Abstract
Human diploid fibroblasts (HDF) immortalized by hTERT and simian virus 40 (SV40) early region (ER) exhibit a limited degree of transformation upon the expression of activated H-RAS (H-RAS V12) compared with rat embryonic fibroblasts (REF) immortalized by SV40 ER. Here, we identified FRA1 as a determinant for this difference in RAS-induced transformation. FRA1 was not induced by H-RAS V12 in the immortalized HDF, in contrast to its marked accumulation in the immortalized REF. Ectopic expression of FRA1 significantly enhanced anchorage-independent growth of various HDF expressing hTERT, SV40 ER, and H-RAS V12. More importantly, FRA1 could induce anchorage-independent growth as well as nude mice tumor formation of the immortalized HDF in the absence of H-RAS V12. The results of an in vitro kinase assay clearly showed that the RAS-induced extracellular signal-regulated kinase (ERK) activation, which is responsible for FRA1 induction, was markedly attenuated in the HDF compared with that in the REF, despite no obvious differences in the phosphorylation status of ERK between the species. Our results strongly suggest that HDF negatively regulate the mitogen-activated protein kinase kinase (MEK)/ERK pathway more efficiently than REF, and consequently express less malignant phenotypes in response to H-RAS V12.
Keywords: human diploid fibroblast, mitogen-activated protein kinase kinase/extracellular signal-regulated kinase, simian virus 40 early region, telomerase, rat embryonic fibroblasts
In contrast to rodent cells, human cells have long been very difficult to be transformed in vitro. Normal (nonestablished) rodent cells can be readily transformed by the concomitant expression of two oncogenes, whereas normal human cells have proven to be resistant to the transformation induced by the same combinations of oncogenes, indicating the fundamental differences in cellular requirements for oncogenic transformation between these species. In 1999, Weinberg and colleagues (1) reported that ectopic expression of the catalytic subunit of human telomerase (hTERT) made normal human foreskin fibroblasts and kidney epithelial cells susceptible to transformation by the combined expression of simian virus 40 (SV40) early region (SV40 ER) and activated H-Ras (H-Ras V12). They further succeeded in transforming several different types of normal human cells by the same combinations of genetic elements. Based on these results, they proposed that the telomere maintenance by hTERT, inactivation of p53 and retinoblastoma (Rb) by SV40 large T antigen, suppression of protein phosphatase PP2A activity by SV40 small t antigen, and constitutive activation of mitogenic signaling by activated Ras are sufficient to transform normal human cells (2). However, we recently demonstrated that, even when all these requirements are fulfilled, the transformed phenotypes of human fibroblasts are much less malignant than those of rat fibroblasts in terms of morphological changes, anchorage independence, and tumorigenicity in nude mice (3, 4). Our results strongly suggest that normal human cells have still undefined intrinsic mechanisms rendering them resistant to oncogenic transformation. In the present study, we found that the expression of FRA1, a member of the family of AP-1 transcription factors, was differentially regulated by RAS in human and rat fibroblasts and concluded that this transcription factor is one of the determinant factors for species-specific susceptibility to RAS-induced transformation.
Results
FRA1 Is Highly Induced by RAS in Rodent Fibroblasts but Not in Human Fibroblasts.
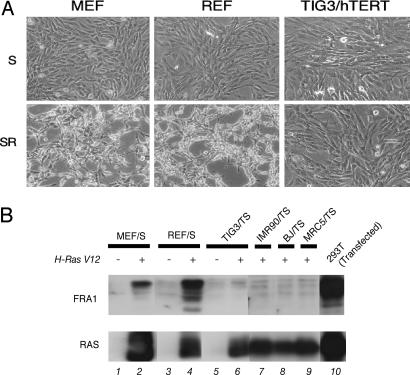
To explore the molecular basis underlying the difference in RAS-induced transformation phenotypes between rodent and human fibroblasts, we performed gene expression profiling by using a CodeLink microarray. Rat embryonic fibroblasts (REF), mouse embryonic fibroblasts (MEF) and a strain of human diploid fibroblasts (HDF), TIG3 ectopically expressing hTERT (TIG3/T), were analyzed. We made comparisons between cells expressing SV40 ER (S) alone and cells coexpressing SV40 ER and H-Ras V12 (SR), because the drastic phenotypic differences between rodent and human fibroblasts were observed in response to the expression of H-RAS V12 (Fig. 1A; ref. 4). The number of genes showing significant (>2-fold) up- or down-regulation by H-RAS V12 was 3,097 genes of 22,045 (14.1%) in REF/S cells, 2,571 genes of 17,418 (14.7%) in MEF/S cells, and 1,106 genes of 16,638 (6.7%) in TIG3/TS cells. This finding clearly indicates that human fibroblasts were more refractory than rodent fibroblasts to RAS-induced changes in gene expression, implying significant differences in the modulation of transcription factors. Then, to identify the candidate genes contributing to species differences in transformation phenotypes, we picked up the genes that were consistently up-regulated by H-RAS V12 in REF/S cells and MEF/S cells but not in TIG3/TS cells (Table 1). Among 23 such genes, we focused on Fosl1 encoding FRA1, a member of the family of AP-1 transcription factors, which is well established as a downstream target of RAS signaling and is also known to play essential roles in RAS-induced transformation in several rodent cell systems. We confirmed this microarray data at the protein expression level by immunoblotting (Fig. 1B). H-RAS V12 markedly induced FRA1 protein in MEF/S and REF/S cells, whereas such strong induction was not observed in any of the HDF examined (TIG3/TS, IMR90/TS, BJ/TS, and MRC5/TS cells).
Fig. 1.
FRA1 is accumulated in rodent cells but not in human cells. (A) Cell morphologies of primary fibroblasts, infected with retroviral vectors expressing indicated genes, are shown. T, hTERT; S, SV40 ER; R, H-RAS V12. (B) Total cell lysates from REF, MEF, and four HDF were subjected to immunoblot analysis with anti-FRA1 antibody (Upper) or anti-RAS antibody (Lower). In lane 10, 293T cells were transiently transfected with human Fra1 cDNA together with H-Ras V12 to verify the reactivity of the antibody against human FRA1 protein that was used.
Table 1.
List of genes induced by H-Ras V12 in rodent fibroblasts but not in human fibroblasts
| Gene | Description | Level of up-regulation |
||
|---|---|---|---|---|
| REF | MEF | TIG3/T | ||
| Ank | Progressive ankylosis | 6.6 | 5.0 | −1.7 |
| Ccnd2 | Cyclin D2 | 77.5 | 2.8 | −1.5 |
| Clcn3 | Chloride channel 3, transcript variant a | 2.2 | 8.1 | 1.3 |
| Cldn11 | Claudin 11 | 9.8 | 3.2 | 1.6 |
| Edg2 | Endothelial differentiation, lysophosphatidic acid G protein-coupled receptor, 2 | 3.2 | 2.3 | 1.6 |
| Fosl1 | Fra1, fos-like antigen 1 | 2.9 | 2.8 | 1.1 |
| Gng2 | Guanine nucleotide-binding protein (G protein), γ2 subunit | 2.8 | 2.4 | 1.6 |
| Got1 | Glutamate oxaloacetate transaminase 1, soluble | 2.3 | 2.2 | −1.1 |
| Gpd2 | Glycerol phosphate dehydrogenase 2, mitochondrial | 2.5 | 3.0 | 1.1 |
| Hmga1 | High-mobility group AT-hook 1 | 3.5 | 6.8 | −1.0 |
| Itga6 | Integrin α6 | 4.6 | 13.6 | −1.0 |
| Itga7 | Integrin α7 | 7.5 | 3.2 | 1.2 |
| Kcnab1 | Potassium voltage-gated channel, shaker-related subfamily, β member 1 | 11.8 | 3.7 | −1.2 |
| Mmp10 | Matrix metalloproteinase 10 | 1,097.3 | 4.5 | −1.0 |
| Ppap2c | Phosphatidic acid phosphatase type 2c | 2.7 | 2.1 | 1.5 |
| Prps2 | Phosphoribosyl pyrophosphate synthetase 2 | 2.3 | 2.3 | −1.1 |
| Pter | Phosphotriesterase-related | 2.5 | 2.7 | 1.2 |
| Rtn2 | Reticulon 2 (Z-band associated protein) | 2.1 | 2.2 | 1.3 |
| Sh3kbp1 | SH3-domain kinase binding protein 1 | 5.0 | 2.7 | −1.2 |
| Slc16a1 | Solute carrier family 16 (monocarboxylic acid transporters), member 1 | 2.3 | 2.8 | −1.0 |
| Slc2a1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 4.4 | 2.8 | 1.2 |
| Slc38a1 | Solute carrier family 38, member 1 | 2.1 | 2.3 | 1.1 |
| Syn1 | Synapsin I | 3.5 | 2.3 | −1.5 |
Gene expression profiles were analyzed by using a CodeLink microarray. MEF, REF, and TIG3, a strain of HDF ectopically expressing hTERT (TIG3/T), were examined. Cells expressing SV40 ER (S) and those coexpressing S and H-Ras V12 (R) were compared for each cell type. The genes that were up-regulated by H-RAS V12 >2-fold in both REF/S cells and MEF/S cells but not in TIG3/TS cells are listed. Only annotated genes were analyzed and are listed alphabetically.
FRA1 Induces Transformation of Human Fibroblasts.
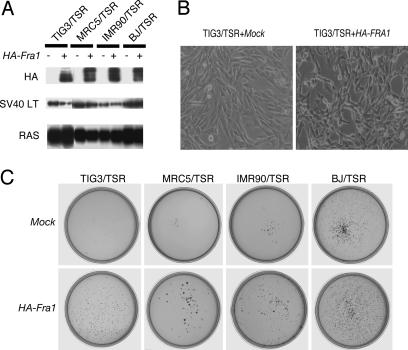
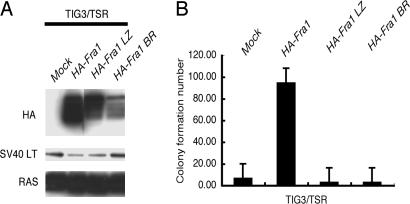
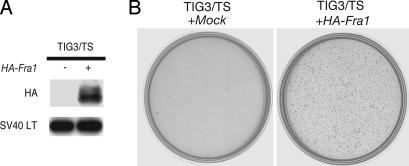
To evaluate the relevance of the FRA1 expression to the transformation phenotypes, we ectopically expressed hemagglutinin (HA)-tagged human Fra1 in several HDF that already expressed hTERT, SV40 ER, and H-Ras V12 (HDF/TSR cells; Fig. 2A). We also confirmed that the ectopically expressed FRA1 in HDF/TSR cells where at a level comparable with that of endogenous FRA1 induced in REF/SR cells (data not shown). After the introduction of FRA1, the TIG3/TSR cells became a little bit smaller, disorganized, and refractile (Fig. 2B). Similar morphological changes were observed in all other HDF/TSR cells examined (data not shown). More importantly, FRA1 expression induced a significant increase in colony formation by all these HDF/TSR cells (Fig. 2C). In the following experiments, TIG3 was used as a representative of HDF. Neither the Fra1 mutant defective in its leucine zipper domain (HA-Fra1 LZ) nor in basic region (HA-Fra1 BR) induced soft agar colony formation in TIG3/TSR cells, indicating that both of these domains are required for transformation (Fig. 3A and B). Interestingly, we found that FRA1 can induce anchorage-independent growth of TIG3/TS cells in the absence of H-RAS V12 as shown by the formation of colonies in soft agar (Fig. 4A and B) and tumorigenicity in nude mice (Fig. 9, which is published as supporting information on the PNAS web site). These results clearly showed the transforming activity of FRA1 in human fibroblasts and suggest that the difference in the expression of FRA1 can, at least in part, explain the difference in susceptibility to RAS-induced transformation between human and rat fibroblasts.
Fig. 2.
Ectopic expression of FRA1 can enhance anchorage-independent growth in HDF/TSR cells. (A) Total cell lysates from four HDF/TSR cells expressing HA-tagged human Fra1 were subjected to immunoblot analysis with antibodies indicated on the left. LT, large T antigen. (B) Cell morphologies were monitored by phase-contrast microscopy at ×100 magnification. (C) Anchorage-independent growth properties of the four HDF/TSR cells used in A. These cells were subjected to a soft agar colony formation assay. Colonies were stained with MTT and photographed 3 weeks after plating.
Fig. 3.
DNA binding domain and dimerization domain of FRA1 are essential for transformation. Wild-type FRA1 (HA-Fra1), an FRA1 mutant defective in its leucine zipper domain (HA-Fra1 LZ), or an FRA1 mutant defective in its basic region (HA-Fra1 BR) were expressed in TIG3/TSR cells. The resulting infectants were subjected to immunoblot analysis with the antibodies indicated on the left (A) or to the soft agar colony formation assay (B).
Fig. 4.
FRA1 can induce anchorage-independent growth of HDF in the absence of H-RAS V12. HA-Fra1 was ectopically expressed in TIG3/TS cells. The resulting cells were subjected to immunoblot analysis with antibodies indicated on the left (A) or to the soft agar colony-formation assay. Colonies were stained with MTT and photographed 2 weeks after plating (B).
Accumulation of FRA1 Is Totally Dependent on Mitogen-Activated Protein Kinase Kinase (MEK)/Extracellular Signal-Regulated Kinase (ERK) Pathway in REF/SR Cells.
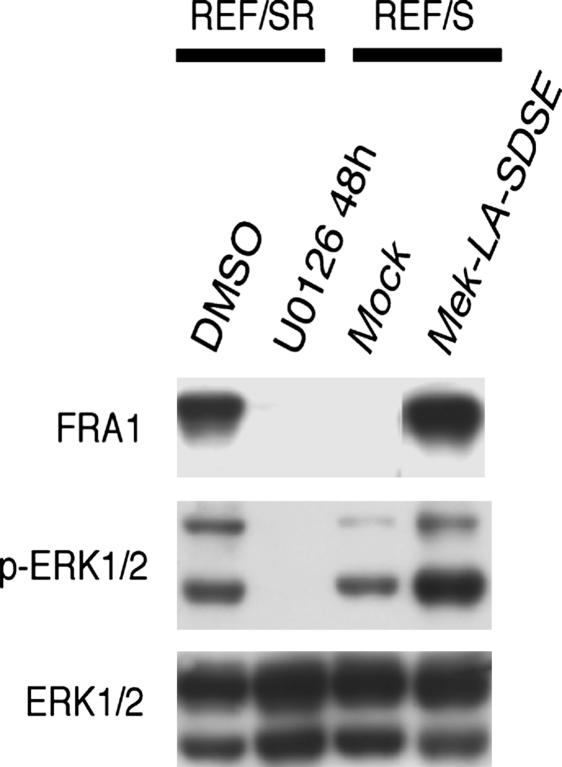
To address why the FRA1 expression regulated by H-RAS V12 was so different between human and rat fibroblasts, we first tried to clarify the signaling pathway responsible for FRA1 accumulation in REF cells. As has been reported (5–8), treatment with a MEK inhibitor, U0126, almost completely suppressed the FRA1 accumulation in REF/SR cells (Fig. 5). Conversely, a constitutively active mutant of MEK, MEK-LA-SDSE, induced FRA1 accumulation in REF/S cells at almost the same level as found with H-RAS V12 (Fig. 5). These results clearly indicate that the FRA1 accumulation induced by H-RAS V12 in our rat fibroblasts immortalized by SV40 ER was MEK/ERK pathway dependent.
Fig. 5.
RAS-induced FRA1 accumulation in rodent cells is totally dependent on the MEK/ERK pathway. REF/SR cells were treated with 25 μM U0126 for 48 h, and total cell lysates were then prepared and subjected to immunoblot analysis with the antibodies indicated on the left. A constitutive active MEK mutant, MEK-LA-SDSE, was expressed in REF/S cells and then subjected to the immunoblot analysis with the same antibodies.
Attenuation of RAS-Induced ERK Activity in Human Fibroblasts.
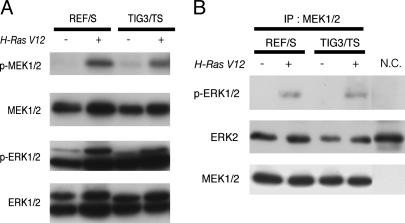
Based on the observation that the MEK/ERK pathway was responsible for the FRA1 accumulation in REF/SR cells, we examined whether there were any differences in the activation of MEK/ERK between rat and human cells. First, we analyzed the phosphorylation status of these kinases by immunoblotting with antibodies specific for their phosphorylated form. As has been reported (3, 4), there was no significant difference between REF/SR and TIG3/TSR cells in the phosphorylation of both MEK and ERK (Fig. 6A). To further explore the possible differences in MEK/ERK pathway between rat and human cells, we performed an in vitro kinase assay. Endogenous MEK1/2 was immunoprecipitated from each cell type and then incubated with recombinant kinase-deficient ERK2 protein as a substrate. As shown in Fig. 6B, the expression of H-RAS V12 clearly increased the MEK activity in both rat and human cells. Next, we tried to examine ERK activity by conducting an in vitro kinase assay. Initially, attempts were made to measure endogenous ERK activity, but we were unable to immunoprecipitate equivalent amounts of ERK from each cell type with several anti-ERK antibodies examined. Therefore, we changed our strategy and introduced HA-tagged rat ERK1 into both rat and human cells. We found that this did not change the expression pattern of FRA1 in either type of cells (Fig. 7B). Ectopically expressed ERK was immunoprecipitated with anti-HA antibody, and then the in vitro kinase assay was performed with recombinant ELK1 protein as a substrate. In this case, we could successfully immunoprecipitate similar amounts of HA-ERK from all of the cells examined. As shown in Fig. 7A, slower migrated ELK1 proteins, which represented the multiply phosphorylated forms, could be detected only in REF/SR cells, indicating that the kinase activity of ERK in REF/SR cells was much higher than that in TIG3/TSR cells, even though there was no significant difference in the ERK phosphorylation status. To further obtain evidence indicating that the in vivo ERK activity was actually higher in REF/SR cells than in TIG3/TSR cells, we examined the phosphorylation status of Thr-359 and Ser-363 residues in p90RSK1, which are well known to be phosphorylated by ERK1/2. Phosphorylation of these residues in p90RSK1 was clearly increased in REF/SR cells but not in TIG3/TSR cells (Fig. 8A). Then we tried to monitor the phosphorylation status of a broader range of proteins supposed to be ERK substrates. To enrich for putative ERK substrates, we used the recently reported (9) GST-PIN1 (peptidylprolyl isomerase 1) fusion protein as a probe. PIN1 specifically binds proteins with phosphorylated Ser or Thr preceding Pro (pSer/Thr-Pro), which conforms to the consensus motif for phosphorylation by ERK. Total cell lysates were subjected to a GST-PIN1 pull-down assay and then to immunoblotting with antibody specific for phoshphorylated Ser and Thr residues (p-Ser/p-Thr). Protein species containing phosphorylated Ser/Thr-Pro motif clearly increased upon expression of H-RAS V12 in rat fibroblasts, whereas no such significant RAS-induced changes were observed in human fibroblasts (Fig. 8B). We confirmed that the treatment of REF/SR cells with U0126 significantly reduced the amounts of p-Ser/p-Thr containing proteins precipitated with GST-PIN1 (data not shown). These results strongly suggest that ERK1/2 in human fibroblasts was not as efficiently activated by H-RAS V12 as that in rat fibroblasts. Given the primary importance of the MEK/ERK pathway in RAS-induced accumulation of FRA1 in rat cells, this attenuation of ERK activity is supposedly the reason for the failure of FRA1 induction in human fibroblasts expressing H-RAS V12.
Fig. 6.
MEK activity is increased by H-RAS V12 in both rodent and human cells. (A) Total cell lysates from REF/S and TIG3/TS cells with or without the expression of H-RAS V12 were subjected to immunoblot analysis with anti-phospho-specific MEK1/2 and ERK1/2 antibodies. (B) Endogenous MEK1/2 proteins were immunoprecipitated from each type of cell lysate and then subjected to an in vitro kinase assay with kinase-deficient ERK2 protein as a substrate as described in Materials and Methods. The reaction mixtures were analyzed by immunoblotting with the antibodies indicated on the left. The immunoprecipitated sample obtained with normal IgG was also subjected to the in vitro kinase assay as a negative control (N.C.).
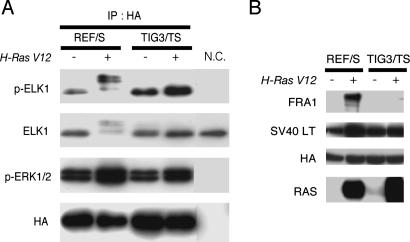
Fig. 7.
RAS-induced ERK activity is much higher in REF cells than in TIG3 cells. (A) HA-tagged rat ERK1 was ectopically overexpressed in REF/S and TIG3/TS cells with or without the expression of H-RAS V12 and were immunnoprecipitated with anti-HA antibody. The immunoprecipitated HA-rat ERK1 proteins were subjected to the in vitro kinase assay with recombinant ELK1 fusion protein as a substrate, as described in Materials and Methods. The reaction mixtures were analyzed by immunoblotting with anti-phospho-specific ELK1 antibody. The immunoprecipitated sample obtained with normal IgG was also subjected to the kinase assay as a negative control (N.C.). (B) Total cell lysates prepared from the cells described in A were subjected to immunoblotting with the antibodies indicated on the left.
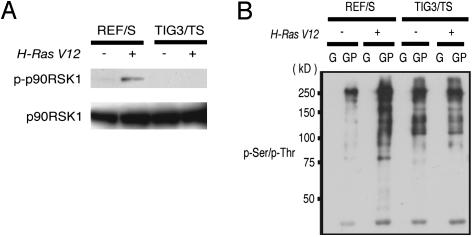
Fig. 8.
Phosphorylation of downstream targets of ERK is enhanced by H-RAS V12 in REF cells but not in TIG3 cells. (A) Total cell lysates from REF/S cells and TIG3/TS cells with or without the expression of H-RAS V12 were subjected to immunoblot analysis with antibody against phospho-p90RSK (Thr-359/Ser-363). (B) Total cell lysates from the same cells as in A were subjected to the GST pull-down assay described in Materials and Methods. Then the GST pull-down samples were analyzed by immunoblotting with anti-phospho-serine/threonine antibody. G represents sample pulled down by GST alone, and GP represents that pulled down by GST-PIN1 fusion proteins.
Discussion
In this study, we investigated the molecular basis underlying the difference in transformation phenotypes between hTERT-expressing human diploid fibroblasts and rodent fibroblasts induced by the combined expression of SV40 ER and H-Ras V12. This oncogene combination seems to be the most powerful one and has been reported to similarly transform both normal mouse fibroblasts and human fibroblasts expressing hTERT (1, 10). However, our studies using several strains of HDF revealed that the transformed phenotypes of the human fibroblasts were much less malignant than those of REF cells (3). Although REF cells are a little bit more susceptible to oncogene-mediated transformation than MEF cells (11), we observed that the phenotypic changes in MEF cells caused by the expression of SV40 ER and H-Ras V12 were more aggressive compared with those in the human fibroblasts in terms of morphology, anchorage-independence, and tumorigenicity (Fig. 1A; K. Sasai and T.A., unpublished results). Moreover, as described herein, changes in the gene expression induced by H-Ras V12 were similarly more evident in MEF cells and REF cells than in TIG3 cells (Table 1). From these results, we concluded that human fibroblasts were more refractory to RAS-induced oncogenic transformation than rodent fibroblasts.
To gain insight into the mechanism underlying this refractory nature of human cells, we explored RAS-mediated gene expression profiles of rodent and human cells. Consequently, we found that FRA1 was strongly induced by H-RAS V12 in rodent fibroblasts but not in human fibroblasts (Fig. 1B). The important roles of FRA1 accumulation during RAS-induced transformation have already been well documented in several rodent cell systems (12–17), where FRA1 is reported to be accumulated through both transcriptional up-regulation and posttranslational stabilization by escaping from proteaosomal degradation (6, 18). We showed that the ectopic expression of FRA1 significantly enhanced the anchorage-independent growth of various HDF that already expressed hTERT plus SV40 ER plus H-Ras V12 (Fig. 2), clearly indicating that the failure of the FRA1 induction, at least in part, explains the resistance to RAS-induced transformation observed in human fibroblasts. More importantly, FRA1 could induce anchorage-independent growth and tumorigenicity in TIG3 cells immortalized by hTERT and SV40 ER in the absence of H-RAS V12 (Figs. 4 and 9). The same results were obtained with BJ cells (data not shown). To our knowledge, this is the first report showing the transformation of normal human fibroblasts without the action of the RAS oncogene, thus underscoring the oncogenic potential of FRA1 toward human cells. Elevated expression of FRA1 has been reported in various human cancers including thyroid tumor, breast cancer, esophageal cancer, methothelioma, glioma, colorectal cancer, and head and neck squamous cell carcinoma (19–23). In several of these cases, the knockdown experiments using siRNA have proven the critical roles of FRA1 in the malignant features of these cancer cells in terms of cell morphology, motility, invasiveness, proliferation, and anchorage independence (21, 24). Because FRA1 supposedly exerts its transforming activity by acting as a transcription factor (13), it is very intriguing to identify the target genes involved in human cell transformation. Our system, in which FRA1 induces the transformation of HDF-expressing hTERT and SV40 ER, provides a valuable experimental setting to address this question.
Consistent with previous reports on NIH 3T3 and rat thyroid cells (6, 25) RAS-induced FRA1 accumulation in our rat fibroblasts was totally dependent on the MEK/ERK pathway. Vial et al. (24) demonstrated that the elevated expression of FRA1 in human colon carcinoma cells bearing mutated RAS was due to the activation of the MEK/ERK pathway. Therefore, it is plausible that the reduced level of FRA1 induction observed in H-RAS V12-expressing human fibroblast was caused by impaired ERK activation. The results of the in vitro kinase assay clearly showed that the RAS-induced ERK activation was markedly attenuated in human fibroblasts compared with that in rat fibroblasts (Fig. 7), despite no obvious differences in the phosphorylation status of this kinase between the two species (Fig. 6A). We also obtained evidence that the direct downstream events of ERK activation exemplified by the phosphorylation of RSK Thr-359/Ser-363 did not take place in human fibroblasts expressing H-RAS V12, supporting the notion that the in vivo activation of ERK was actually down-regulated in these cells. Because the activation of rat ERK1 ectopically expressed in human fibroblasts was suppressed, the difference in the regulatory machinery rather than one in the primary structure of ERK between rat and human is supposedly critical for the observed difference in the response of this kinase. Although it is generally believed that the phosphorylation of ERK at its Thr-202/Tyr-204 represents its activated state, several instances showed that this phosphorylation event does not necessarily ensure the activation of its downstream target (23, 26, 27). Kim et al. (28) reported that ERK is constitutively phosphorylated in senescent human fibroblasts but is inactivated by the binding with a protein phosphatase, MKP3, whose activity is reduced by oxidation. Purcell et al. (29) also reported that the phosphorylated form of ERK2 interacts with and is negatively regulated by the LIM-only protein FHL2 in cardiomyocytes. There also are several other proteins that can bind to and cause the mislocalization of phosphorylated ERK, such as PEA-15 and Sef (30, 31). We have preliminary data suggesting the association of some molecule with the phosphorylated ERK in human fibroblasts but not in rat fibroblasts (K.K. and T.A., unpublished results), and speculate that this molecule might act as an inhibitor to interfere with the kinase activity as well as with the correct localization of the phosphorylated form of ERK. We are currently trying to verify this hypothesis. Although the precise molecular mechanism still remains elusive, our results clearly demonstrate that human fibroblasts have a more efficient system to prevent the overactivation of the MEK/ERK pathway, and thereby fail to induce FRA1 and exhibit less malignant phenotypes in response to H-RAS V12. We consider that the recently reported highly resistant nature of human fibroblasts against RAS-induced morphological changes (4) can also be attributed to this negative regulation of the MEK/ERK pathway. Because the phenotypes of FRA1-expressing HDF/TSR are still milder than those of REF/SR, other genes, such as those listed in Table 1, should contribute to the observed phenotypic difference.
It is quite conceivable that the disruption of the human cell-specific negative regulatory mechanism of the MEK/ERK pathway suggested by this study might actually be involved in human cancer. Elucidating the molecular nature of this mechanism would be of great importance to fully understand the difference in susceptibility to oncogenic transformation between human and rodent cells.
Materials and Methods
Cells and Cell Culture.
TIG3, MRC5, and IMR90 cells are HDFs derived from embryonic lung. BJ cells are HDF derived from the foreskin of a newborn. REFs were prepared from 16-day-old Fisher rat embryos, and MEFs, from 14-day-old F1 embryos obtained from a cross between C57BL/6 and BALB/c. All HDF, REF, and MEF were cultured in DMEM supplemented with 10% FBS, as described (3).
Retroviral-Mediated Gene Transfer.
All of the gene transfer experiments were carried out by using retroviral vectors. Retroviral vectors encoding SV40 ER (including large T and small T antigens), hTERT (human telomerase catalytic subunit), and H-Ras V12 (activated form of H-Ras) have been described (3). cDNA of LA-SDSE MEK (MAPKK), a constitutively active mutant of Mek (32), was generously provided by E. Nishida (Kyoto University, Kyoto) and subcloned into CX4bleo. Full-length human Fra1 cDNA was amplified by PCR, and then tagged with the HA epitope at its N terminus and subcloned into CX4.1hisD. Fra1 mutants defective in their leucine zipper domain (HA-Fra1 LZ) or in their basic region (HA-Fra1 BR) were made by mimicking the corresponding mutants of c-Fos, which have proven to be deficient in DNA complex formation at the AP-1-recognition sequence (33, 34). The HA-Fra1 LZ mutant was generated by inserting two amino acids (FA) between amino acids 160 and 161; and the HA-Fra1 BR one, by inserting two amino acids between amino acids (AA) 129 and 130. These insertional mutants were made by using a QuikChange mutagenesis kit (Stratagene). For the HA-tagged rat Erk1 construct, full-length rat Erk1 cDNA was amplified by PCR, then tagged with HA epitope at N terminus and subcloned into CX4bleo. Production of and infection with retroviral vectors were performed as described (3).
Microarray Gene Expression Analyses.
Total RNA was extracted by using RNeasy kit (Qiagen, Valencia, CA) and then hybridized to CodeLink Mouse Whole Genome (MEF/S and MEF/SR cells), Rat Whole Genome (REF/S and REF/SR cells), or UniSet Human 20K I (TIG3/TS and TIG3/TSR cells) chips (Amersham Pharmacia). The methods for labeling, hybridization, scanning, and data analyses are available at http://www1.amershambiosciences.com.
Immunoblot Analysis.
Immunoblotting was performed as described (35). The following antibodies were used: Mouse monoclonal anti-SV40 T antigen (BD PharMingen), mouse monoclonal anti-RAS and mouse monoclonal anti-phospho-serine/threonine (BD Transduction Laboratories), rabbit polyclonal anti-FRA1 (R-20) and rabbit polyclonal anti-p90RSK1 (Santa Cruz Biotechnology), mouse monoclonal anti-phospho-p44/42 MAP kinase (Thr-202/Tyr-204), rabbit polyclonal anti-phospho-MEK1/2 (Ser-217/221), rabbit polyclonal anti-MEK1/2 rabbit polyclonal phospho-ELK1 (Ser-383), rabbit polyclonal ELK1, and phospho-p90RSK (Thr-359/Ser-363; Cell Signaling Technology), rat monoclonal anti-HA High-Affinity (Roche), and rabbit polyclonal anti-MAP kinase 1/2-CT (Upstate Biotechnology).
Soft-Agar Colony-Formation Assays.
Single-cell suspensions of 2 × 104 cells were plated per 60-mm culture dish in 3 ml of DMEM containing 10% FCS and 0.36% agar on a layer of 5 ml of the same medium containing 0.7% agar. Plates were fed weekly with 0.5 ml of DMEM/10% FCS. Two weeks after plating, colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and photographs of the stained colonies were taken.
In Vitro Kinase Assay.
Cells were lysed in lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate,1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 μg/μl leupeptin, and 1 mM PMSF. Protein concentrations were determined by using a BCA protein assay kit (Pierce) and were adjusted to 1 μg/μl. For in vitro MEK assays, endogenous MEK protein was immunoprecipitated from 500 μg of precleared cell lysate by using 1 μg of anti-MEK1 NT (Upstate Biotechnology) and coupled to protein G-Sepharose for 2 h at 4°C. Then the precipitates were washed three times with 1 ml of lysis buffer, after which almost equivalent amounts of MEK1 protein were subjected to an in vitro kinase assay in 50 μl of kinase buffer (100 mM 4-morpholinepropanesulfonic acid (Mops), pH 7.2/125 mM β-glycerophosphate/25 mM EGTA/5 mM sodium orthovanadate/5 mM DTT/75 mM magnesium chloride/1 mM ATP) with 0.4 μg of inactive recombinant ERK2 protein as a substrate (Cell Signaling Technology). The above reaction mixtures were incubated for 30 min at 30°C. Then the samples were boiled with 3× sample buffer and subjected to immunoblot analysis with anti-phospho-ERK antibody. For the ERK in vitro kinase assay, ectopically expressed HA-rat ERK proteins were immunoprecipitated from 500 μg of precleared cell lysates with anti-HA antibody (Roche) and then incubated for 30 min at 30°C with ELK1 fusion protein (Cell Signaling Technology) in a 50-μl reaction buffer containing 25 mM Tris (pH 7.5), 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM sodium orthovanadate, 10 mM magnesium chloride, and 10 mM ATP. Then the samples were boiled with 3× sample buffer and subjected to immunoblot analysis with anti-phospho-ELK antibody.
GST Pull-Down Assay.
pGEX-4T-2-Pin1 was constructed by inserting PCR-amplified full-length human Pin1 cDNA into pGEX-4T2. GST and GST-PIN1 fusion proteins were prepared as described (36). The GST pull-down assay was performed as described (37). Cell lysate (500 μl) was incubated for 4 h at 4°C with 10 μg of GST or GST-PIN1 recombinant protein bound to glutathione-Sepharose beads. Precipitates were then washed three times with 600 μl of lysis buffer and subjected to immunoblot analysis.
Tumorigenicity Assays.
For details, see Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We are grateful to H. Inoue (Shiga University Medical Science, Shiga, Japan), E. Nishida (Kyoto University, Kyoto), T. Shishido (Nara Institute of Science and Technology, Kansai Science City, Japan), P. Sharp (Massachusetts Institute of Technology, Cambridge, MA), and R. Weinberg (Massachusetts Institute of Technology) for providing the reagents. We also thank T. Iwahara for discussion of this study. This work was supported by a grant from the program grants-in-aid for Specially Promoted Research of the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by funding from Osaka City and Sony Corporation.
Abbreviations
- SV40
simian virus 40
- ER
early region
- REF
rat embryonic fibroblast
- MEF
mouse embryonic fibroblast
- HA
hemagglutinin
- HDF
human diploid fibroblast
- ERK
extracellular signal-regulated kinase
- MEK
mitogen-activated protein kinase kinase.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 2.Hahn W. C., Weinberg R. A. Nat. Rev. Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- 3.Akagi T., Sasai K., Hanafusa H. Proc. Natl. Acad. Sci. USA. 2003;100:13567–13572. doi: 10.1073/pnas.1834876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sukezane T., Oneyama C., Kakumoto K., Shibutani K., Hanafusa H., Akagi T. Oncogene. 2005;24:5648–5655. doi: 10.1038/sj.onc.1208724. [DOI] [PubMed] [Google Scholar]
- 5.Young M. R., Nair R., Bucheimer N., Tulsian P., Brown N., Chapp C., Hsu T. C., Colburn N. H. Mol. Cell. Biol. 2002;22:587–598. doi: 10.1128/MCB.22.2.587-598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casalino L., De Cesare D., Verde P. Mol. Cell. Biol. 2003;23:4401–4415. doi: 10.1128/MCB.23.12.4401-4415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burch P. M., Yuan Z., Loonen A., Heintz N. H. Mol. Cell. Biol. 2004;24:4696–4709. doi: 10.1128/MCB.24.11.4696-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann E., Thiefes A., Buhrow D., Dittrich-Breiholz O., Schneider H., Resch K., Kracht M. J. Biol. Chem. 2005;280:9706–9718. doi: 10.1074/jbc.M407071200. [DOI] [PubMed] [Google Scholar]
- 9.Ley R., Ewings K. E., Hadfield K., Howes E., Balmanno K., Cook S. J. J. Biol. Chem. 2004;279:8837–8847. doi: 10.1074/jbc.M311578200. [DOI] [PubMed] [Google Scholar]
- 10.Rangarajan A., Hong S. J., Gifford A., Weinberg R. A. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Land H., Parada L. F., Weinberg R. A. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 12.Wisdom R., Verma I. M. Mol. Cell. Biol. 1993;13:2635–2643. doi: 10.1128/mcb.13.5.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergers G., Graninger P., Braselmann S., Wrighton C., Busslinger M. Mol. Cell. Biol. 1995;15:3748–3758. doi: 10.1128/mcb.15.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson R., Spiegelman B., Hanahan D., Wisdom R. Mol. Cell. Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki T., Murakami M., Onai N., Fukuda E., Hashimoto Y., Sonobe M. H., Kameda T., Ichinose M., Miki K., Iba H. J. Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfarr C. M., Mechta F., Spyrou G., Lallemand D., Carillo S., Yaniv M. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 17.Saez E., Rutberg S. E., Mueller E., Oppenheim H., Smoluk J., Yuspa S. H., Spiegelman B. M. Cell. 1995;82:721–732. doi: 10.1016/0092-8674(95)90469-7. [DOI] [PubMed] [Google Scholar]
- 18.Vial E., Marshall C. J. J. Cell Sci. 2003;116:4957–4963. doi: 10.1242/jcs.00812. [DOI] [PubMed] [Google Scholar]
- 19.Chiappetta G., Tallini G., De Biasio M. C., Pentimalli F., de Nigris F., Losito S., Fedele M., Battista S., Verde P., Santoro M., Fusco A. Clin. Cancer Res. 2000;6:4300–4306. [PubMed] [Google Scholar]
- 20.Ramos-Nino M. E., Timblin C. R., Mossman B. T. Cancer Res. 2002;62:6065–6069. [PubMed] [Google Scholar]
- 21.Belguise K., Kersual N., Galtier F., Chalbos D. Oncogene. 2005;24:1434–1444. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
- 22.Debinski W., Gibo D. M. Mol. Cancer Res. 2005;3:237–249. doi: 10.1158/1541-7786.MCR-05-0004. [DOI] [PubMed] [Google Scholar]
- 23.Pollock C. B., Shirasawa S., Sasazuki T., Kolch W., Dhillon A. S. Cancer Res. 2005;65:1244–1250. doi: 10.1158/0008-5472.CAN-04-1911. [DOI] [PubMed] [Google Scholar]
- 24.Vial E., Sahai E., Marshall C. J. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 25.Treinies I., Paterson H. F., Hooper S., Wilson R., Marshall C. J. Mol. Cell. Biol. 1999;19:321–329. doi: 10.1128/mcb.19.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith E. R., Smedberg J. L., Rula M. E., Hamilton T. C., Xu X. X. J. Biol. Chem. 2001;276:32094–32100. doi: 10.1074/jbc.M105009200. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Zhang B., Wang M., Carr B. I. J. Biol. Chem. 2003;278:11138–11144. doi: 10.1074/jbc.M209108200. [DOI] [PubMed] [Google Scholar]
- 28.Kim H. S., Song M. C., Kwak I. H., Park T. J., Lim I. K. J. Biol. Chem. 2003;278:37497–37510. doi: 10.1074/jbc.M211739200. [DOI] [PubMed] [Google Scholar]
- 29.Purcell N. H., Darwis D., Bueno O. F., Muller J. M., Schule R., Molkentin J. D. Mol. Cell. Biol. 2004;24:1081–1095. doi: 10.1128/MCB.24.3.1081-1095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Formstecher E., Ramos J. W., Fauquet M., Calderwood D. A., Hsieh J. C., Canton B., Nguyen X. T., Barnier J. V., Camonis J., Ginsberg M. H., Chneiweiss H. Dev. Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 31.Torii S., Kusakabe M., Yamamoto T., Maekawa M., Nishida E. Dev. Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 32.Fukuda M., Gotoh I., Adachi M., Gotoh Y., Nishida E. J. Biol. Chem. 1997;272:32642–32648. doi: 10.1074/jbc.272.51.32642. [DOI] [PubMed] [Google Scholar]
- 33.Neuberg M., Adamkiewicz J., Hunter J. B., Muller R. Nature. 1989;341:243–245. doi: 10.1038/341243a0. [DOI] [PubMed] [Google Scholar]
- 34.Busch S. J., Sassone-Corsi P. Trends Genet. 1990;6:36–40. doi: 10.1016/0168-9525(90)90071-d. [DOI] [PubMed] [Google Scholar]
- 35.Akagi T., Murata K., Shishido T., Hanafusa H. Mol. Cell. Biol. 2002;22:7015–7023. doi: 10.1128/MCB.22.20.7015-7023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tani K., Sato S., Sukezane T., Kojima H., Hirose H., Hanafusa H., Shishido T. J. Biol. Chem. 2003;278:21685–21692. doi: 10.1074/jbc.M301447200. [DOI] [PubMed] [Google Scholar]
- 37.Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T., et al. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.