Abstract
The host restriction factor TRIM5α mediates species-specific, early blocks to retrovirus infection; susceptibility to these blocks is determined by viral capsid sequences. Here we demonstrate that TRIM5α variants from Old World monkeys specifically associate with the HIV type 1 (HIV-1) capsid and that this interaction depends on the TRIM5α B30.2 domain. Human and New World monkey TRIM5α proteins associated less efficiently with the HIV-1 capsid, accounting for the lack of restriction in cells of these species. After infection, the expression of a restricting TRIM5α in the target cells correlated with a decrease in the amount of particulate capsid in the cytosol. In some cases, this loss of particulate capsid was accompanied by a detectable increase in soluble capsid protein. Inhibiting the proteasome did not abrogate restriction. Thus, TRIM5α restricts retroviral infection by specifically recognizing the capsid and promoting its rapid, premature disassembly.
Keywords: B30.2(SPRY) domain; tripartite motif; HIV-1; RING, B-box, and coiled-coil protein; innate immunity
Primate cells express dominant factors that can determine the species-specific tropism of a particular retrovirus (1–3). TRIM5α mediates the early block to HIV (HIV type 1, HIV-1) infection in Old World monkey cells (4). TRIM5α is a tripartite motif protein, with RING, B-box, and coiled-coil (RBCC) domains (5). Only the longest TRIM5 isoform, TRIM5α, possesses a C-terminal B30.2 domain and exhibits antiretroviral activity (4, 6–9). Variation in TRIM5α protein sequences among primate species accounts for the observed patterns of species-specific restrictions to retrovirus infection (10, 11). In some cases, variation in the B30.2 domain has been implicated in the virus-specific restricting activity of TRIM5α proteins from different species (9, 12, 13).
TRIM5α prevents retrovirus infection by an unknown mechanism. Reverse transcripts of the restricted virus fail to accumulate in the blocked cells, indicating that an early, postentry event is disrupted by TRIM5α (4). Several lines of evidence hint that TRIM5α associates directly or indirectly with the restricted retroviral capsid. First, the viral determinants of susceptibility to restriction map to the capsid protein (14–17). Second, when introduced into cells, assembled and proteolytically processed capsids of virus-like particles, but not monomeric capsid proteins expressed in the cells, can compete for the restriction factor(s) (14, 16, 18, 19). Third, TRIMCyp is an HIV-1-restricting factor in owl monkeys consisting of the RBCC domains of TRIM5 fused with cyclophilin A, a known capsid ligand (14, 18 –23). Finally, an association of TRIM5αhu with capsids prepared by detergent treatment of a restricted murine leukemia virus (MLV) has been observed (24).
The events in retrovirus infection that occur between entry into the host cell and reverse transcription are poorly understood. The uncoating of the HIV-1 capsid is thought to precede reverse transcription, whereas MLV capsid proteins remain associated with the reverse transcription and preintegration complexes (25–28). The study of HIV-1 capsid mutants suggests that capsid uncoating may be a temporally regulated process, with either too rapid or too slow disassembly compromising virus infectivity (29). Host cell factors involved in uncoating are unknown (21, 30, 31). Given this sparse state of knowledge, TRIM5α-mediated restriction could conceivably involve prevention of capsid uncoating, acceleration of capsid uncoating, or destruction of the capsid.
Here we investigate the ability of TRIM5α to interact in a specific manner with the HIV-1 capsid. We establish a previously undescribed assay to follow the fate of the retroviral capsid in the cytosol of newly infected cells and examine the effect of expression of a restricting TRIM5α protein on the particulate and soluble forms of the capsid.
Results
B30.2 Domain-Dependent Association of TRIM5αrh with HIV-1 Capsids.
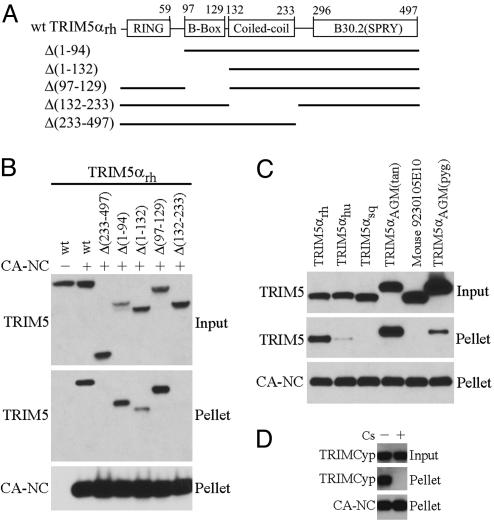
To investigate whether rhesus monkey TRIM5αrh associates with HIV-1 capsids, we used HIV-1 capsid complexes assembled from purified recombinant capsid–nucleocapsid (CA–NC) protein (32–35) to detect the binding of either WT or mutant TRIM5α proteins (Fig. 1A) from cell lysates. We verified by electron microscopy that our CA–NC preparations formed cylindrical and conical structures, which have been shown to recapitulate the authentic surface lattice structure of native HIV-1 capsids (33, 35, 36). After incubation of the HIV-1 CA–NC complexes with TRIM5αrh-containing cell lysates, the mixtures were layered onto 70% sucrose cushions. The WT TRIM5αrh protein did not sediment through 70% sucrose in the absence of added CA–NC complexes (Fig. 1B). After incubation with HIV-1 CA–NC complexes, the WT TRIM5αrh protein was detected in the pellet. By contrast, the Δ(233–497) TRIM5αrh mutant, which retains only the RBCC domains of TRIM5αrh, did not cosediment with the CA–NC complexes (Fig. 1B). Apparently, HIV-1 capsid association depends on carboxyl-terminal TRIM5α sequences, including the B30.2 domain.
Fig. 1.
Specific association of TRIM5α variants with HIV-1-CA–NC complexes. (A) The diagram shows the domain structure of TRIM5αrh. The sequences included in each of the mutant proteins are indicated by the solid lines. All of the proteins contain a C-terminal HA epitope tag. (B) In vitro-assembled CA–NC complexes were mixed with 293T lysates containing WT TRIM5αrh-HA or the indicated TRIM5αrh-HA deletion mutants and layered onto 70% sucrose before centrifugation. Immediately before mixing, an aliquot of the cell lysate was removed and blotted with α-HA antibodies to determine the steady-state expression levels of the different TRIM5 variants (input). After centrifugation, the pellet was resuspended in SDS sample buffer and analyzed by Western blotting with an anti-HA antibody (to detect TRIM5) or an anti-p24 antibody (to detect CA-NC). (C) An experiment similar to that described in B was carried out, except that the 293T cell lysates contained the indicated TRIM proteins. (D) An experiment similar to that described in B was carried out, except that the 293T cell lysates contained TRIMCyp-HA and were mixed with CA–NC complexes in the presence of 0.1% DMSO (−) or 25 μM cyclosporine (Cs) (+).
Contribution of RBCC Domains to TRIM5αrh-Capsid Association.
The ability of TRIM5αrh variants lacking one or more of the RBCC domains (Fig. 1A) to interact with the HIV-1 CA–NC complexes was examined. The Δ(1–94) and Δ(97–129) TRIM5αrh mutants, which lack the RING and B-box 2 domains, respectively, associated efficiently with the CA–NC complexes (Fig. 1B). The Δ(1–132) mutant lacking both RING and B-box 2 domains also retained the ability to cosediment with the CA–NC complexes. By contrast, the Δ(132–233) TRIM5αrh mutant, which lacks the coiled-coil domain, did not cosediment with HIV-1 CA–NC complexes (Fig. 1B). Thus, the coiled-coil and B30.2 domains, but not the RING or B-box 2 domains, contribute to TRIM5αrh association with the HIV-1 capsid.
Association of TRIM5-Related Proteins from Different Species with HIV-1 Capsids.
We examined the ability of TRIMCyp and primate TRIM5α variants to cosediment with HIV-1 CA–NC complexes. None of the TRIM5-related proteins sedimented through 70% sucrose in the absence of CA–NC complexes (data not shown). TRIM5α proteins from two species of Old World monkeys, rhesus macaques and African green monkeys, strongly bound the HIV-1 CA–NC complexes (Fig. 1C). Human TRIM5α (TRIM5αhu), which only moderately restricts HIV-1 (4), only weakly associated with the HIV-1 CA–NC complexes. TRIM5αsq from squirrel monkeys, a New World monkey species, does not restrict HIV-1 (11) and demonstrated no detectable binding to the HIV-1 CA–NC complexes. Mice lack a TRIM5 ortholog (10); the closest mouse relative to TRIM5, 9230105E10Rik, which does not inhibit HIV-1 infection (11), did not cosediment with the HIV-1 CA–NC complexes. The TRIMCyp protein associated with the HIV-1 CA–NC complexes (Fig. 1D). This interaction was inhibited by cyclosporine A, which allows HIV-1 to escape restriction by TRIMCyp (21, 22, 37, 38). Thus, there is a good correlation between the ability of TRIM5 relatives from different species to restrict HIV-1 infection and the observed association with HIV-1 capsids.
An Assay to Measure Particulate and Soluble Capsid Proteins in the Cytosol of Infected Cells.
To gain insight into the mechanism of TRIM5α restriction, we examined the effect of TRIM5α binding on the fate of the retroviral capsid in infected cells. The measurement of total intracellular capsid protein cannot reliably monitor infectious capsids because of the very large fraction of defective retroviruses that bind target cells and become trapped nonproductively in intracellular compartments such as endosomes and lysosomes (39). We designed a sedimentation assay that (i) distinguishes nonspecifically endocytosed capsids from the cytosolic capsids that are potentially on the infection pathway and (ii) separates particulate capsid proteins, presumably retaining a higher-order structure, from soluble capsid proteins. The first aim was achieved by taking advantage of the difference in density between endosomes/lysosomes and retroviral capsids (29, 40–43). The second aim was achieved by velocity sedimentation, separating the particulate capsids with high sedimentation rates from the soluble proteins with much lower sedimentation rates. The assay is summarized in Fig. 2A. Fig. 2B shows the results of a pilot experiment to determine a sucrose concentration that allows detection of HIV-1 capsids only when functional vesicular stomatitis virus (VSV)-G envelope glycoproteins are present on the infecting virions. The control HIV-1 virions without envelope glycoproteins [HIV-1(Env−)] can nonspecifically bind target cells and be endocytosed, but these capsids cannot enter the cytosol (39). VSV-G-pseudotyped HIV-1 [HIV-1(VSV-G)] and HIV-1(Env−) virions were allowed to attach to cells at 4°C; after shifting the temperature to 37°C to allow virus entry, cells were lysed in a detergent-free buffer. After low-speed centrifugation to pellet nuclei and cell debris, the cytosolic extracts were layered onto cushions of different sucrose concentrations. A 50% sucrose cushion was optimal for discriminating between intracellular capsids associated with entry-competent viruses and those nonspecifically associated with cells (Fig. 2B). By contrast, a cushion of 45% sucrose did not allow discrimination between HIV-1(VSV-G) and HIV-1(Env−) virions, and a cushion of 55% sucrose did not allow detection of any particulate capsid, even in the lysates of cells exposed to entry-competent viruses. These results suggest that the intracellular particulate capsids detected in our assay exhibit densities between those of 45% sucrose (1.20 g/ml) and 55% sucrose (1.26 g/ml), a result that is in good agreement with previous estimates of the density of cores (1.24–1.26 g/ml) prepared by detergent treatment of HIV-1 virions (29, 41, 43). These results are also consistent with the expectation that much of the capsid protein nonspecifically taken into cells is trapped in endosomes or lysosomes, which have reported densities of <1.20 g/ml (40, 42). Indeed, a visible layer that formed at the boundary of the cell lysate and 50% sucrose cushion during centrifugation contained large amounts of p24 capsid protein after incubation of cells with either HIV-1(VSV-G) or HIV-1(Env−) virions (data not shown). Because this material forms a discrete layer at the lysate-cushion interface, we could collect a sample from the top of the gradient for the purpose of measuring the soluble capsid protein in the cell lysate. The HIV-1 capsid protein was detectable in this supernatant sample only when entry-competent virions were used for infection (see below), suggesting that these soluble proteins were derived from viral components that had entered the cytosol. Thus, this assay allows us to measure both particulate and soluble forms of the HIV-1 capsid protein in the cytosol of cells exposed to infectious viruses.
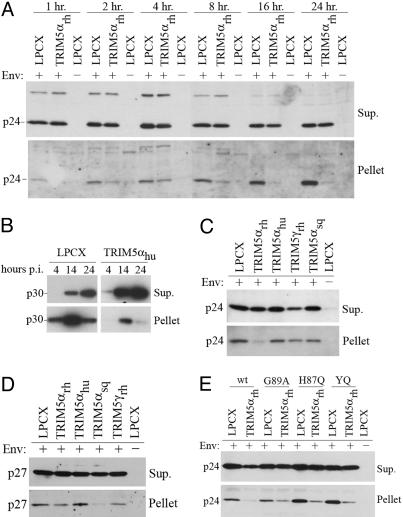
Fig. 2.
Assay to follow the fate of the retroviral capsid in infected cells. (A) Lysates from HeLa cells incubated with HIV-1(VSV-G) or HIV-1(Env−) viruses were cleared and analyzed on sucrose gradients, as described in Materials and Methods. A 100-μl sample (the “supernatant”) was collected from the very top of the gradient, avoiding the layer of endosomes/lysosomes at the interface of lysate and cushion. The pellet was also collected and resuspended directly in SDS sample buffer. (B) HeLa cells were incubated with HIV-1(VSV-G) or HIV-1(Env−) viruses at 4°C for 30 min and then at 37°C for 4 h. Cell lysates were prepared and analyzed on gradients consisting of the indicated concentrations of sucrose. The pellet was resuspended in sample buffer and Western blotted by using an anti-p24 antibody.
Decreases in the Amount of Particulate Capsids in Cells Expressing a Restricting TRIM5α Protein.
The assay described above was used to examine the fate of the HIV-1 capsid after infection of HeLa cells expressing the restricting TRIM5αrh protein and control cells transduced with the empty LPCX vector. As an additional control, the LPCX vector-transduced cells were exposed to HIV-1(Env−) virions. The virions were incubated with the target cells at 4°C to permit virus attachment, and the temperature was shifted to 37°C to allow the infection process to proceed. At different time points, the cells were lysed and the cell lysates analyzed by sedimentation on 50% sucrose cushions, as described above. The amount of HIV-1 p24 capsid protein in the interface between cell lysate and sucrose cushion was similar in the TRIM5αrh-expressing and control cells, regardless of the presence of envelope glycoproteins on the HIV-1 virions (data not shown); this result confirms that similar amounts of virus were incubated with the target cells, and that the interfacial layer contains virions that are nonspecifically endocytosed into the cells. As expected, no p24 capsid protein was detected in either the supernatants or pellets derived from cells incubated with the HIV-1(Env−) virions (Fig. 3A). The steady-state levels of p24 capsid protein in the supernatants from TRIM5αrh-expressing and control cells incubated with entry-competent viruses were comparable at all time points examined. By contrast, at all time points, the amounts of pelletable capsid in the cells expressing TRIM5αrh were lower than those detected in the control LPCX-transduced cells. This decrease in particulate capsid in TRIM5αrh-expressing cells was evident as early as 1 h after the 37°C temperature shift. The amount of HIV-1 p24 capsid protein in the pellets derived from the control LPCX cells increased from the time of the temperature shift until 24 h later, after which the levels decreased (Fig. 3A and data not shown). Because the infections are not synchronized and our assay measures total steady-state levels of cytosolic capsid proteins, these observations suggest that virus entry-dependent processes continue to introduce new capsids into the cytosol for several hours after the shift to 37°C. We conclude that the presence of TRIM5αrh in target cells results in a decrease in the particulate HIV-1 capsid, but not in the soluble capsid protein, in the cytosol.
Fig. 3.
Effect of TRIM5α expression on the fate of the retroviral capsid in infected cells. (A) HeLa cells expressing an empty control vector (LPCX) or TRIM5αrh-HA were incubated with HIV-1(VSV-G) or HIV-1(Env−) at 4°C for 30 min, and then at 37°C for the indicated times. Cell lysates were analyzed as described in Fig. 2A. Supernatants and pellets were Western blotted with an anti-p24 antibody. (B) NIH3T3 cells expressing an empty control vector (LPCX) or TRIM5αhu-HA were incubated with N-MLV at 4°C for 30 min, and then at 37°C for the indicated times. Cell lysates were analyzed as described in Fig. 2A. Supernatants and pellets were Western blotted with an antibody directed against the p30 capsid protein. (C) HeLa cells expressing an empty control vector (LPCX) or the indicated TRIM5α proteins were incubated with HIV-1(VSV-G) or HIV-1(Env−) vectors at 37°C for 4 h, washed, and allowed to incubate for an additional 4 h at 37°C. Cell lysates were analyzed as in Fig. 2A. The supernatant and pellet were probed with anti-p24 antibodies. (D) HeLa cells expressing the indicated TRIM5 variants were incubated with SIVmac(VSV-G) and SIVmac(Env−) vectors and analyzed as described in C. The supernatant and pellet were probed with an antibody directed against the SIV p27 capsid protein. (E) HeLa cells expressing an empty control vector (LPCX) or TRIM5αrh-HA were incubated as described in C with either WT HIV-1(VSV-G), WT HIV-1(Env−), or HIV-1(VSV-G) viruses with the G89A, H87Q, and Q50Y/T54Q (YQ) capsid proteins. Cell lysates were analyzed as in Fig. 2A, and the pellet and supernatant were probed with an anti-p24 antibody.
To investigate the generality of the above observations, we studied the fate of N-tropic MLV (N-MLV) capsids in mouse NIH 3T3 cells that express TRIM5αhu, which can potently block N-MLV infection (6–8, 44), and in control cells transduced with the empty LPCX vector. N-MLV capsids were recovered more efficiently after sucrose gradient centrifugation than HIV-1 capsids, allowing us to examine the effects of target cell expression of TRIM5αhu on the conversion of particulate capsids to soluble capsid proteins. At each time point examined, the particulate N-MLV p30 capsid protein in the pellet derived from the TRIM5αhu-expressing cells was significantly less than that from the control LPCX-transduced cells (Fig. 3B). The decrease in particulate N-MLV capsid protein in the TRIM5αhu-expressing cells coincided with a concomitant increase in soluble p30 capsid protein in the supernatant. Thus, the total amount of N-MLV capsid protein does not significantly differ in the cytosol of the TRIM5αhu-expressing and control LPCX cells. Rather, the conversion of the capsid protein from a particulate form to a soluble form is accelerated in the TRIM5αhu-expressing cells.
Of note, in both TRIM5αhu-expressing and control NIH 3T3 cells, the migration on SDS-polyacrylamide gels of the N-MLV p30 capsid proteins derived from the pellets and supernatants was indistinguishable (data not shown). This observation argues against a major covalent modification (for example, ubiquitylation, sumoylation, or proteolytic cleavage) of the majority of the N-MLV capsid, at steady state, during the course of normal uncoating or as a result of TRIM5αhu activity.
Correlation Between Decreases in the Level of Particulate Capsids and Restriction of Infection.
We examined the fate of the HIV-1 capsid in HeLa cells expressing TRIM5 variants from different primates. HeLa cells expressing comparable levels of TRIM5αrh, TRIM5αhu, TRIM5γrh, and TRIM5αsq, as well as control cells transduced with the LPCX vector, were incubated with HIV-1(VSV-G) and HIV-1(Env−) virions. Cell lysates were prepared at 8 h after the 37°C temperature shift and analyzed on 50% sucrose gradients, as described above. In cells expressing TRIM5αrh, we observed a decrease in the amount of HIV-1 p24 capsid protein in the pellet relative to the amount of capsid protein seen in the LPCX cells (Fig. 3C). No significant differences in the level of soluble capsid protein were observed in the supernatants of TRIM5αrh-expressing and LPCX cells. By contrast, the expression of TRIM5αhu, TRIM5γrh, and TRIM5αsq, which do not potently restrict HIV-1 infection (4, 11), did not result in a reproducible decrease in the levels of particulate capsid relative to those observed in the control cells. Thus, there is a correlation between the ability of TRIM5 variants to restrict HIV-1 infection and the conversion of the particulate HIV-1 capsid to nonpelletable forms.
The same set of cells expressing the above-described TRIM5 variants was incubated with VSV G-pseudotyped or Env− SIVmac virions. A dramatic loss of particulate SIV p27 capsid protein was observed in the cytosol of cells expressing TRIM5αsq, which strongly restricts SIVmac infection (11), relative to the p27 levels in the pellets of the other cells (Fig. 3D). The p27 capsid proteins in the pellets derived from cells expressing TRIM5αrh, TRIM5αhu, and TRIM5γrh, which do not potently block SIVmac infection (11), were readily detectable. No differences in the levels of soluble SIVmac p27 capsid proteins were observed in supernatants derived from the cells expressing the different TRIM5 variants. As was seen above for HIV-1, a correlation was observed between the TRIM5-mediated restriction of SIVmac infection and the loss of particulate forms of the capsid protein in the cytosol of cells exposed to the virus.
The Fate of Mutant HIV-1 Capsids that Escape TRIM5α Restriction.
Some changes in the HIV-1 capsid have been shown to allow HIV-1 to escape partially from the restrictions present in the cells of various Old World monkeys (15, 37, 38, 45). Partial escape from TRIM5αrh-mediated restriction is also associated with the inability of the HIV-1 capsid to bind cyclophilin A (37, 38, 45, 46). We examined the fate of cytosolic capsids in TRIM5αrh-expressing cells 8 h after incubation with VSV G-pseudotyped HIV-1 containing capsid changes associated with partial escape from restriction: (i) G89A, which decreases the binding of cyclophilin A to the capsid (47), (ii) H87Q, altering the cyclophilin A-binding loop (37), and (iii) the YQ alteration affecting helix 3 (37). In the TRIM5αrh-expressing cells, the amounts of particulate capsid in the cytosol were greater for all three mutants than for the WT virus (Fig. 3E). In the LPCX control cells, the pelletable capsid levels were higher for the H87Q and YQ mutants than for the WT virus. We conclude that HIV-1 capsid changes that allow partial escape from the effects of Old World monkey TRIM5α proteins confer greater resistance to TRIM5α-mediated conversion of particulate capsids to nonpelletable forms.
Independence of TRIM5αrh Restriction of HIV-1 and Proteasomal Function.
Because some RING domain-containing proteins are involved in ubiquitylation and proteasomal degradation (48–52), we tested the importance of the proteasome to TRIM5αrh-mediated restriction of HIV-1. HeLa cells were incubated with VSV G-pseudotyped HIV-1-GFP, which expresses GFP upon successful infection of cells (53), in the absence or presence of a proteasome inhibitor, clasto-Lactacystin β-lactone. As reported in refs. 54 and 55, a small but reproducible increase in the efficiency of HIV-1 infection in the presence of the proteasomal inhibitor was observed (Fig. 4, which is published as supporting information on the PNAS web site). However, proteasome inhibition did not significantly alter the block to HIV-1 infection mediated by TRIM5αrh.
Discussion
Old World monkey TRIM5α proteins specifically bound HIV-1 capsid-like structures. All of the TRIM5 variants tested that block HIV-1 infection associated with the HIV-1 CA–NC complexes, supporting the importance of capsid binding for restriction. Human TRIM5α bound the HIV-1 capsid-like complexes more weakly than the Old World monkey TRIM5α proteins, explaining the lower potency of human TRIM5α in blocking HIV-1 infection (4, 9, 13). The TRIM5α protein from the squirrel monkey, a New World monkey species, did not associate with the HIV-1 capsid complexes, consistent with the inability of most New World monkey TRIM5α molecules to block HIV-1 infection (11). One exceptional New World monkey, the owl monkey, encodes a TRIM5-cyclophilin A fusion protein called TRIMCyp (21, 22). We show that TRIMCyp binds the HIV-1 capsids in a manner inhibitable by cyclosporine, which is known to disrupt cyclophilin A–HIV-1 capsid interactions and relieve the block to HIV-1 infection in owl monkey cells (23, 38, 56). Like TRIMCyp, TRIM5α may use its carboxyl terminus for interaction with the targeted capsid, because the association of TRIM5αrh with the HIV-1 CA–NC complexes depended on the B30.2 domain. The interaction of the human TRIM5α protein with the N-MLV capsid has also been reported to require an intact B30.2 domain (24). Determinants of differences in anti-HIV-1 potency between species-specific variants of TRIM5α map to the B30.2 domain (9, 13, 57), raising the possibility that these affect capsid recognition. Significant interspecies variation in the TRIM5α B30.2 domain and the evidence for strong positive selection acting on this domain (10, 57) support a direct interaction of TRIM5α with potentially rapidly evolving viral capsids.
The TRIM5αrh coiled coil was also necessary for the interaction with the HIV-1 CA–NC complexes. The TRIM5α coiled coil mediates trimerization, which may allow TRIM5α to interact with sites on the surface lattice of the retroviral capsid that exhibit trimeric pseudosymmetry (28). Significant gains in avidity would accrue to the interactions of two oligomeric complexes with compatible symmetry.
Our results suggest that the ability of a TRIM5α protein to bind the HIV-1 capsid is not sufficient to achieve restriction of viral infection. TRIM5αrh mutants lacking the RING and/or B-box 2 domains associated with the HIV-1 CA–NC complexes but were partially or completely defective in establishing a block to HIV-1 infection (12, 58). Thus, these amino-terminal TRIM5α elements possibly serve as effector domains for restriction, contributing to higher-order interactions with other TRIM5α molecules or with necessary cofactors.
We devised an assay to follow the steady-state levels of retroviral capsids in the cytosol of infected cells. We took advantage of the low density of endosomes and lyosomes to eliminate the large quantities of nonspecifically endocytosed retroviral particles (39) from consideration in this assay. The small fraction of cytosolic capsid proteins was further separated based on sedimentation velocity into particulate capsids and soluble capsid proteins. The observed total steady-state levels of capsid proteins reflect both the introduction of the capsids into the cytosol by the viral entry process and the turnover of the capsid proteins. The total cytosolic level of capsid protein was not altered by the expression of a restricting TRIM5α, indicating that restriction involves neither a blockade of the introduction of the capsid into the cytosol nor the promotion of capsid degradation. Consistent with the latter point, proteasome inhibition did not significantly affect the restriction of HIV-1 infection by TRIM5αrh. Moreover, previous studies (4, 12, 58) demonstrated that the TRIM5αrh RING domain contributes to the potency of, but is not absolutely required for, HIV-1 restriction. Finally, we did not observe any indication of a TRIM5α-associated modification (e.g., ubiquitylation) of the restricted retroviral capsids in the cytosol. Together, these observations argue against a model of bulk capsid degradation after recognition by the TRIM5α protein.
Expression of a restricting TRIM5α protein resulted in a loss of the pelletable capsid in the cytosol of infected cells. This diminution in the amount of particulate capsid strongly correlated with the ability of TRIM5α variants from different species to restrict HIV-1 and SIV infection and with the susceptibility of HIV-1 capsid mutants to restriction. Because the entry of viruses in our system is not synchronized, we cannot, as yet, use this assay to estimate the absolute rates of uncoating or capsid turnover in infected cells. However, differences in the level of particulate capsid in the cytosol of TRIM5α-expressing and control cells were apparent as early as 1 h after the initiation of virus infection. The recovery of N-MLV capsids from the 50% sucrose gradients was more efficient than that of HIV-1 capsids, allowing us to observe directly the conversion of particulate N-MLV capsids into soluble capsid proteins. The expression of the N-MLV-restricting protein, TRIM5αhu, in the target cells accelerated this conversion. The decreased stability of HIV-1 capsids in concentrated sucrose solutions (29, 43) lowers the recovery of capsids in our pellet, leading to an underestimation of the intact cytosolic capsid and an overestimation of the disassembled HIV-1 capsid proteins in our system. Therefore, although the loss of particulate, cytosolic HIV-1 capsid as a consequence of TRIM5α expression is clearly evident in our assay, the conversion of this capsid protein to soluble forms makes an undetectably small contribution to the total capsid protein in the supernatant.
Consistent with the model that TRIM5α mediates restriction by accelerating the uncoating of the viral capsid are our observations that, compared with the WT HIV-1, HIV-1 capsid mutants partially resistant to the Old World monkey restriction exhibited higher levels of particulate capsid in the cytosol of TRIM5α-expressing cells. One of the HIV-1 capsid mutants, G89A, does not bind cyclophilin A, which has been shown to augment TRIM5αrh restriction of HIV-1 by an unknown mechanism (37, 38, 46). Two other HIV-1 capsid mutants, H87Q and YQ, exhibited levels of particulate capsids higher than those of the WT HIV-1 virus not only in TRIM5αrh-expressing cells, but also in the control HeLa cells. These mutant capsids may be intrinsically more stable, or they may be less susceptible to the modest anti-HIV-1 activity of the endogenous human TRIM5α protein in the HeLa cells (4). Indeed, these mutants exhibit better infectivity than the WT virus in HeLa cells as well as Old World monkey cells (37). The mechanisms whereby the H87Q and YQ changes exert their phenotypes are unknown. The H87Q change has been shown to result in a 4- to 5-fold decrease in the affinity of cyclophilin A binding (59). The changes in the YQ mutant may directly influence capsid stability, because the corresponding residues in the MLV capsid (60) form important hydrogen bonds across adjacent hexamers. Thus, H87Q and YQ may achieve decreased sensitivity to TRIM5α action by different mechanisms (37).
Additional research is required to determine how TRIM5α promotes rapid uncoating of the restricted virus capsid and why accelerated disassembly of the capsid is detrimental to infection. Changes in the HIV-1 capsid that either increase or decrease capsid stability decrease the efficiency of early events in infection (29). Accelerated disassembly of the retroviral capsid may prematurely expose the viral RNA or viral enzymes to degradative processes or disrupt capsid associations with the remainder of the retroviral core that are directly or indirectly critical for reverse transcription. Understanding the TRIM5α antiviral mechanism may help to elucidate the process of retroviral uncoating.
Materials and Methods
Cells and Viruses.
HeLa cells expressing HA epitope-tagged TRIM5α variants (4, 8, 11) and recombinant viruses are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Capsid-Binding Assay.
An assay measuring the cosedimentation of TRIM5 variants with assembled HIV-1 capsid complexes is described in Supporting Materials and Methods.
Fate-of-Capsid Assay.
HeLa (6 × 106) or NIH 3T3 (1.5 × 106) cells stably expressing different TRIM5 variants were seeded in 80-cm2 flasks, and the next day, incubated with recombinant viruses prepared as described in Supporting Materials and Methods. Cells were incubated with either 10 ml HIV-1(VSV-G) [≈5 × 106 reverse transcriptase (RT) counts] (53), 10 ml SIVmac (VSV-G) (≈106 RT counts), or 10 ml N-MLV(VSV-G) (≈5 × 105 RT counts) for 30 min at 4°C. Then, the cells were shifted to 37°C until they were harvested at various time points. If the cells were going to be harvested >4 h after infection, the virus suspension was removed at the 4-h time point and replaced with fresh medium. The cells were washed three times with ice-cold PBS and detached by incubating with 1 ml of pronase (7 mg/ml in DMEM) for 5 min at 4°C. The cells were washed once in DMEM containing 10% FBS and two times in PBS. The pellet was resuspended in 2.5 ml hypotonic lysis buffer (10 mM Tris·HCl, pH 8.0/10 mM KCl/1 mM EDTA) and placed on ice for 15 min. The cells were lysed by using a 7-ml Dounce homogenizer and 15 strokes with pestle B. Cell debris was removed by centrifugation for 3 min at 2,000 × g. After centrifugation, 2 ml of lysate was layered onto a 7-ml 50% sucrose cushion (made in PBS) and centrifuged at 125,000 × g for 2 h at 4°C in a Beckman SW41 rotor. After centrifugation, 100 μl from the top-most part of the supernatant was collected and made 1× in SDS sample buffer. The pellet was resuspended in 100 μl of 1× SDS sample buffer. The samples were subjected to SDS/PAGE and Western blotting for capsid proteins.
Immunoblotting.
HA-tagged TRIM5 variants and capsid proteins were detected as described in Supporting Materials and Methods.
Proteasome Inhibition and HIV-1 Infection.
See Supporting Materials and Methods for more information.
Supplementary Material
Acknowledgments
We thank Ms. Yvette McLaughlin and Sheri Farnum for manuscript preparation. This work was supported by National Institutes of Health Grants AI063987, HL54785, and AI45405 and Center for AIDS Research Award AI28691, the International AIDS Vaccine Initiative, the Bristol-Myers Squibb Foundation, the William A. Haseltine Foundation for the Arts and Sciences, and the late William F. McCarty-Cooper. M.S. was supported by a National Defense Science and Engineering Fellowship and is a Fellow of the Ryan Foundation.
Abbreviations
- CA–NC
capsid–nucleocapsid
- HIV-1(Env−)
control HIV-1 virions without envelope glycoproteins
- HIV-1(VSV-G)
vesicular stomatitis virus-G-pseudotyped HIV-1
- MLV
murine leukemia virus
- N-MLV
N-tropic MLV
- RBCC
RING, B-box and coiled-coil
- VSV
vesicular stomatitis virus.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
See Commentary on page 5249.
References
- 1.Bieniasz P. D. Trends Microbiol. 2003;11:286–291. doi: 10.1016/s0966-842x(03)00123-9. [DOI] [PubMed] [Google Scholar]
- 2.Goff S. P. Mol. Cell. 2004;16:849–859. doi: 10.1016/j.molcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Stoye J. P. Proc. Natl. Acad. Sci. USA. 2002;99:11549–11551. doi: 10.1073/pnas.192449399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 5.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., Ballabio A. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2004;101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keckesova Z., Ylinen L. M. J., Towers G. J. Proc. Natl. Acad. Sci. USA. 2004;101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perron M. J., Stremlau M., Song B., Ulm W., Mulligan R. C., Sodroski J. Proc. Natl. Acad. Sci. USA. 2004;101:11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap M. W., Nisole S., Stoye J. P. Curr. Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 10.Song B., Gold B., O’Huigin C., Javanbakht H., Li X., Stremlau M., Winkler C., Dean M., Sodroski J. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song B., Javanbakht H., Perron M., Park D. H., Stremlau M., Sodroski J. J. Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Caballero D., Hatziioannou T., Yang A., Cowan S., Bieniasz P. D. J. Virol. 2005;79:8969–8978. doi: 10.1128/JVI.79.14.8969-8978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stremlau M., Perron M., Welikala S., Sodroski J. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan S., Hatziioannou T., Cunningham T., Muesing M. A., Gottlinger H. G., Bieniasz P. D. Proc. Natl. Acad. Sci. USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kootstra N. A., Munk C., Tonnu N., Landau N. R., Verma I. M. Proc. Natl. Acad. Sci. USA. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munk C., Brandt S. M., Lucero G., Landau N. R. Proc. Natl. Acad. Sci. USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens C. M., Yang P. C., Gottlinger H., Sodroski J. J. Virol. 2003;77:726–731. doi: 10.1128/JVI.77.1.726-731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besnier C., Takeuchi Y., Towers G. Proc. Natl. Acad. Sci. USA. 2002;99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodding M. P., Bock M., Yap M. W., Stoye J. P. J. Virol. 2005;79:10571–10577. doi: 10.1128/JVI.79.16.10571-10577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luban J., Bossolt K. L., Franke E. K., Kalpana G. V., Goff S. P. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 21.Nisole S., Lynch C., Stoye J. P., Yap M. W. Proc. Natl. Acad. Sci. USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayah D. M., Sokolskaja E., Berthoux L., Luban J. Nature. 2004;430:569–573. [Google Scholar]
- 23.Thali M., Bukovsky A., Kondo E., Rosenwirth B., Walsh C. T., Sodroski J., Gottlinger H. G. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 24.Sebastian S., Luban J. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fassati A., Goff S. P. J. Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fassati A., Goff S. P. J. Virol. 1999;73:8919–8925. doi: 10.1128/jvi.73.11.8919-8925.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farnet C. M., Haseltine W. A. J. Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller M. D., Farnet C. M., Bushman F. D. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forshey B. M., von Schwedler U., Sundquist W. I., Aiken C. J. Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auewarakul P., Wacharapornin P., Srichatrapimuk S., Chutipongtanate S., Puthavathana P. Virology. 2005;337:93–101. doi: 10.1016/j.virol.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 31.Gurer C., Hoglund A., Hoglund S., Luban J. J. Virol. 2005;79:5557–5567. doi: 10.1128/JVI.79.9.5557-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehrlich L. S., Agresta B. E., Carter C. A. J. Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganser B. K., Li S., Klishko V. Y., Finch J. T., Sundquist W. I. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 34.Gross I., Hohenberg H., Krausslich H. G. Eur. J. Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 35.Li S., Hill C. P., Sundquist W. I., Finch J. T. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 36.Briggs J. A., Wilk T., Welker R., Krausslich H. G., Fuller S. D. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens C. M., Song B., Perron M. J., Yang P. C., Stremlau M., Sodroski J. J. Virol. 2004;78:5423–5437. doi: 10.1128/JVI.78.10.5423-5437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towers G. J., Hatziioannou T., Cowan S., Goff S. P., Luban J., Bieniasz P. D. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 39.Marechal V., Clavel F., Heard J. M., Schwartz O. J. Virol. 1998;72:2208–2212. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Findlay J. B. C., Evans W. H. Biological Membranes: A Practical Approach. Oxford: IRL; 1987. [Google Scholar]
- 41.Kotov A., Zhou J., Flicker P., Aiken C. J. Virol. 1999;73:8824–8830. doi: 10.1128/jvi.73.10.8824-8830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price C. A. Methods Enzymol. 1974;31:501–519. [PubMed] [Google Scholar]
- 43.Welker R., Hohenberg H., Tessmer U., Huckhagel C., Krausslich H. G. J. Virol. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap M. W., Nisole S., Lynch C., Stoye J. P. Proc. Natl. Acad. Sci. USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hatziioannou T., Cowan S., Von Schwedler U. K., Sundquist W. I., Bieniasz P. D. J. Virol. 2004;78:6005–6012. doi: 10.1128/JVI.78.11.6005-6012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berthoux L., Sebastian S., Sokolskaja E., Luban J. Proc. Natl. Acad. Sci. USA. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bukovsky A. A., Weimann A., Accola M. A., Gottlinger H. G. Proc. Natl. Acad. Sci. USA. 1997;94:10943–10948. doi: 10.1073/pnas.94.20.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatziioannou T., Perez-Caballero D., Cowan S., Bieniasz P. D. J. Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kallijarvi J., Lahtinen U., Hamalainen R., Lipsanen-Nyman M., Palvimo J. J., Lehesjoki A. E. Exp. Cell Res. 2005;308:146–155. doi: 10.1016/j.yexcr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Kudryashova E., Kudryashov D., Kramerova I., Spencer M. J. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 51.Meroni G., Diez-Roux G. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 52.Xu L., Yang L., Moitra P. K., Hashimoto K., Rallabhandi P., Kaul S., Meroni G., Jensen J. P., Weissman A. M., D’Arpa P. Exp. Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 53.Hofmann W., Schubert D., LaBonte J., Munson L., Gibson S., Scammell J., Ferrigno P., Sodroski J. J. Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz O., Marechal V., Friguet B., Arenzana-Seisdedos F., Heard J. M. J. Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei B. L., Denton P. W., O’Neill E., Luo T., Foster J. L., Garcia J. V. J. Virol. 2005;79:5705–5712. doi: 10.1128/JVI.79.9.5705-5712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braaten D., Franke E. K., Luban J. J. Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawyer S. L., Wu L. I., Emerman M., Malik H. S. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Javanbakht H., Diaz-Griffero F., Stremlau M., Si Z., Sodroski J. J. Biol. Chem. 2005;280:26933–26940. doi: 10.1074/jbc.M502145200. [DOI] [PubMed] [Google Scholar]
- 59.Yoo S., Myszka D. G., Yeh C., McMurray M., Hill C. P., Sundquist W. I. J. Mol. Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 60.Mortuza G. B., Haire L. F., Stevens A., Smerdon S. J., Stoye J. P., Taylor I. A. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.