiac muscle compared with control (Fig. 5C). Interestingly, γ-actin protein levels were also elevated in DNase I-enriched supernatants of cardiac muscle from the Fiona line of transgenic mdx mice (Fig. 5C). Expression of the utrophin transgene in the Fiona line is driven by the human skeletal muscle α-actin promoter (22), which is only weakly active in adult cardiac muscle. The data in Fig. 5 A and C indicate that the specific absence of dystrophin from cardiac muscle is sufficient to induce gross elevation of γ-actin protein levels. Because regeneration is minimal in cardiac muscle (25), we conclude that increased γ-actin levels in mdx heart and skeletal muscle are a specific compensatory response to the destabilized mechanical linkage between the costamere and sarcolemma in dystrophin-deficient muscle.
Discussion
Our results provide new insight into the function of dystrophin and how muscle cells adapt when dystrophin is ablated through genetic defect. Our analysis of peeled sarcolemma from several lines of transgenic mdx mice indicate that either one of two known actin binding domains plus the CR are minimally required to anchor costameric actin filaments to mechanically peeled sarcolemma.
Using a newly generated mAb raised against a mixed antigen of purified bovine brain γ-actin and a synthetic peptide corresponding to its unique amino terminus, we found γ-actin to be most predominantly localized to the sarcolemma and more faintly within an internal reticular lattice in normal muscle (Fig. 2). Our results closely match γ-actin localization in chicken skeletal muscle (16) and also the staining pattern observed in human skeletal muscle with antibodies to Aplysia actin that reacted with mammalian cytoplasmic actins but not muscle actin (26). Others have reported γ-actin immunoreactivity most predominantly associated with the Z-disk (27, 28). We cannot explain this difference other than to note that predominant sarcolemmal staining was consistently observed with antibodies raised against whole protein (Fig. 2) (16, 26), whereas Z-disk staining was observed exclusively with antibodies to synthetic peptides (27, 28).
Most interestingly, we demonstrate that γ-actin levels were increased 10-fold in skeletal muscle cells of dystrophin-deficient mdx mice (Figs. 3–5). Moreover, γ-actin levels were restored to normal in mdx muscle expressing full-length utrophin or functional dystrophin constructs but remained elevated in animals expressing nonfunctional dystrophins (Fig. 5). In vitro, cytoplasmic actin synthesis is controlled at the level of transcription and mRNA stability (29–31). However, microarray studies indicated that cytoplasmic γ-actin transcript was elevated only 2-fold in dystrophin-deficient muscle (32, 33), whereas we measured a 10-fold increase in protein levels. This discrepancy suggests that γ-actin translation or protein stability may be altered in dystrophin-deficient muscle. Moreover, the concomitant elevation of cytoplasmic γ-actin transcript (32, 33) and protein (Figs. 3–5) in dystrophin-deficient muscle runs contrary to the autoregulatory feedback mechanism elucidated from in vitro studies (29–31, 34, 35).
Because dystrophin has been shown to retard actin filament depolymerization in vitro (7, 8) and anchor costameric actin to the sarcolemma in vivo (11), we hypothesize that cytoplasmic γ-actin levels may be elevated in dystrophin-deficient muscle as part of a compensatory remodeling program. The dystrophin-deficient muscle cell may sense sarcolemmal instability and respond by elevating γ-actin levels to cause an increase in the number of filaments associated with the sarcolemma in mdx muscle (Fig. 4 C and D). Previous studies have reported 2- to 4-fold up-regulation of several costameric proteins in dystrophin-deficient muscle including γ-filamin, plectin, talin, vinculin, and the laminin receptor α7β1 integrin (36–40). Although not present at normal costameres, the dystrophin homologue utrophin is up-regulated and recruited to costameres in dystrophin-deficient muscle (11, 12). Because all but one of the up-regulated proteins bind filamentous actin, we conclude that increased levels of γ-actin may provide a common and central connecting point in remodeling the costamere to partially reinforce the mechanically weakened sarcolemma in the absence of dystrophin.
Forced expression of either cytoplasmic β- or γ-actin was previously shown to ablate contractile function in isolated cardiac myocytes (41). When smooth muscle γ-actin was overexpressed in cardiac muscle, its incorporation into sarcomeric thin filaments resulted in altered myocyte Ca2+ sensitivity and economy of force production (42). Both of these results suggest that increased cytoplasmic γ-actin could inhibit force production by dystrophin-deficient muscle (43). However, we detected no γ-actin in washed myofibrils prepared from control or mdx muscle. Furthermore, transgenic expression of the Dp260 construct in mdx muscle restored γ-actin levels back to normal (Fig. 5A), yet the force deficit was not corrected (13). Our data indicate that elevated γ-actin levels do not contribute to the force deficit observed in mdx muscle.
Finally, excess cytoplasmic γ-actin may explain how loss of dystrophin leads to abnormalities in cell signaling or gene expression. The actin depolymerizing agent latrunculin B was shown to activate the stress-activated protein kinase pathway in yeast (44) whereas the mitogen-activated protein kinase pathways mediated by c-Jun amino-terminal kinase, p38, and extracellular signal-regulated kinase are all reportedly activated in dystrophin-deficient muscle (45–49). Monomeric actin levels have also been shown to negatively regulate gene expression mediated by serum response factor (50) in part through direct interaction with the myocardin-related family of transcriptional coactivators (51). Based on decreased transcription in response to elevated monomeric actin levels (50) and/or serum response factor ablation (52, 53), one would expect decreased expression of cytoplasmic γ-actin, cardiac α-actin, β1 integrin, talin, and vinculin in dystrophin-deficient muscle. However, all of these serum response factor-mediated genes exhibit increased transcript and protein levels in dystrophin-deficient muscle (32, 33, 38–40, 54). Clearly, the pathways regulating gene expression in dystrophin-deficient muscle are more complex than can be extrapolated from current model systems. Analysis of animal models overexpressing cytoplasmic γ-actin in skeletal muscle will help define the contribution of elevated γ-actin to the diverse phenotypes caused by dystrophin deficiency.
Materials and Methods
Animals.
Control C57BL/6J, dystrophin-deficient C57BL/10mdx, and C57BL/6J-Lama2dy mice were purchased from The Jackson Laboratory. Fiona transgenic mdx mice expressing full-length utrophin (22) were provided by Kay Davies (Oxford University, Oxford). Transgenic mdx expressing Dp260 or constructs deleted for dystrophin exons 71–78 (Δ71–78) or hinge 2 through spectrin repeat 19 (ΔH2-R19) were previously described (13, 55, 56) and were provided by Jeffrey Chamberlain (University of Washington, Seattle). Transgenic mdx mice expressing Dp71 or a construct deleted for exons 64–67 (ΔCR) were previously described (13–15, 55, 56).
Purified Actins.
Purified rabbit skeletal muscle α-actin and human platelet actin (a 4:1 mixture of β- and γ-cytoplasmic actins) were purchased from Cytoskeleton (Denver, CO). Bovine brain γ-actin was purified by DNase I affinity chromatography as described (57).
Antibodies.
pAbs specific for γ-actin were generated (Genemed Synthesis, San Francisco) by immunizing rabbits with a synthetic peptide corresponding to the unique amino terminus of γ-actin (acetyl-EEEIAALVIDNGSGC) coupled to keyhole limpet hemocyanin and affinity-purified against immobilized bovine brain γ-actin. mAbs specific for γ-actin were generated by Harlan Bioproducts for Science (Madison, WI). Mice were immunized with a combination of purified bovine brain cytoplasmic γ-actin and the keyhole limpet hemocyanin-conjugated amino-terminal peptide of γ-actin. Hybridoma supernatants were screened by ELISA and Western blotting, and clones were selected based on specificity for cytoplasmic γ-actin over α-sarcomeric or platelet β/γ-actins. Five mAbs specific for γ-actin were isolated, and mAb 2–4 was further characterized and used in all subsequent experiments. Antibodies specific for sarcomeric α-actin, β-actin, and mouse laminin-1 were purchased from Sigma. The mAb McB2 against the α2-subunit of the Na+/K+-ATPase was previously characterized (58). Peroxidase-labeled secondary antibodies against rabbit and mouse Igs were purchased from Chemicon International (Temecula, CA) and Roche (Indianapolis), respectively. Alexa Fluor 488 anti-mouse, Alexa Fluor 568 anti-rabbit Igs, and Alexa Fluor 568-conjugated phalloidin were purchased from Molecular Probes.
Tissue Preparations.
KCl-washed microsomes and washed myofibrils were prepared from skeletal muscle of control and mdx mice as described (21, 59). For soluble muscle protein extracts, skeletal muscle was dissected from mice immediately after killing and snap-frozen in liquid nitrogen. Frozen muscle was pulverized by using a liquid nitrogen-cooled mortar and pestle and gently mixed for 1 h at 4°C in 10 ml/g cold 0.15 M NaCl/50 mM Tris·HCl (pH 7.4) (TBS) also containing a mixture of protease inhibitors. The homogenate was centrifuged at 100,000 × g for 40 min at 4°C, and the supernatant was supplemented with ATP, DTT, and MgCl2 to final concentrations of 0.1 mM, 0.2 mM, and 2.0 mM, respectively. A total of 400 μl of supernatant and 100 μl of DNase I affigel (57) were incubated for 2 h at 4°C with mixing. After washing with 2 mM Tris·HCl (pH 8.0)/0.2 mM CaCl2/2 mM MgCl2/1 mM NaN3/0.1 mM ATP/0.5 mM DTT, bound proteins were eluted with 1% SDS sample buffer for SDS gel electrophoresis and Western blot analysis. Organs from control and mdx mice were dissected immediately after killing, snap-frozen in liquid nitrogen, and pulverized in a liquid nitrogen-cooled mortar and pestle. The pulverized tissue was solubilized for 2 min in 1% SDS, 5 mM EGTA, and a mixture of protease inhibitors at 100°C. After centrifugation at 12,000 × g to remove insoluble material, the protein concentration of the supernatants was determined with the Bio-Rad DC protein assay kit using BSA as standard.
Immunofluorescence Microscopy.
Mechanical isolation and fluorescence imaging of peeled sarcolemma were performed as described (11–13). For cryostat sectioning, skeletal muscles were dissected from age-matched C57BL/6 control and mdx mice and frozen in liquid nitrogen-cooled isopentane. Cryosections (10 μm) were fixed for 10 min with 4% paraformaldehyde in PBS and blocked with 5% goat serum in PBS for 1 h at room temperature. Sections were incubated with primary antibodies diluted in PBS overnight at 4°C, followed by fluorescent secondary antibody for 30 min at 37°C. Confocal microscopy was performed by using a Bio-Rad MRC1000 scan head mounted transversely to an inverted Nikon Diaphot 200 microscope with a ×40 oil immersion objective (W. M. Keck Laboratory for Biological Imaging at the University of Wisconsin). Separate single-channel serial optical sections were collected by using a krypton/argon mixed-gas air-cooled laser set to 488-nm and 568-nm excitation wavelengths with 10% power and an iris of 5.9, gain of 1,403, and black level of 6. The corresponding digital images were then merged by using confocal assistant, and the figures were prepared in illustrator (Adobe Systems, San Jose, CA).
SDS/PAGE and Western Blot Analysis.
Samples were resolved on 3–12% SDS polyacrylamide gels and stained with Coomassie blue or transferred to nitrocellulose membranes as described (7). 2D electrophoresis was performed by Kendrick Laboratories (Madison, WI) by using 1% pH 4–6 ampholines (Amersham Pharmacia Biosciences) and 1% pH 4–8 ampholines (Gallard-Schlesinger Industries) for 9,600 volt-hours, and slab gels were transferred onto poly(vinylidene difluoride) membranes. For the peptide blocking experiments, 0.1 mg of γ-actin peptide (acetyl-EEEIAALVIDNGSGC) or β-actin peptide (acetyl-DDDIAALVVDNGSGC) was added to the γ-actin primary antibodies before incubation with blots. Immunoreactivity was detected with peroxidase-labeled secondary antibodies and SuperSignal West Pico chemiluminescence purchased from Pierce. The poly(vinylidene difluoride) transfers of 2D gels were sequentially stained with mAbs and pAbs to γ-actin followed by β- or α-actin mAbs, and the blots were stripped with 50 mM glycine (pH 2.5) between trials. For quantitative Western blotting, control and mdx DNase I eluates prepared from a measured amount of total muscle homogenate protein were resolved by SDS/PAGE, transferred to nitrocellulose, and stained with γ-actin mAb 2–4 in parallel with a standard curve of purified brain γ-actin. γ-Actin immunoreactivity was detected with 125I-labeled goat anti-mouse IgG (PerkinElmer) and quantified densitometrically by using uvp labworks imaging and analysis software. γ-Actin as a percentage of total muscle protein was converted to cellular concentration based on a muscle protein density of 0.2 g/ml wet weight.
Supplementary Material
Acknowledgments
We thank Drs. Jeffrey Chamberlain and Kay Davies for providing transgenic mice. This work was supported by the Muscular Dystrophy Association, National Institutes of Health Grants AR042423 and AR049899, and National Research Service Award T32 HL07936 from the University of Wisconsin (Madison) Cardiovascular Research Center.
Abbreviations
- pAb
polyclonal antibody
- CR
cysteine-rich domain.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.O’Brien K. F., Kunkel L. M. Mol. Genet. Metab. 2001;74:75–88. doi: 10.1006/mgme.2001.3220. [DOI] [PubMed] [Google Scholar]
- 2.Cohn R. D., Campbell K. P. Muscle Nerve. 2000;23:1456–1471. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Blake D. J., Weir A., Newey S. E., Davies K. E. Physiol. Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 4.Ervasti J. M. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki A., Yoshida M., Yamamoto H., Ozawa E. FEBS Lett. 1992;308:154–160. doi: 10.1016/0014-5793(92)81265-n. [DOI] [PubMed] [Google Scholar]
- 6.Jung D., Yang B., Meyer J., Chamberlain J. S., Campbell K. P. J. Biol. Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 7.Rybakova I. N., Amann K. J., Ervasti J. M. J. Cell Biol. 1996;135:661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rybakova I. N., Ervasti J. M. J. Biol. Chem. 1997;272:28771–28778. doi: 10.1074/jbc.272.45.28771. [DOI] [PubMed] [Google Scholar]
- 9.Amann K. J., Renley B. A., Ervasti J. M. J. Biol. Chem. 1998;273:28419–28423. doi: 10.1074/jbc.273.43.28419. [DOI] [PubMed] [Google Scholar]
- 10.Amann K. J., Guo W. X. A., Ervasti J. M. J. Biol. Chem. 1999;274:35375–35380. doi: 10.1074/jbc.274.50.35375. [DOI] [PubMed] [Google Scholar]
- 11.Rybakova I. N., Patel J. R., Ervasti J. M. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybakova I. N., Patel J. R., Davies K. E., Yurchenco P. D., Ervasti J. M. Mol. Biol. Cell. 2002;13:1512–1521. doi: 10.1091/mbc.01-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner L. E., DelloRusso C., Crawford R. W., Rybakova I. N., Patel J. R., Ervasti J. M., Chamberlain J. S. Hum. Mol. Genet. 2002;11:1095–1105. doi: 10.1093/hmg/11.9.1095. [DOI] [PubMed] [Google Scholar]
- 14.Cox G. A., Sunada Y., Campbell K. P., Chamberlain J. S. Nat. Genet. 1994;8:333–339. doi: 10.1038/ng1294-333. [DOI] [PubMed] [Google Scholar]
- 15.Rafael J. A., Cox G. A., Corrado K., Jung D., Campbell K. P., Chamberlain J. S. J. Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig S. W., Pardo J. V. Cell Motil. 1983;3:449–462. doi: 10.1002/cm.970030513. [DOI] [PubMed] [Google Scholar]
- 17.Spencer M. J., Walsh C. M., Dorshkind K. A., Rodriguez E. M., Tidball J. G. J. Clin. Invest. 1997;99:2745–2751. doi: 10.1172/JCI119464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd C. M., Berendse M., Lloyd D. G., Schevzov G., Grounds M. D. Exp. Cell Res. 2004;297:82–96. doi: 10.1016/j.yexcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 19.McHugh K. M., Crawford K., Lessard J. L. Dev. Biol. 1991;148:442–458. doi: 10.1016/0012-1606(91)90263-3. [DOI] [PubMed] [Google Scholar]
- 20.Yates L. D., Greaser M. L. J. Mol. Biol. 1983;168:123–141. doi: 10.1016/s0022-2836(83)80326-x. [DOI] [PubMed] [Google Scholar]
- 21.Yates L. D., Greaser M. L. J. Biol. Chem. 1983;258:5770–5774. [PubMed] [Google Scholar]
- 22.Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J. M., Davies K. Nat. Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 23.Erba H. P., Eddy R., Shows T., Kedes L., Gunning P. Mol. Cell. Biol. 1988;8:1775–1789. doi: 10.1128/mcb.8.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokunaga K., Takeda K., Kamiyama K., Kageyama H., Takenaga K., Sakiyama S. Mol. Cell. Biol. 1988;8:3929–3933. doi: 10.1128/mcb.8.9.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regula K. M., Rzeszutek M. J., Baetz D., Seneviratne C., Kirshenbaum L. A. Cardiovasc. Res. 2004;64:395–401. doi: 10.1016/j.cardiores.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Lubit B. W., Schwartz J. H. J. Cell Biol. 1980;86:891–897. doi: 10.1083/jcb.86.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakata T., Nishina Y., Yorifuji H. Biochem. Biophys. Res. Commun. 2001;286:156–163. doi: 10.1006/bbrc.2001.5353. [DOI] [PubMed] [Google Scholar]
- 28.Kee A. J., Schevzov G., Nair-Shalliker V., Robinson C. S., Vrhovski B., Ghoddusi M., Qiu M. R., Lin J. J., Weinberger R., Gunning P. W., et al. J. Cell Biol. 2004;166:685–696. doi: 10.1083/jcb.200406181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bershadsky A. D., Gluck U., Denisenko O. N., Sklyarova T. V., Spector I., Ben Ze’ev A. J. Cell Sci. 1995;108:1183–1193. doi: 10.1242/jcs.108.3.1183. [DOI] [PubMed] [Google Scholar]
- 30.Lyubimova A., Bershadsky A. D., Ben Ze’ev A. J. Cell. Biochem. 1997;65:469–478. [PubMed] [Google Scholar]
- 31.Lyubimova A., Bershadsky A. D., Ben Ze’ev A. J. Cell. Biochem. 1999;76:1–12. doi: 10.1002/(sici)1097-4644(20000101)76:1<1::aid-jcb1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y.-W., Zhao P., Borup R., Hoffman E. P. J. Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter J. D., Khanna S., Kaminski H. J., Rao J. S., Merriam A. P., Richmonds C. R., Leahy P., Li J., Guo W., Andrade F. H. Hum. Mol. Genet. 2002;11:263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd C., Schevzov G., Gunning P. J. Cell Biol. 1992;117:787–797. doi: 10.1083/jcb.117.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schevzov G., Lloyd C., Gunning P. DNA Cell Biol. 1995;14:689–700. doi: 10.1089/dna.1995.14.689. [DOI] [PubMed] [Google Scholar]
- 36.Thompson T. G., Chan Y. M., Hack A. A., Brosius M., Rajala M., Lidov H. G., McNally E. M., Watkins S., Kunkel L. M. J. Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroder R., Mundegar R. R., Treusch M., Schlegel U., Blumcke I., Owaribe K., Magin T. M. Eur. J. Cell Biol. 1997;74:165–171. [PubMed] [Google Scholar]
- 38.Law D. J., Allen D. L., Tidball J. G. J. Cell Sci. 1994;107:1477–1483. doi: 10.1242/jcs.107.6.1477. [DOI] [PubMed] [Google Scholar]
- 39.Vachon P. H., Xu H., Liu L., Loechel F., Hayashi Y., Arahata K., Reed J. C., Wewer U. M., Engvall E. J. Clin. Invest. 1997;100:1870–1881. doi: 10.1172/JCI119716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodges B. L., Hayashi Y. K., Nonaka I., Wang W., Arahata K., Kaufman S. J. J. Cell Sci. 1997;110:2873–2881. doi: 10.1242/jcs.110.22.2873. [DOI] [PubMed] [Google Scholar]
- 41.von Arx P., Bantle S., Soldati T., Perriard J. C. J. Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin A. F., Phillips R. M., Kumar A., Crawford K., Abbas Z., Lessard J. L., de Tombe P., Solaro R. J. Am. J. Physiol. 2002;283:H642–H649. doi: 10.1152/ajpheart.00890.2001. [DOI] [PubMed] [Google Scholar]
- 43.Gillis J. M. J. Muscle Res. Cell Motil. 1999;20:605–625. doi: 10.1023/a:1005545325254. [DOI] [PubMed] [Google Scholar]
- 44.Gachet Y., Tournier S., Millar J. B. A., Hyams J. S. Nature. 2001;412:352–355. doi: 10.1038/35085604. [DOI] [PubMed] [Google Scholar]
- 45.Kolodziejczyk S. M., Walsh G. S., Balazsi K., Seale P., Sandoz J., Hierlihy A. M., Rudnicki M. A., Chamberlain J. S., Miller F. D., Megeney L. A. Curr. Biol. 2001;11:1278–1282. doi: 10.1016/s0960-9822(01)00397-9. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura A., Harrod G. V., Davies K. E. Neuromusc. Disord. 2001;11:251–259. doi: 10.1016/s0960-8966(00)00201-7. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura A., Yoshida K., Takeda S., Dohi N., Ikeda S. FEBS Lett. 2002;520:18–24. doi: 10.1016/s0014-5793(02)02739-4. [DOI] [PubMed] [Google Scholar]
- 48.Kumar A., Khandelwal N., Malya R., Reid M. B., Boriek A. M. FASEB J. 2004;18:102–113. doi: 10.1096/fj.03-0453com. [DOI] [PubMed] [Google Scholar]
- 49.Lang J. M., Esser K. A., Dupont-Versteegden E. E. Exp. Biol. Med. (Maywood, NJ) 2004;229:503–511. doi: 10.1177/153537020422900608. [DOI] [PubMed] [Google Scholar]
- 50.Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- 51.Miralles F., Posern G., Zaromytidou A. I., Treisman R. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 52.Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S., Czubryt M. P., McAnally J., Bassel-Duby R., Richardson J. A., Wiebel F. F., Nordheim A., Olson E. N. Proc. Natl. Acad. Sci. USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng B. S., Zhao P., Pattison J. S., Gordon S. E., Granchelli J. A., Madsen R. W., Folk L. C., Hoffman E. P., Booth F. W. J. Appl. Physiol. 2002;93:537–545. doi: 10.1152/japplphysiol.00202.2002. [DOI] [PubMed] [Google Scholar]
- 55.Crawford G. E., Faulkner J. A., Crosbie R. H., Campbell K. P., Froehner S. C., Chamberlain J. S. J. Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper S. Q., Hauser M. A., DelloRusso C., Duan D., Crawford R. W., Phelps S. F., Harper H. A., Robinson A. S., Engelhardt J. F., Brooks S. V., et al. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 57.Renley B. A., Rybakova I. N., Amann K. J., Ervasti J. M. Cell Motil. Cytoskeleton. 1998;41:264–270. doi: 10.1002/(SICI)1097-0169(1998)41:3<264::AID-CM7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 58.Urayama O., Heather S., Sweadner K. J. J. Biol. Chem. 1989;264:8271–8280. [PubMed] [Google Scholar]
- 59.Ohlendieck K., Ervasti J. M., Snook J. B., Campbell K. P. J. Cell Biol. 1991;112:135–148. doi: 10.1083/jcb.112.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.