Abstract
Sequence divergence in cis-regulatory elements is an important mechanism contributing to functional diversity of genes during evolution. Gene duplication and divergence provide an opportunity for selectively preserving initial functions and evolving new activities. Many vertebrates have 39 Hox genes organized into four clusters (Hoxa–Hoxd); however, some ray-finned fishes have extra Hox clusters. There is a single Hoxa2 gene in most vertebrates, whereas fugu (Takifugu rubripes) and medaka (Oryzias latipes) have two coparalogous genes [Hoxa2(a) and Hoxa2(b)]. In the hindbrain, both genes are expressed in rhombomere (r) 2, but only Hoxa2(b) is expressed in r3, r4, and r5. Multiple regulatory modules directing segmental expression of chicken and mouse Hoxa2 genes have been identified, and each module is composed of a series of discrete elements. We used these modules to investigate the basis of differential expression of duplicated Hoxa2 genes, as a model for understanding the divergence of cis-regulatory elements. Therefore, we cloned putative regulatory regions of the fugu and medaka Hoxa2(a) and -(b) genes and assayed their activity. We found that these modules direct reporter expression in a chicken assay, in a manner corresponding to their endogenous expression pattern in fugu. Although sequence comparisons reveal many differences between the two coparalogous genes, specific subtle changes in seven cis elements of the Hoxa2(a) gene restore segmental regulatory activity. Therefore, drift in subsets of the elements in the regulatory modules is responsible for the differential expression of the two coparalogous genes, thus providing insight into the evolution of cis elements.
Keywords: Hox gene regulation, hindbrain, vertebrate development, fugu
Many vertebrates, including humans, mice, and chickens, have 39 Hox genes organized into four clusters (Hoxa–Hoxd), each cluster on a different chromosome (1). These genes are arranged in 13 paralogous groups on the basis of their relative position within each cluster and the sequence similarity of their encoded proteins (2). The vertebrate Hox clusters are proposed to have evolved from an ancestral homeobox gene cluster (3–5) by successive genome-wide duplication events (the 2R hypothesis) ≈500 million years (Myr) ago, followed by divergence (6–8). It has been postulated that there was an additional “fish-specific genome duplication” (3R) ≈320 Myr ago (6, 9) that led to a further expansion in the number of Hox clusters in certain fish, as compared with other vertebrates (10–13). During evolution, the paralogous genes can diverge, resulting in a gain or loss of function due to changes in the coding sequences or regulatory elements. As a consequence, these duplicated genes may eventually subdivide the functions of the original ancestral gene or evolve new activities. An individual gene also may degenerate to a pseudogene or be completely lost from the genome because of functional compensation by a paralog. It is widely believed that the small size and modular nature of regulatory elements makes them an effective target for change, contributing to morphological diversity during evolution (14).
The vertebrate hindbrain is organized into segmental units termed “rhombomeres” (r) (shown schematically in Fig. 1B), and segmental expression of Hox genes is essential for patterning regional identity (15). Hoxa2 is expressed in r2 and in posterior regions of the hindbrain in mice, chickens, and zebrafish (16–19) and has been shown to play multiple roles in head development (20–22). As a result of duplication, Takifugu rubripes (fugu, or pufferfish) and Oryzias latipes (medaka) have coparalogous genes designated Hoxa2(a) and Hoxa2(b); in zebrafish, Hoxa2(a) is a pseudogene (Fig. 1A and refs. 10–13). In fugu, Hoxa2(a) and Hoxa2(b) display different expression patterns in the hindbrain; Hoxa2(b) is expressed in r2–r7, as in mouse, chicken, and zebrafish embryos, whereas fugu Hoxa2(a) is seen only in a subset of cells in r1 and r2 (see Fig. 1B and ref. 11).
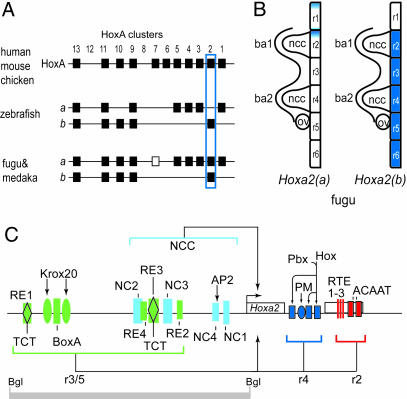
Fig. 1.
Genomic organization, expression, and regulatory modules of Hoxa2 genes. (A) Schematic diagram of Hoxa cluster organization in various species among selected vertebrates, illustrating differing copy numbers in amniotes and fish. Bat, Carollia perspillate; shark, Heterodontus francessi. The positions of the Hoxa2 genes are outlined in blue. Note that Hoxa2(a) has become a diverged pseudogene in zebrafish. The open box indicates that, although the Hoxa7 gene is present in the fugu cluster, it is absent in the medaka. (B) Schematic diagram of the expression domains in the hindbrain of the two fugu coparalogous genes, Hoxa2(a) and -(b). Hoxa2(a) shows weak, restricted expression in r1 and r2, whereas Hoxa2(b) shows strong expression in r2–r6 (11), as seen for the single Hoxa2 gene in amniotes. ba, branchial arch; ncc, neural crest cells; ov, otic vesicle. (C) Schematic diagram of the four distinct regulatory modules directing Hoxa2 expression in r2, r4, r3/5, and neural crest cells during hindbrain development. The gray bar marks the position of an 809-bp BglII fragment in the mouse locus that contains the r3/r5 and neural crest cell enhancer modules. NCC, neural crest cells; RE, rhombomeric element; TCT, element containg TCT triplet; NC, neural crest; AP2, AP2 binding site; RTE, rhombomere 2 element; PM, Prep/Meis site; Bgl, BglII site; Pbx/Hox, bipartite binding sites for Hox and Pbx proteins; Krox20, Krox20 binding site; BoxA, putative Sox binding site.
Using functional assays in chicken and mouse embryos, we have identified three conserved modules in the Hoxa2 locus (Fig. 1C) that direct segmental expression in the hindbrain; these include a module in the intergenic region mediating expression of Hoxa2 in r3 and r5 (r3/5), an r4 module located in the intron, and an r2 module located in the second exon (refs. 23 and 24 and our unpublished data). Overlapping with the r3/5 module, an additional enhancer has been found that directs Hoxa2 expression in cranial neural crest (25). On the basis of this mechanistic knowledge of segmental regulation, it was determined that the duplicated Hoxa2 genes in fugu provide an excellent model system for examining evolutionary changes that lead to altered expression patterns. Therefore, we investigated the differential expression of these genes by analyzing the fugu cis-regulatory elements controlling rhombomeric expression in chicken and mouse embryos. We found that subtle sequence drift in specific regulatory elements is responsible for the differential expression of the two coparalogous genes, Hoxa2(a) and -(b).
Results
Analysis of the Basis for Differential r3/5 Expression.
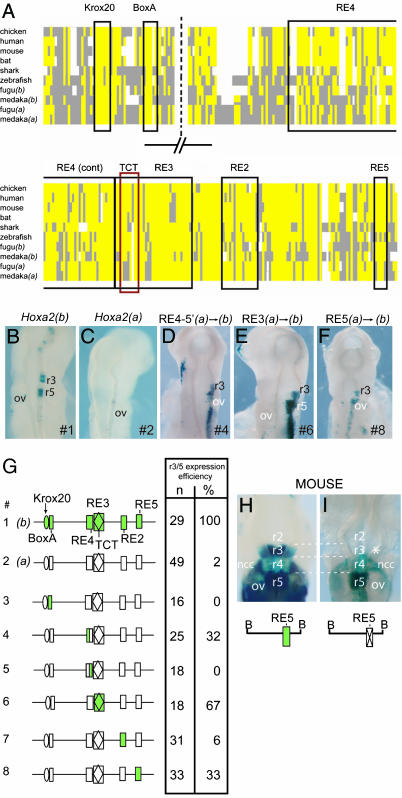
The Hoxa2 r3/5 regulatory module in mice consists of multiple cis elements [rhombomeric element (RE) 1–RE4, Krox20, and BoxA] embedded in an 809-bp BglII fragment in the intergenic region of Hoxa2/3 (Fig. 1C and refs. 23 and 24). In fugu, it has previously been shown that Hoxa2(b) is expressed in r3 and r5, whereas Hoxa2(a) is not (11). Multispecies sequence alignments were used to identify the regions in fugu and medaka that are equivalent to the murine r3/5 enhancer (Fig. 2A; and Fig. 6, which is published as supporting information on the PNAS web site). These regions were cloned from the fugu Hoxa2(a) and -(b) genes, linked to a lacZ reporter, and electroporated into chicken embryos to evaluate the modules regulatory potential. The Hoxa2(b) enhancer mediates strong expression in the hindbrain in r3 and r5 (Fig. 2 B and G), whereas embryos carrying the Hoxa2(a) enhancer display either no expression in the hindbrain or only a small number of positive cells in r5 (Fig. 2 C and G). These regulatory data in chicken embryos directly correlate with the differential endogenous expression of the two coparalogs in fugu (11). A similar correlation in regulatory activity of Hoxa2(a) and -(b) genes was obtained by using the medaka r3/5 enhancer regions (data not shown).
Fig. 2.
Analysis of the basis of differential r3/5 expression of the fugu Hoxa2(a) and Hoxa2(b) genes. (A) Schematic representation of multispecies alignment of the Hoxa2 r3/5 regulatory module (original sequence alignment is shown in Fig. 6). A consensus sequence was derived based on sequence identity in >50% of the species. Yellow boxes indicate identical sequences, as compared with the consensus; gray boxes indicate diverged sequence; and white spaces are gaps introduced to maintain maximal alignment. The positions of the conserved elements are indicated by outlined boxes, with the name of each element given above the box. (B–F) Representative transgene expression in chicken embryos electroporated with reporter constructs under control of the r3/5 enhancer elements from fugu Hoxa2(b) (B) and Hoxa2(a) (C), with specific conversions between the two coparalogous sequences noted above D–F. The specific constructs used in each case are indicated at the bottom right in each panel. (G) Constructs (#) tested are diagrammed (Left), and the quantitative results of their analysis in chicken embryo electroporation studies are tabulated (Right). The Hoxa2(b) elements are shown in green; the elements of Hoxa2(a) are in white. In modified constructs (#3–8), the colored element indicates the specific region that has been changed from Hoxa2(a) to Hoxa2(b), in the context of the remaining Hoxa2(a) sequence. In the table, n is the total number of electroporated embryos examined, and the efficiency of the elements’ ability to direct r3/5 restricted expression is given as a percentage. (H and I) Transgene expression in mouse embryos injected with reporter constructs under the control of a wild-type Hoxa2 BglII fragment (see Fig. 1C for location of the fragment) (23) that directs staining in r3, r5, and neural crest cells (H), and a version that carries a mutation in the newly discovered RE5 motif (I). Note the specific loss of reporter staining in r3 (∗) when RE5 is mutated (I). ov, otic vesicle; ncc, neural crest cells; B, BglII site.
Despite the fact that the r3/5 enhancer from the fugu Hoxa2(a) gene does not direct segmental expression in chicken embryos, we are able to identify sequences that correspond to the critical cis components of the mouse enhancer (Krox20, BoxA, RE4, TCT motif, RE3, and RE2). The presence of these motifs does not reflect a high degree of general sequence conservation between the mouse and fugu Hoxa2(a) and Hoxa2(b) r3/5 enhancers because sequences outside of these motifs are highly diverged. The Krox20 sites from both fugu Hoxa2 genes are identical and align perfectly with those observed in the chicken, mouse, bat, and human r3/5 enhancers (Fig. 2A). We did observe a number of sequence changes in the putative cis components of the Hoxa2(a) enhancer, as compared with its Hoxa2(b) coparalog. Potentially important small changes in the spacing between the Krox20 binding site and the BoxA motifs between Hoxa2(a) and -(b) are also present. Therefore, the difference in regulatory activity between the two fugu Hoxa2 enhancers could reflect the overall sequence divergence in the enhancers, or could arise because of specific changes in the known cis elements.
To distinguish between these possibilities, and to experimentally define which changes contribute to the differential expression of the coparalogs, we designed a series of constructs in which diverged cis elements from the fugu Hoxa2(a) module were replaced by sequences from Hoxa2(b) (Fig. 2G). In this context, we have preserved all of the sequences of the entire Hoxa2(a) enhancer, with the exception of the specific base-pair changes that convert an individual motif to that of Hoxa2(b). Changes in the RE2 and BoxA motifs, and their spacing, had no effect on regulatory activity (Fig. 2G and constructs 3 and 7). Intriguingly, two constructs that individually swapped the RE3 or RE4 elements resulted in a restoration of r3/5 enhancer activity (Fig. 2 D and E). The RE4 region spans ≈70 bp, with a number of sequence differences between species and coparalogs (Fig. 2A), which makes it difficult to pinpoint specific functional changes between Hoxa2(a) and -(b). Therefore, we divided this element into halves and swapped them individually. Changing the 5′ half of the RE4 element (construct 4) partially restored function of the fugu Hoxa2(a) r3/5 enhancer, as demonstrated by the fact that 32% of the electroporated embryos showed specific reporter staining in r3/5 (Fig. 2 D and G). Changes in the other half of the RE4 motif (construct 5) had no effect on enhancer activity (Fig. 2G). In the RE3 element, a prominent change occurred in the embedded TCT motif, a sequence previously shown to play an important role in r3/5 activity (24). The first three highly conserved nucleotides, TCT, have evolved to TGC in the r3/5 regulatory module of both fugu and medaka Hoxa2(a). Replacing the Hoxa2(a) RE3 element with that of Hoxa2(b) (construct 6) results in r3/5-specific lacZ expression in the hindbrain of the majority of embryos (67%) electroporated with this construct (Fig. 2 E and G).
In our sequence alignments with other species, we observed several other regions of conservation in the fugu r3/5 enhancers in addition to the known cis elements. Some of these new regions had an identical sequence between all species and were, therefore, unlikely to contribute to the differential activity of the fugu r3/5 enhancers. However, one region displayed sequence divergence in the Hoxa2(a) genes of fugu and medaka, compared with their Hoxa2(b) coparalogs; we termed this region RE5 (Fig. 2A). The RE5 consensus sequence TTTCC has been changed to CTTCT in fugu and medaka Hoxa2(a). To test whether these sequence differences are important in terms of regulatory activity, we generated a construct (construct 8) in which the RE5 motif in fugu Hoxa2(a) was converted to that of Hoxa2(b). Interestingly, this change also partially restored Hoxa2(a) enhancer activity; 33% of the electroporated embryos showed r3/5-specific reporter staining (Fig. 2 F and G). This result suggests that, in fugu, RE5 is a previously unrecognized cis component of r3/5 enhancer activity. To determine whether this motif also plays an important role in regulating mouse Hoxa2, we specifically deleted the RE5 element in the 809-bp murine r3/5 enhancer, linked it to a lacZ reporter gene, and scored for regulatory activity in transgenic mouse embryos (Fig. 2 H and I). Although the wild-type fragment directed strong reporter staining in r3, r5, and neural crest cells (Fig. 2H, see also ref. 23), the variant in which RE5 was deleted consistently (3/3) resulted in a loss of expression in r3 and a reduction in r5 (Fig. 2I). This outcome demonstrates that RE5 has an important and conserved input into r3/5 enhancer activity.
Our results show that specific changes in the RE3, RE4, or RE5 cis elements of the enhancer are sufficient to partially restore r3/5 activity. Hence, despite the high degree of overall divergence, these motifs appear to play an important role in the differential expression of the Hoxa2(a) and -(b) genes in fugu.
Analysis of Differential Expression in r4.
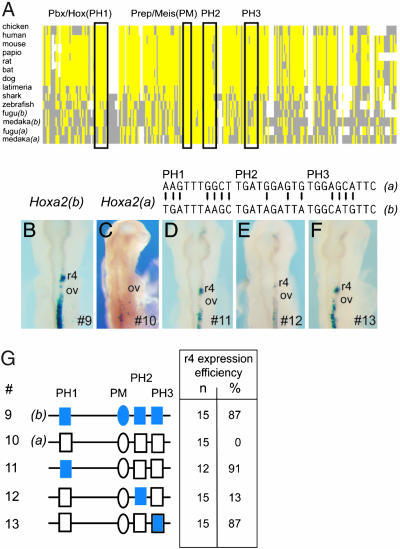
We next made similar comparisons to examine the functional basis of differential regulation in r4. Fugu Hoxa2(a) is not expressed in r4, whereas Hoxa2(b) is strongly expressed in r4 (Fig. 1B and ref. 11). The r4 expression of Hoxa2 in mice and chickens is mediated by auto- and crossregulatory inputs of Hox proteins by means of the presence of four cis elements embedded within the Hoxa2 intron (Fig. 3A; Fig. 7, which is published as supporting information on the PNAS web site; and our unpublished data). There are three bipartite Pbx/Hox binding sites (PH1–PH3) and a single Pbx/Prep/Meis site. The Hoxa2(b) intron, when linked to a lacZ reporter gene and assayed for enhancer activity in the developing chicken hindbrain, directs strong reporter expression in r4 (Fig. 3B), whereas the equivalent sequence from Hoxa2(a) lacks r4 activity (Fig. 3C). The r4 regulatory potential of the enhancers correlates directly with the differential endogenous r4 expression of these two coparalogs in fugu (11).
Fig. 3.
Analysis of the differential regulation of fugu Hoxa2(a) and Hoxa2(b) in r4. (A) Schematic representation of multispecies alignment of the Hoxa2 r4 regulatory module (original sequence alignment is shown in Fig. 7). A consensus sequence was derived based on sequence identity in >50% of the species. Yellow boxes indicate identical sequences, as compared with the consensus; gray boxes indicate diverged sequence; and white spaces are gaps introduced to maintain maximal alignment. The positions of the conserved elements are indicated by outlined boxes, with the name of each element given above the box. (B–F) Representative transgene expression in chicken embryos electroporated with reporter constructs under control of the r4 enhancer elements from fugu Hoxa2(b) (B) and Hoxa2(a) (C), with specific conversions between the two coparalogous sequences noted above D–F. The specific constructs used in each case are indicated at the bottom right in each panel. (G) Constructs (#) tested are diagrammed (Left), and the quantitative results of their analysis in chicken electroporation studies are tabulated (Right). The Hoxa2(b) elements are shown in blue; the elements of Hoxa2(a) are in white. In modified constructs (#11–13), the colored element indicates the specific region that has been changed from Hoxa2(a) to Hoxa2(b), in the context of the remaining Hoxa2(a) sequence. In the table, n is the total number of electroporated embryos examined, and the efficiency of the elements’ ability to direct r3/5 restricted expression is given as a percentage. PH, Prep/Hox; PM, Prep/Meis; ov, otic vesicle.
Sequence alignments reveal that all three of the Pbx/Hox binding sites (PH1–PH3) are diverged between Hoxa2(a) and -(b), whereas the Prep/Meis site is completely conserved in all species examined (Figs. 3A and 7). The first four bases of the bipartite Pbx/Hox consensus sequence (5′-TGATNNATGC-3′) are key determinants for binding Pbx/Exd, and the G in position 2 is critical for activity (26, 27). In the intronic enhancer of both fugu and medaka Hoxa2(a), there are seven differences in PH1, three in PH2, and four in PH3, as compared with the equivalent regions in Hoxa2(b) (see Fig. 7). The differences in PH1 of fugu Hoxa2(a) include an A in position 2, suggesting that PH1 is not functional. We generated constructs that converted each PH element in the Hoxa2(a) sequence to that of Hoxa2(b), and examined the effect on r4 activity (Fig. 3 D–G). The modifications in PH1 and PH3 restored strong reporter activity in r4 (Fig. 3 D, F, and G; and constructs 11 and 13). In contrast, the changes in PH2 (construct 12) had little or no effect on r4 activity, although one embryo did display weak patchy staining in r4, suggesting that the Hoxa2 r4 module may be working at a low level (Fig. 3 E and G). As seen with the r3/5 enhancer, specific changes in cis-regulatory motifs of the r4 enhancer are capable of restoring activity, even in the background of the highly diverged Hoxa2(a) intronic sequence.
Analysis of Differential Expression of Hoxa2(a) and -(b) in r2.
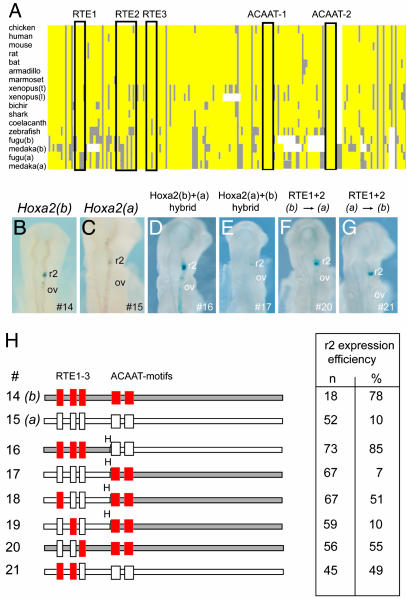
Unlike the absence of segmental expression of Hoxa2(a) in r3, r4, and r5, in situ analysis of fugu Hoxa2(a) and -(b) shows that both genes are expressed in r2 (11). Hoxa2(b) is expressed robustly throughout r2, whereas Hoxa2(a) is expressed weakly in a small subset of cells (Fig. 1B, see also ref. 11). The r2 regulatory module of Hoxa2 is embedded in the second exon and consists of five cis elements [r2 element (RTE) 1–3, and ACAAT 1 and 2] (Fig. 4A; Fig. 8, which is published as supporting information on the PNAS web site; and our unpublished data). We tested the regulatory potential of the respective r2 enhancers from fugu Hoxa2(a) and -(b) by linking them to a lacZ reporter and electroporating them into chicken embryos. The fragment from Hoxa2(b) (construct 14) mediated robust and efficient expression (78%) in r2 (Fig. 4 B and G), whereas the region from Hoxa2(a) (construct 15) was less efficient (10%) and directed staining in only a small number of cells in r2 (Fig. 4 C and G). This difference in enhancer activity correlates with the endogenous expression of the respective genes in r2.
Fig. 4.
Analysis of differential r2 enhancer activity of fugu Hoxa2(a) and Hoxa2(b). (A) Schematic representation of multispecies alignment of the Hoxa2 r2 regulatory module (original sequence alignment is shown in Fig. 8). A consensus sequence was derived based on sequence identity in >50% of the species. Yellow boxes indicate identical sequences, as compared with the consensus; gray boxes are diverged sequence; and white spaces are gaps introduced to maintain maximal alignment. The positions of the conserved elements are indicated by outlined boxes, with the name of each element given above the box. (B–G) Representative transgene expression in chicken embryos electroporated with reporter constructs under control of the r2 enhancer elements from fugu Hoxa2(b) (B) and Hoxa2(a) (C), with specific conversions between the two coparalogous sequences noted above D–G. The specific constructs used in each case are indicated at the bottom right in each panel. (G) Constructs (#) tested are diagrammed (Left), and the quantitative results of their analysis in chicken electroporation studies are tabulated (Right). The Hoxa2(b) elements are shown in red and gray; the elements of Hoxa2(a) are in white. In modified constructs (#16–21), the colored element indicates the specific region that has been changed from Hoxa2(a) to Hoxa2(b), in the context of the remaining Hoxa2(a) sequence. In the table, n is the total number of electroporated embryos examined, and the efficiency of the elements’ ability to direct r2 restricted expression is given as a percentage. ov, otic vesicle; H, HincII site.
The r2 enhancer is located entirely within the highly conserved coding region. Interspecies sequence alignments of the Hoxa2 r2 modules revealed that the ACAAT 1 and ACAAT 2 motifs are conserved in all species (Figs. 4A and 8), and hence are unlikely to contribute to differential enhancer activity. To determine whether differences in activity map to the regions containing the RTE1–3 motifs, we swapped these respective domains between Hoxa2(a) and -(b) (Fig. 4H and constructs 16 and 17). The fragment spanning RTE1–3 from Hoxa2(b) rescues full activity of the Hoxa2(a) enhancer in r2 (Fig. 4 D and H). In contrast, the corresponding fragment from Hoxa2(a) reduces the activity of the Hoxa2(b) enhancer. Therefore, the differential activity of these enhancers resides in the region spanning the RTE1–3 elements. To more precisely define the differences, we generated a construct (construct 18) that replaced the Hoxa2(a) version of RTE1. This change increased enhancer activity to 51%, and a construct (construct 21) swapping both RTE1 and RTE2 displayed an increase in activity to 49% (Fig. 4 G and H). However, replacing the Hoxa2(a) RTE2 with that of Hoxa2(b) (construct 19) (Fig. 4H) did not rescue any enhancer activity. Similarly, converting the RTE1 and RTE2 elements in the Hoxa2(b) enhancer (construct 20) to those of Hoxa2(a) results in a decrease in activity from 78% to 50% (Fig. 4 F and H). Together, these findings indicate that the changes in RTE1 play an important role in the differential activity of the fugu Hoxa2(a) and -(b) r2 enhancers.
Discussion
The goal of this study was to understand the regulatory basis that generates the differential expression of the coparalogous fugu Hoxa2(a) and -(b) genes in the developing hindbrain. Hoxa2(b) is expressed in r2–r7, whereas Hoxa2(a) is detected only in a subset of cells in r1 and r2 (11). Our systematic analysis of evolutionary changes in the cis-regulatory regions of the duplicated fugu Hoxa2 genes accounts for their different segmental expression patterns in the hindbrain. Within the r2, r4, and r3/5 regulatory modules, a number of the key cis elements essential for activity are conserved between the fugu Hoxa2(a) and -(b) genes. However, we also identified subtle sequence changes in specific regulatory elements within each module in Hoxa2(a). In functional assays, we demonstrated that activity could be partially restored to the Hoxa2(a) control modules by changing these subtle sequence differences to those present in Hoxa2(b). This outcome strongly suggests that these alterations are responsible for the differential expression of these two genes. Our results provide insight into the evolution of regulation and function of duplicated genes and reveal a number of interesting findings with respect to the control of Hox genes and hindbrain patterning.
The 5′ flanking (intergenic) region and intron of fugu Hoxa2(a) and -(b) are highly diverged from each other and from other vertebrates. Hence, the differences in the two genes’ segmental expression in the hindbrain could simply reflect a widespread degree of sequence variation that has eliminated hindbrain regulatory modules from the Hoxa2(a) gene. On the basis of characterization in chicken and mouse transgenic assays, a minimum of 15 different cis elements have been shown to participate in the activity of the r2, r4, and r3/5 enhancers. Our sequence comparisons reveal that variants of all 15 of these cis elements are present in both fugu coparalogs (Fig. 5). Some elements, such as the Krox20, Prep/Meis, and ACAAT sites, are nearly identical in all species and display no differences between fugu Hoxa2(a) and -(b), indicating that these elements are unlikely to contribute to differential regulation. In the other 11 cis elements, differences correspond to subtle or small numbers of base-pair changes in the respective motifs between the coparalogs or other species, rather than a complete absence or deletion of the respective element. In four of these more-diverged cis elements, we have shown that the differences between the coparalogs apparently do not account for altered expression because conversion of the sequences from Hoxa2(a) to Hoxa2(b) has no effect on enhancer activity. However, in the remaining seven cis elements, converting the motifs from Hoxa2(a) to those of Hoxa2(b) partially restored regulatory activity. In each of the three segmental regulatory modules examined, the loss of enhancer activity from the Hoxa2(a) gene correlates with changes in multiple cis elements, rather than a single alteration. Together, our results suggest that changes in these seven cis elements are responsible for the differential expression of the coparalogous Hoxa2 genes in fugu. In further support of this idea, the majority of sequence changes in the same seven regulatory elements from the fugu coparalogs were present in the respective medaka Hoxa2(a) and -(b) genes. We found that the r2, r4, and r3/5 enhancers from the medaka Hoxa2(b) gene were also active in the chicken transgenic assays, whereas those from Hoxa2(a) were not (data not shown). This finding implies that the sequence drift in the regulatory region of Hoxa2(a) occurred before the evolutionary split that led to the lineages of the spiny-ray fishes, medaka and fugu. Interestingly, in the zebrafish lineage, the Hoxa2(a) diverged into an unexpressed, nonfunctional pseudogene.
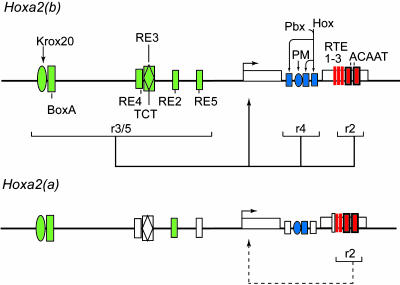
Fig. 5.
Summary of the activity of the rhombomeric regulatory modules of fugu Hoxa2(a) and -(b). Shown are schematic representations of the fugu Hoxa2(b) regulatory modules (Upper) and the fugu Hoxa2(a) modules (Lower). In Hoxa2(b), the solid-colored shapes represent active functional elements involved in segmental regulation. In Hoxa2(a), the open (white) shapes illustrate the diverged inactive elements, whereas the colored shapes are conserved and have the potential to be active. The solid lines below Hoxa2(b) highlight active modules; and the dashed line illustrates that the r2 module in Hoxa2(a) is partially active, as defined by our analyses. PM, Prep/Meis site; Pbx/Hox, bipartite binding sites for Hox and Pbx proteins; Krox20, Krox20 binding site, BoxA, putative Sox binding site.
Our work reinforces the view of the modular nature of segmental Hox expression in the vertebrate hindbrain. It is interesting that, despite the large degree of sequence divergence, we find modules and motifs in Hoxa2(a) partially conserved, even though they appear to be nonfunctional in relation to segmental expression. This finding implies that there might have been selective pressure to maintain the integrity of the modules. This situation could arise because these motifs may have other functions at later stages or in other tissues, even though they do not mediate rhombomeric expression. Alternatively, the motifs may be positioned near cis elements that regulate other domains of expression of the Hoxa2 gene. In this regard, the Hoxa2 r3/5 enhancer is embedded in a region that regulates expression of the gene in neural crest cells, and some motifs may be involved in both activities (24, 25). The fact that expression from Hoxa2(a) is only partially lost in r2, whereas other segmental domains are completely absent, may reflect its location in the coding region, where additional constraints on base-pair changes are operating to maintain this sequence.
The findings in this study directly support the idea that changes in cis-regulatory modules are a major contributing factor in generating diversity of expression, and presumably function, of duplicated genes (14, 28). Although the Hoxa2(a) enhancers lacked activity in our chicken transgenic assay, it is important to note that the coding region of this gene is fully intact and presumably participates in other functional activities. Fugu is not a good laboratory system for probing this question, but the conservation between the fugu and medaka Hoxa2 coparalogs suggests that morpholino knockdown experiments in medaka might provide insight into the distinct roles of these genes. In mouse mutants, Hoxa2 has been shown to play a functional role in hindbrain segments and cranial neural crest (20, 21). Rhombomeric expression and regulation correlates with the fugu Hoxa2(b) gene, but in testing its enhancers we found no regulatory activity in neural crest cells. This activity may reside with the Hoxa2(a) gene or may have been adopted by another Hox gene. In zebrafish, analysis of the Hoxa2 and Hoxb2 genes has indicated that both are necessary in patterning cranial crest (29), whereas in the mouse only Hoxa2 is required (20, 21). This difference might have arisen as a result of changes in the neural crest regulatory modules. Segregation of cis-regulatory elements between duplicated Hoxb1 genes has also been observed (30, 31), and it may be a general feature of duplicated Hox genes. In summary, we have used these interspecies comparisons, combined with knowledge of functionally relevant cis-regulatory modules of Hoxa2, to probe the basis of differential expression of two fugu coparalogs. However, we also found that it is possible to discover new regulatory elements in modules by using this approach, which may be helpful for dissecting the cis components of other genes.
Materials and Methods
Chicken Embryo Electroporation.
Chicken embryos were electroporated as described in ref. 32. Circular plasmid DNA (0.75–2 μg/μl) was injected into the neural tube of HH9-stage embryos. DNA was subjected to electroporation, and the embryos were allowed to develop for a further 15 h in ovo before staining for β-galactosidase activity.
Transgenic Mice.
Transgenic mouse embryos were generated as described in ref. 24. Briefly, the inserts were first released from the vector by digestion with appropriate enzymes. After electrophoretic separation, the inserts were extracted from agarose by using MinElute (Qiagen, Valencia, CA). The DNAs were injected into the pronucleus of fertilized eggs and reimplanted into foster animals. Embryos were then harvested and analyzed 9.5 days postcoitum.
Constructs and Site-Directed Mutagenesis.
Fugu constructs were generated by PCR from genomic DNA (primer sequences are given in Table 1, which is published as supporting information on the PNAS web site) and cloned into TA-cloning vectors (Promega). Site-directed mutagenesis (Table 1) was performed with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The fragments were then cloned into the BGZ40 vector, which contains a lacZ reporter gene linked to a minimal human β-globin promoter.
Sequence Alignments.
Chicken (33) and bat (34) Hoxa2 sequences were determined from plasmid and phage clones. Alignments were generated by using the Hoxa2 regions, including the publicly available sequences for other species. Local alignments were performed with Vector NTI’s integrated clustalw (35) alignment program (Invitrogen). Fugu sequences were obtained from the Comparative Genomics Group’s web site at http://fugu.biology.qmul.ac.uk.
Supplementary Material
Acknowledgments
We thank Chris Cretekos and Richard Behringer (National Science Foundation Grant IBN0220458) for the bat genomic phage library, Makoto Furutani-Seiki for medaka genomic DNA, and Yoshiyuki Imai for medaka embryos. We also value our discussions with Arcady Mushegian. S.T. was supported by Boehringer Ingelheim Funds. This work was funded by the Stowers Institute for Medical Research. S.T. is a Ph.D. student registered with the Open University, U.K., and this work was done to fulfill, in part, requirements for his thesis research.
Abbreviations
- r
rhombomere
- RE
rhombomeric element
- RTE
r2 element.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Krumlauf R. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 2.Krumlauf R. BioEssays. 1992;14:245–252. doi: 10.1002/bies.950140408. [DOI] [PubMed] [Google Scholar]
- 3.Kappen C., Schughart K., Ruddle F. H. Proc. Natl. Acad. Sci. USA. 1989;86:5459–5463. doi: 10.1073/pnas.86.14.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham A., Papalopulu N., Krumlauf R. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- 5.Duboule D., Dollé P. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popovici C., Leveugle M., Birnbaum D., Coulier F. Biochem. Biophys. Res. Commun. 2001;288:362–370. doi: 10.1006/bbrc.2001.5794. [DOI] [PubMed] [Google Scholar]
- 7.Panopoulou G., Poustka A. J. Trends Genet. 2005;21:559–567. doi: 10.1016/j.tig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe K. H. Nat. Rev. Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 9.Vandepoele K., De Vos W., Taylor J. S., Meyer A., Van de Peer Y. Proc. Natl. Acad. Sci. USA. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amores A., Force A., Yan Y.-L., Joly L., Amemiya C., Fritz A., Ho R., Langeland J., Prince V., Wang Y.-L., et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 11.Amores A., Suzuki T., Yan Y.-L., Pomeroy J., Singer A., Amemiya C., Postlethwait J. H. Genome Res. 2004;14:1–10. doi: 10.1101/gr.1717804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoegg S., Meyer A. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Aparicio S., Hawker K., Cottage A., Mikawa Y., Zuo L., Chen E., Krumlauf R., Brenner S. Nat. Genet. 1997;16:79–84. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- 14.Gompel N., Prud’homme B., Wittkopp P. J., Kassner V. A., Carroll S. B. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 15.Lumsden A., Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 16.Hunt P., Gulisano M., Cook M., Sham M. H., Faiella A., Wilkinson D., Boncinelli E., Krumlauf R. Nature. 1991;353:861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- 17.Krumlauf R. Trends Genet. 1993;9:106–112. doi: 10.1016/0168-9525(93)90203-t. [DOI] [PubMed] [Google Scholar]
- 18.Prince V., Lumsden A. Development (Cambridge, U.K.) 1994;120:911–923. doi: 10.1242/dev.120.4.911. [DOI] [PubMed] [Google Scholar]
- 19.Prince V. E., Moens C. B., Kimmel C. B., Ho R. K. Development (Cambridge, U.K.) 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- 20.Rijli F. M., Mark M., Lakkaraju S., Dierich A., Dollé P., Chambon P. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 21.Gendron-Maguire M., Mallo M., Zhang M., Gridley T. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 22.Gavalas A., Davenne M., Lumsden A., Chambon P., Rijli F. M. Development (Cambridge, U.K.) 1997;124:3693–3702. doi: 10.1242/dev.124.19.3693. [DOI] [PubMed] [Google Scholar]
- 23.Nonchev S., Vesque C., Maconochie M., Seitanidou T., Ariza-McNaughton L., Frain M., Marshall H., Sham M. H., Krumlauf R., Charnay P. Development (Cambridge, U.K.) 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 24.Maconochie M. K., Nonchev S., Manzanares M., Marshall H., Krumlauf R. Dev. Biol. 2001;233:468–481. doi: 10.1006/dbio.2001.0197. [DOI] [PubMed] [Google Scholar]
- 25.Maconochie M., Krishnamurthy R., Nonchev S., Meier P., Manzanares M., Mitchell P. J., Krumlauf R. Development (Cambridge, U.K.) 1999;126:1483–1494. doi: 10.1242/dev.126.7.1483. [DOI] [PubMed] [Google Scholar]
- 26.Chan S.-K., Mann R. S. Proc. Natl. Acad. Sci. USA. 1996;93:5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan S.-K., Ryoo H. D., Gould A., Krumlauf R., Mann R. S. Development (Cambridge, U.K.) 1997;124:2007–2014. doi: 10.1242/dev.124.10.2007. [DOI] [PubMed] [Google Scholar]
- 28.Carroll S. B. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter M. P., Prince V. E. Dev. Biol. 2002;247:367–389. doi: 10.1006/dbio.2002.0701. [DOI] [PubMed] [Google Scholar]
- 30.Prince V. E., Pickett F. B. Nat. Rev. Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 31.McClintock J. M., Kheirbek M. A., Prince V. E. Development (Cambridge, U.K.) 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- 32.Itasaki N., Bel-Vialar S., Krumlauf R. Nat. Cell Biol. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- 33.Tümpel S., Maconochie M., Wiedemann L. M., Krumlauf R. Dev. Biol. 2002;246:45–56. doi: 10.1006/dbio.2002.0665. [DOI] [PubMed] [Google Scholar]
- 34.Cretekos C. J., Rasweiler J. J., Behringer R. R. Reprod. Fertil. Dev. 2001;13:691–695. doi: 10.1071/rd01115. [DOI] [PubMed] [Google Scholar]
- 35.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.