Abstract
Subgroup J avian leukosis virus (ALV-J) is a recently identified avian oncogenic retrovirus responsible for severe economic losses worldwide. In contrast with the other ALV subgroups, ALV-J predominantly induces myeloid leukosis in meat-type chickens. Despite significant homology with the other ALV subgroups across most of the genome, the envelope protein of ALV-J (EnvJ) shares low homology with the others. Pathogenicity and myeloid leukosis induction map to the env gene of ALV-J. A chimeric protein composed of the surface domain of EnvJ fused to the constant region of a rabbit IgG and mass spectrometry were used to identify the chicken Na+/H+ exchanger type 1 (chNHE1) as a binding protein for ALV-J. Flow cytometry analysis and coprecipitation experiments demonstrated a specific interaction between EnvJ and chNHE1. When introduced into nonpermissive human 293T cells and quail QT6 cells, chNHE1 conferred susceptibility to EnvJ-mediated infection. Furthermore, 293T cells expressing chNHE1 fused with 293T cells expressing EnvJ in a low-pH-dependent manner. Together, these data identify chNHE1 as a cellular receptor for the highly pathogenic ALV-J.
Keywords: retrovirus, viral receptor, viral envelope
Avian leukosis viruses (ALV) are a group of avian retroviruses that induce tumors in host birds. The viruses that infect chickens are classified into six subgroups (A–E and J) on the basis of the envelope glycoprotein responsible for specific viral interference patterns, virus neutralization, and host range (1). The most recently identified subgroup, ALV-J, predominantly infects meat-type chickens and turkeys (1). ALV-J was identified in 1988 and became widespread in commercial meat-type poultry during the 1990s. The transmission of ALV-J is much higher than other ALV subgroups (2), thus making control and eradication significantly more difficult. The virus also evolves rapidly with sequence variations clustered in hypervariable regions of the envelope protein (Env) (3). In contrast to other subgroups, which primarily cause lymphoma, ALV-J mainly induces myeloid leukosis (4). ALV-J infection causes disease and death in both broiler breeders and egg layers and represents a significant problem for the commercial poultry industry worldwide, with estimated losses of 1.5% per week in excess mortality (5).
Retroviruses infect host cells through specific interactions between viral Env and cell surface receptors. The Env surface subunit (SU) directly binds to the receptor, and subsequent conformational changes in the Env transmembrane (TM) subunit drive fusion of the viral and cellular membranes (6). The receptors for all of the other major ALV subgroups (A–E) have been identified (7–10). Receptor distribution is a major determinant of ALV subgroup tropism. Env is also a major determinant for the induction of lymphoid and myeloid tumors by ALV-A and ALV-J, respectively (4), presumably because the specific receptors for ALV-A and ALV-J are differentially expressed on different cell lineages. This hypothesis is supported by the observation that ALV-J replicates well in monocyte cultures but poorly in lymphoid follicles, whereas ALV-A replicates well in lymphoid but poorly in myeloid lineages (4).
Variation in the distribution of receptors for the diverse ALV subgroups among chicken populations accounts for differences in the susceptibility to ALV infection. Functional receptors for ALV subgroups A–E are not expressed in all lines of chickens (11–13). ALV-J appears to be an exception because all lines of chickens screened to date are susceptible to ALV-J infection (14). However, quail and many other avian species are resistant to infection by ALV-J, suggesting that the virus receptor is not broadly conserved in avian species (1).
Here we use a combined biochemical and genetic approach to identify cell surface proteins that interact with the ALV-J SU and that function to mediate ALV-J infection of cells. These studies identify the chicken N+/H+ exchanger type 1 (chNHE1) as a cellular receptor for ALV-J.
Results
Identification of a 90-kDa Cell Surface Protein That Binds EnvJ.
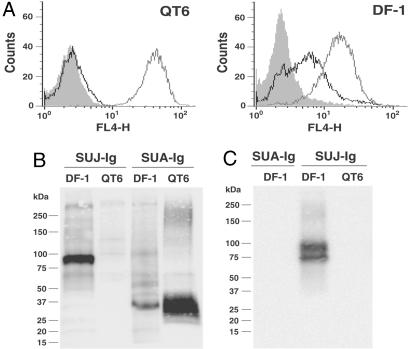
To identify cellular factors involved in ALV-J entry, a chimeric protein composed of the SU domain of EnvJ fused to the constant region fragment (Fc) region of a rabbit IgG was constructed to produce a SUJ-Ig immunoadhesin. Similar immunoadhesins produced with the ALV-A and ALV-B Env were previously shown to specifically bind the cellular receptors for these viruses (8). FACS analysis demonstrated that SUJ-Ig bound cells permissive for ALV-J replication (chicken DF-1) but not cells from quail (QT6) that are resistant (Fig. 1A) (1). Additionally, SUJ-Ig did not bind to several mammalian cell lines that are nonpermissive for ALV-J infection (data not shown). As anticipated, the control SUA-Ig immunoadhesin containing the SU domain of the ALV-A envelope (EnvA) bound DF-1 and QT6 cells because both are permissive for ALV-A (Fig. 1A). Compared with SUA-Ig, the mean fluorescence intensity for the subgroup J immunoadhesin was significantly lower, suggesting either lower expression or lower affinity of the cell surface binding partner for SUJ-Ig.
Fig. 1.
A 90-kDa chicken protein associates with the SU domain of EnvJ. (A) QT6 or DF-1 cells were incubated with SARS S1-Ig (shaded area, negative control), SUA-Ig (gray line), or SUJ-Ig (black line) and then a Cy5-conjugated goat anti-rabbit secondary antibody. Binding of immunoadhesins to cells was measured by flow cytometry. (B) Surface-biotinylated QT6 or DF-1 cell lysates were precipitated with SUJ-Ig or SUA-Ig. Precipitated proteins were subjected to SDS/PAGE and Western blot analysis by using streptavidin horseradish peroxidase. (C) QT6 or DF-1 cell lysates were precipitated with SUJ-Ig or SUA-Ig. Precipitated proteins were subjected to SDS/PAGE and Western blot analysis by using an anti-NHE1 monoclonal antibody. FL4-H denotes fluorescence intensity.
The ability of SUJ-Ig to specifically recognize cells susceptible to ALV-J infection suggested that this reagent might be used to identify the cellular receptor for this virus. Cell surface proteins capable of interacting with ALV-J Env were identified by precipitation using SUJ-Ig after surface biotinylation. A protein of ≈90 kDa was precipitated from permissive DF-1 cells but not restrictive QT6 cells (Fig. 1B). As a control, SUA-Ig precipitated a protein of ≈30 kDa, corresponding to the ALV-A receptor Tva, from both DF-1 and QT6 cell lysates (Fig. 1B).
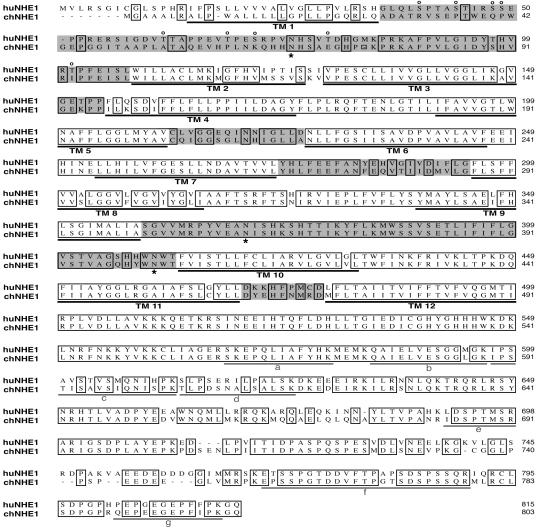
To identify the 90-kDa SUJ-Ig binding band, proteins from several plates of DF-1 cells were pooled and precipitated with SUJ-Ig. After SDS/PAGE and protein staining, two bands in the 90-kDa region of the gel were excised and subjected to trypsin digestion and mass spectrometry. By comparison to the protein database, two proteins, thioredoxin peroxidase and NHE1, were identified. Thioredoxin peroxidase is a 23-kDa protein that does not localize to the cell surface; therefore, we did not analyze it further. In contrast, mammalian NHE1 is a 91-kDa plasma membrane protein containing 12 membrane-spanning segments (15). Seven peptides corresponding to the highly conserved C-terminal region of NHE1 were identified (see Fig. 5). The chNHE1 protein has not been well characterized, and its full-length cDNA has not been cloned; however, NHE1 activity was localized to the plasma membrane in chicken osteoclasts (16–18).
Fig. 5.
Alignment of the amino acid sequences of chNHE1 and huNHE1. Deduced amino acid sequences for huNHE1 and chNHE1 are shown. Identical amino acids are enclosed in boxes. Gaps are indicated by dashes. The 12 predicted TM segments are underlined and designated TM1–TM12. ECLs are highlighted in gray. The three conserved N-linked glycosylation sites are marked by asterisks below the sequence. Potential O-linked glycosylation sites in huNHE1 that are absent in chNHE1 are designated by “o” above the sequence. Peptides identified by mass spectrometry are underlined and designated by the letters a–g in gray.
ALV-J Env and chNHE1 Specifically Interact.
To confirm the ability of chNHE1 to interact with ALV-J Env, monoclonal antibody 4E9, specific for an epitope in the conserved cytoplasmic tail of porcine NHE1, was used (16). SUJ-Ig precipitated NHE1 from lysates of ALV-J-permissive DF-1 but not ALV-J-nonpermissive QT6 cells (Fig. 1C). The upper band correlated with the 90-kDa band in Fig. 1B. The lower-molecular-mass band appears to be a core-glycosylated form of NHE1 that does not localize to the cell surface (19). SUA-Ig served as a control and did not precipitate NHE1 from DF-1 cells.
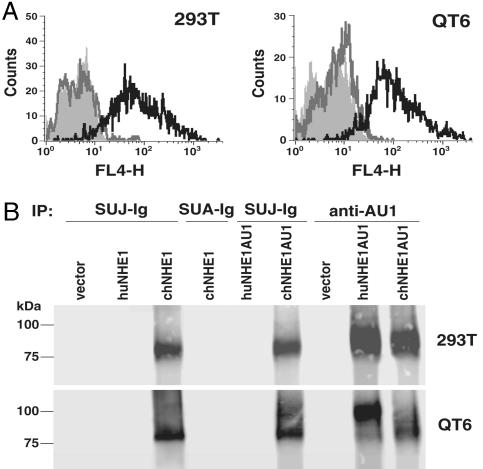
To extend our analysis of the ALV-J Env and NHE1 interaction, we cloned the full-length cDNA encoding chNHE1 from DF-1 cell mRNA by using RT-PCR and a 5′ RACE strategy. A cDNA encoding human NHE1 (huNHE1) was isolated by PCR from a HeLa cDNA library. Human cells are resistant to ALV-J infection (see Fig. 3C) (data not shown); therefore, we hypothesized that chNHE1, but not huNHE1, should interact with SUJ-Ig. FACS analysis demonstrated that 293T cells transiently expressing chNHE1 specifically bound SUJ-Ig (Fig. 2A) but not the control immunoadhesin SUA-Ig (data not shown). In contrast, transfection of huNHE1 cDNA or vector alone into 293T cells did not mediate binding by SUJ-Ig (Fig. 2A). Similarly, QT6 cells expressing chNHE1 were recognized by SUJ-Ig, whereas quail cells expressing huNHE1 were not (Fig. 2A). To further demonstrate the specific interaction between EnvJ and chNHE1, we used SUJ-Ig or SUA-Ig to precipitate NHE1 from the lysates of 293T or QT6 cells transiently expressing high levels of chNHE1, huNHE1, or vector alone. Only chNHE1 coprecipitated with SUJ-Ig (Fig. 2B). As expected, the control SUA-Ig did not precipitate chNHE1 (Fig. 2B).
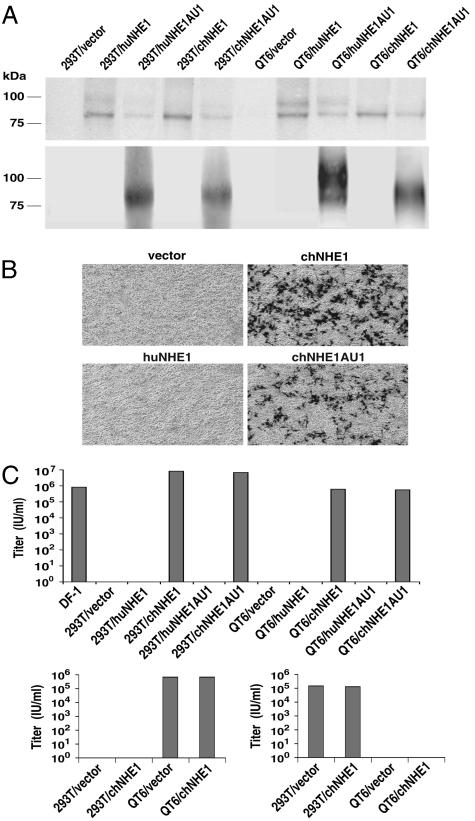
Fig. 3.
chNHE1 allows EnvJ-mediated infection of resistant cells. (A) Lysates of 293T or QT6 cells transfected with vector alone or vector expressing huNHE1, chNHE1, AU1-tagged huNHE1, or AU1-tagged chNHE1 were subjected to SDS/PAGE and Western blot analysis with an anti-NHE1 monoclonal antibody (Upper) or a mouse anti-AU1 antibody (Lower). (B) 293T cells transfected with vector alone or vector expressing huNHE1, chNHE1, or AU1-tagged chNHE1 were infected with HIV(EnvJ) encoding β-galactosidase. Two days after infection, cells were stained for β-galactosidase activity and subjected to light microscopy and photography. (C) (Upper) Cells as in A and DF-1 cells were infected with HIV(EnvJ). (Lower) As controls, cells were infected with HIV(EnvA) (Left) or HIV(SARS S) (Right) pseudotype viruses. Cells were stained 2 days after infection, and titers were determined by enumerating blue foci. The experiment was repeated three times, and results from one representative experiment are shown. Abbreviations are the same as in Fig. 2.
Fig. 2.
Specific interaction between SUJ-Ig and chNHE1. (A) FACS analysis of the SUJ-Ig–NHE1 interaction. 293T or QT6 cells transfected with vector alone (shaded area, no line), vector expressing huNHE1 (thick gray line), or chNHE1 (thick black line) were incubated with SUJ-Ig and then a Cy5-conjugated goat anti-rabbit secondary antibody. Binding of immunoadhesins to cells was measured by flow cytometry. (B) Lysates of 293T or QT6 cells transfected with vector alone or vector expressing huNHE1, chNHE1, AU1-tagged huNHE1 (huNHE1AU1), or AU1-tagged chNHE1 (chNHE1AU1) were precipitated with SUJ-Ig, SUA-Ig, or a rabbit anti-AU1 antibody. Precipitated proteins were subjected to SDS/PAGE and Western blot analysis by using an anti-NHE1 monoclonal antibody. FL4-H denotes fluorescence intensity.
In mammals, NHE1 is a housekeeping protein and expressed in all tissues (15). Both 293T and QT6 cells express low levels of endogenous NHE1 that can be recognized by the 4E9 antibody after long exposure of Western blots (data not shown). To distinguish the transiently expressed NHE1 from endogenous NHE1 and to confirm expression of the exogenous human and avian proteins, C-terminally AU1-tagged chNHE1 and huNHE1 proteins were generated and expressed in 293T or QT6 cells. Only AU1-tagged chNHE1 was specifically precipitated by SUJ-Ig (Fig. 2B). In contrast, both AU1-tagged proteins could be immunoprecipitated by an anti-AU1 antibody (Fig. 2B). Overall, these results demonstrate a specific association between the SU domain of EnvJ and chNHE1.
chNHE1 Confers Susceptibility to EnvJ-Mediated Infection.
Next, we investigated the ability of chNHE1 to support viral infection by using viruses pseudotyped with EnvJ. Fig. 3A confirmed the expression of chNHE1 and huNHE1, with or without the AU1 tag, in 293T and QT6 cells. The 4E9 antibody detected tagged and untagged NHE1 (Fig. 3A Upper), whereas an anti-AU1 antibody only detected the AU1-tagged proteins (Fig. 3A Lower). These cells expressing huNHE1 or chNHE1 were challenged with HIV-1 pseudotypes carrying EnvJ on their surface. Both tagged and untagged chNHE1, but not huNHE1, conferred susceptibility to EnvJ-pseudotyped virus [HIV(EnvJ)] (Fig. 3B). Results from infection experiments are summarized in Fig. 3C. When expressed in 293T and QT6 cells, tagged and untagged chNHE1 permitted HIV(EnvJ) infection at titers similar to or higher than those seen with DF-1 cells as targets (Fig. 3C Upper). As expected from the binding studies, huNHE1 did not facilitate entry by EnvJ. chNHE1 expression in 293T cells did not confer susceptibility to EnvA-pseudotyped virus, HIV(EnvA), nor did it increase the infectious titer of HIV(EnvA) on QT6 cells (Fig. 3C Lower Left). Similarly, it did not confer susceptibility to a control severe acute respiratory syndrome coronavirus S (SARS S)-pseudotyped virus, HIV(SARS S), in QT6 cells, or increase the infectious titer of this pseudotype on 293T cells (Fig. 3C Lower Right). These infection data, coupled with the binding data presented above, support the hypothesis that chNHE1 is a cellular receptor for ALV-J.
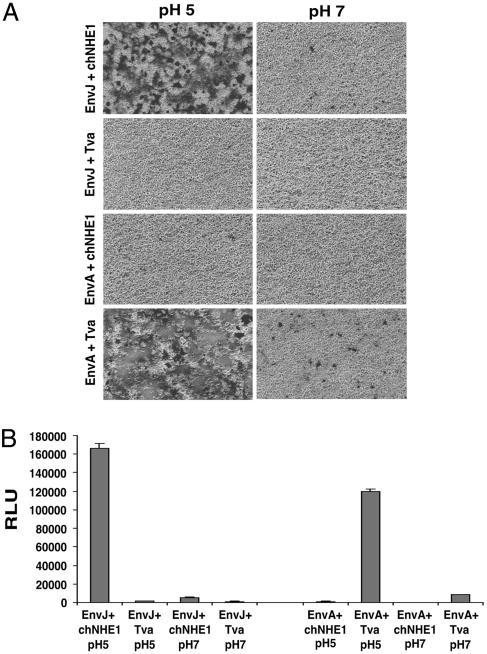
EnvJ-Mediated Membrane Fusion Requires chNHE1 and Low pH.
Analysis of EnvJ pseudotypes revealed that lysosomotropic agents that raise the endosomal pH blocked infection (P.B., unpublished observation). Thus, it is possible that chNHE1 acts solely as an attachment factor and facilitates viral entry by trafficking the viral Env to the endosome where low pH-triggered membrane fusion takes place. To address whether chNHE1 is specifically required for membrane fusion, we performed cell–cell fusion assays by using a β-galactosidase α-complementation system (20). Cells expressing EnvJ formed numerous, large syncytia with target cells expressing chNHE1 at pH 5 (Fig. 4A). EnvJ fusion was strictly dependent on chNHE1 expression in the target cells and was not seen with cells expressing the ALV-A receptor Tva at low or neutral pH. EnvJ-mediated cell–cell fusion was also highly pH-dependent, and only a few tiny syncytia and low β-galactosidase activity were seen at pH 7 (Figs. 4 A and B). Fusion of the control EnvA-expressing cells depended on Tva and was strongly enhanced by low pH as reported in refs. 6 and 21. These results demonstrate a specific requirement of chNHE1 for EnvJ-mediated membrane fusion and provide further evidence that chNHE1 is a functional receptor for ALV-J; they are also consistent with a pH-dependent mechanism for ALV-J entry.
Fig. 4.
Cell–cell fusion of EnvJ- and chNHE1-expressing cells. 293T cells were transfected with a plasmid expressing the α-subunit of β-galactosidase and a plasmid expressing EnvJ or EnvA. These effector cells were mixed at a 1:1 ratio with target 293T cells transfected with a plasmid expressing the ω-subunit of β-galactosidase and a plasmid expressing chNHE1 or Tva. Cell mixtures were pulsed in pH 5 or pH 7 media for 10 min at 37°C. Cell–cell fusion was analyzed by X-gal staining, light microscopy, and photography (A) or by a β-galactosidase activity assay (B). In B, triplicate samples were analyzed. Means and standard deviations of random light units (RLU) are shown on the histogram. Abbreviations are the same as in Fig. 2.
Comparison of the chNHE1 and huNHE1 Amino Acid Sequences.
The deduced amino acid sequence for chNHE1 in comparison with that of huNHE1 is shown in Fig. 5. Overall, the two molecules are 79% identical, with chNHE1 12 residues shorter than its human homolog. All of the previously reported residues and regions involved in ion translocation and pH sensing (15, 22) are conserved, indicating that chNHE1 is likely a functional Na+/H+ exchanger. Moreover, the cytoplasmic tails of chNHE1 and huNHE1 share 80% identity, and residues and domains involved in phosphorylation and activation of NHE1 and in binding signaling complexes (15) are conserved, suggesting that the activity of chNHE1 is regulated similarly to that of huNHE1. The 12 putative TM segments shown for chNHE1 were deduced based on a topology model of huNHE1 that proposes the presence of a large exofacial reentrant loop between TM9 and TM10 (15, 23). This model was supported by the defined structure of a bacterial homolog of NHE1 (22). Based on this model, we propose that the N and C termini of chNHE1 are located in the cytosol, whereas all three conserved N-linked glycosylation sites (residues 68, 362, and 402 of chNHE1) are extracellular.
Despite the overall high homology between chNHE1 and huNHE1, three of the six predicted extracellular loops (ECLs), where one might predict interactions with EnvJ to occur, are not highly conserved. The first ECL (ECL1) between the first and second membrane-spanning domains of chNHE1 is only 39% identical to that of huNHE1. Within ECL1, the human protein contains numerous O-glycosylation sites (19) that are absent in chNHE1. ECL3 and ECL6 are two short loops that demonstrate 53% and 55% identity with huNHE1. The other three extracellular regions are more highly conserved between the chicken and human proteins with identities between 71% and 96%.
Discussion
We have identified chNHE1 as a functional receptor for ALV-J through a combined biochemical purification and functional cloning approach. chNHE1 specifically bound to, and was purified with, an EnvJ SU domain immunoadhesin. Expression of chNHE1 in nonsusceptible avian or mammalian cells permitted binding of EnvJ SU to these cells and conferred susceptibility to ALV-J envelope-mediated infection. Mammalian cells expressing chNHE1 formed syncytia with cells expressing EnvJ in a pH-dependent manner.
Analysis of the chicken genome database revealed that sequences encoding chNHE1 are located on chromosome 23. Previous studies detected a NHE1 transcript in chicken embryonic fibroblasts (17) and intestinal brush border membrane (18) by Northern blot analysis with a probe from the highly conserved cytoplasmic tail sequence but failed to recognize the NHE1 protein with a polyclonal antibody against huNHE1. Additionally, a NHE1-like activity was demonstrated in chicken embryonic fibroblasts (17), and NHE1 protein and transcripts were reported in chicken osteoclasts (16). In these studies, we detected the chNHE1 protein in chicken DF-1 fibroblasts and produced a full-length cDNA clone for chNHE1. Analysis of the deduced amino acid sequence demonstrated that the features required for ion translocation and pH sensing by huNHE1 (15, 22) are conserved in the chicken homolog, suggesting that the chNHE1 we identified is a functional Na+/H+ exchanger.
The discovery of the subgroup J receptor completes the identification of the cellular receptors for the major α-retroviruses. In contrast to chNHE1, the receptors for ALV subgroups A–E are all single-pass membrane proteins encoded by the genes tva, tvb, and tvc (7–10). ALV-J appears to be derived by recombination of an exogenous ALV with ancient endogenous avian viral sequences in the chicken genome and acquisition of an endogenous avian viral env gene (24, 25). The Env protein of ALV-J is only weakly related to the Env proteins of the subgroup A–E viruses (24, 26). Significant variation has been reported in the sequence of the envelope genes of ALV-J field isolates (5, 27). Nonetheless, receptor interference studies using a DF-1/J-resistant cell line indicate that all subgroup J isolates use the same receptor (28). In general, related retroviruses use similar types of molecules as receptors. The human, simian, and feline immunodeficiency viruses all use a receptor and a multipass coreceptor, whereas the divergent ungulate lentivirus, equine infectious anemia virus, uses a single-pass receptor (29). Similarly, γ-retroviruses, such as murine leukemia virus, feline leukemia virus, and gibbon ape leukemia virus, with envelopes similar to the murine leukemia virus prototype use divergent receptors that are all multipass proteins (30).
For ALV subgroups A–E, nonfunctional receptor alleles contribute to genetic resistance of chicken lines to infection (11–13). For example, two different forms of the ALV-A receptor Tva, with either a cysteine-to-tryptophan change that abrogates EnvA binding or a frameshift that abolishes receptor expression, lead to genetic resistance to infection (31). Similarly, sequence variation in Tvb accounts for differential interactions with the subgroup B, D, and E Env proteins and susceptibility to the viruses carrying them (32). In contrast, all chicken lines analyzed to date are susceptible to ALV-J (14). Mammalian NHE1 is a housekeeping protein that regulates intracellular pH and cell volume. Accordingly, NHE1 is ubiquitously expressed in all tissues (15). chNHE1 probably carries out housekeeping functions similar to mammalian NHE1, and its expression is likely required in all lines of chickens, possibly accounting for the broad susceptibility of chickens to ALV-J. Analysis of ALV-J tropism in infected chickens by in situ hybridization demonstrates infection of many tissues (33, 34) consistent with the presumed extensive expression of chNHE1. High levels of viral RNA were seen in the heart, kidney, pituitary, adrenal, and thyroid glands. However, in both of these studies, no infection was seen in the bone marrow and only cells with dendritic morphology were stained in the bursa and thymus (33). It is possible that NHE-1 is abundantly expressed on a small subset of differentiating myeloid cells, causing them to be more effectively targeted by ALV-J, but the low abundance or distribution of these cells does not allow detection during in situ analysis. Supporting this hypothesis is the finding that avian osteoclasts (a myeloid-derived lineage) significantly up-regulate NHE1 during differentiation (16). In addition, a common nonneoplastic effect of ALV-J infection in chickens is cardiomyopathy (35). In mammals, NHE1 has been found to be abundantly expressed in heart and to play an important role in hypertrophy and cardiac failure (reviewed in ref. 36). These findings imply a possible direct role for NHE1 in cardiac pathogenesis during ALV-J infection.
Quail and mammalian cells express endogenous NHE1 but are not permissive for ALV-J infection (Fig. 3). The general requirement for NHE1 suggests that resistance of other avian species (with the exception of turkey) to ALV-J infection likely reflects sequence variation rather than absence of the receptor. The large first ECL of chNHE1 is the most divergent, with only 39% identity compared with huNHE1, whereas the other large loop, ECL5, is 96% identical. The remaining four ECLs are relatively small, and only ECL3 and ECL6 display <70% identity to huNHE1. This analysis suggests that ECL1 may be a focus for further studies on NHE1–EnvJ interaction. Isolation of the cDNAs encoding NHE1 from resistant (quail) and susceptible (turkey) avian species will help identify the receptor sequence determinants for viral infection and provide the basis for developing genetic and biochemical approaches to screen for resistant chickens, to impair ALV-J infection, or to facilitate ALV-J detection in field samples.
Results from a cell–cell fusion assay (Fig. 4) suggest that ALV-J entry requires receptor binding and low pH activation, as has been demonstrated for the other ALV subgroups studied to date (6). However, unlike ALV-A envelope entry, ALV-J Env-mediated entry is effectively inhibited by brief ammonium chloride treatment (P.B., unpublished results). Moreover, microscopic examination of the fusion assay reveals that there is some leakiness to EnvA-mediated fusion at pH 7, whereas EnvJ is more pH-dependent (Fig. 4A). A two-step entry mechanism in which Env–receptor interaction renders the viral glycoprotein sensitive to low-pH-induced activation has been proposed for ALV-A and ALV-B (6). Current models propose that, in the first step, the receptor triggers structural rearrangement and insertion of the fusion peptide into the target membrane (21, 37, 38). Peptide inhibitors bind to this form, suggesting that it is a prehairpin intermediate (21, 37). Low pH appears to act later and promote steps leading to membrane fusion, perhaps by driving six-helix bundle formation (21, 37, 38). It will be important to determine whether EnvJ, whose sequence is quite divergent and has only 32% and 55% identity in the SU and TM subunits, respectively, compared with the other ALV Env proteins, utilizes a similar two-step entry mechanism.
Recent evidence suggests that, in addition to ion translocation and pH regulation, NHE1 also functions as a scaffold to assemble signaling complexes and as a plasma membrane anchor for actin-based cytoskeleton network (reviewed in ref. 39). Its long cytoplasmic tail binds different families of signaling molecules and is phosphorylated upon activation (15). NHE1 activity is modulated by diverse extracellular and intracellular stimuli. Increased NHE1 activity promotes cell cycle progression, cell proliferation, and cell survival; reduces cell adhesion and increases cell migration; and is necessary for oncogenic transformation, tumor development, and tumor invasion (39). Analysis of chimeric viruses with exchanged envelope genes demonstrated that EnvJ is a major determinant for the myeloid lineage-specific oncogenicity induced by ALV-J (4). It is interesting to speculate that binding and infection with ALV-J activates chNHE1 and contributes to oncogenic transformation of myeloid cells possibly by affecting the activation properties and downstream signaling pathways of NHE1.
In summary, the identification of chNHE1 as a receptor for ALV-J sheds light on the mechanism by which ALV-J infects cells. Further studies may help limit ALV-J infection and the severe economic losses engendered by this virus. Discovery of the ALV-J receptor also provides an additional model system to study retroviral Env–receptor interactions and to analyze determinants of ALV-induced myeloid versus lymphoid oncogenicity.
Methods
Cell Lines and Plasmids.
Human 293T cells, chicken embryonic fibroblast DF-1 cells, and quail QT6 cells were maintained in DMEM supplemented with 10% FCS.
Expression plasmids pCB6-EnvA (40), pCB6-SUA-Ig (8), pCB6-Tva (7), pCAGGS-SARS S (41), pCAGGS-S1-Ig (42), pCMV-α, and pCMV-ω (20) were described previously. pCB6-EnvJ was constructed by removing the EnvJ insert from pBSR54FL (containing the proviral clone of the ADOL-R5–4 ALV-J isolate) with KpnI and BalI and cloning it into pCB6. pCB6-SUJ-Ig was constructed by replacing the SU sequence of EnvA in pCB6-SUA-Ig (8) with the SU sequence of EnvJ by using the KpnI and BamHI sites. pCAGGS-chNHE1 was constructed by isolating chNHE1 cDNA from DF-1 mRNA with RT-PCR and a 5′ RACE kit (Invitrogen) and cloning it into the pCAGGS expression plasmid. pCAGGS-huNHE1 was constructed by PCR amplification of huNHE1 cDNA from a HeLa cDNA library and cloning it into pCAGGS. pCAGGS-chNHE1AU1 and pCAGGS-huNHE1AU1 were constructed by PCR amplification with primers appending an AU1-coding sequence to the C terminus of NHE1. All primer sequences are listed in the Supporting Methods, which is published as supporting information on the PNAS web site.
Immunoadhesin Production and Pseudotype Preparation.
SUJ-Ig, SUA-Ig (8), and S1 (the S1 domain of SARS S protein)-Ig (42) were produced from 293T cells and purified with protein A agarose as described in ref. 8. HIV-LacZ reporter viruses pseudotyped with EnvJ [HIV(EnvJ)], EnvA [HIV(EnvA)], or SARS S [HIV(SARS S)] were prepared with a three-plasmid cotransfection system as in ref. 41.
Protein Precipitation and Mass Spectrometry.
DF-1 or QT6 cells were surface-labeled with 1 mg/ml sulfo-NHS-LC-biotin (Pierce) in PBS (pH 8) for 30 min and lysed in 1% Brij 97 (Sigma)/150 mM NaCl/20 mM Tris, pH 8/5 mM iodoacetamide (Brij lysis buffer). Cell lysate was incubated overnight with 4 μg of SUJ-Ig or SUA-Ig bound to protein A agarose at 4°C. Precipitates were washed twice in Brij lysis buffer and once in PBS. Bound proteins were eluted in Laemmli sample buffer and separated by SDS/PAGE. The resulting Western blot was probed with streptavidin–horseradish peroxidase (Invitrogen). By using this approach, ≈8 × 107 unlabeled DF-1 cells were used to generate two distinct bands of ≈90 kDa that could be visualized by Sypro Ruby staining (Bio-Rad). These bands were excised from the gel and subjected to trypsin digestion and MALDI mass spectrometry.
293T or QT6 cells were transfected with pCAGGS, pCAGGS-chNHE1, pCAGGS-huNHE1, pCAGGS-chNHE1AU1, or pCAGGS-huNHE1AU1 by using calcium phosphate precipitation and lysed in Brij lysis buffer 48 h after transfection. Lysates were incubated overnight with 4 μg of protein A agarose-bound SUJ-Ig, SUA-Ig, or a rabbit anti-AU1 antibody (Bethyl Laboratories, Montgomery, TX). Precipitates were washed, eluted, and subjected to SDS/PAGE as above. The resulting Western blot was probed with 4E9, a monoclonal antibody against porcine NHE1 (Chemicon) and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce).
Flow Cytometry.
DF-1 or QT6 cells were incubated with 8 μg/ml S1-Ig, SUJ-Ig, or SUA-Ig in PBS plus 5% FCS. Cells were washed in PBS and incubated with a Cy5-conjugated goat anti-rabbit antibody (Chemicon). Cells were washed and analyzed by flow cytometry. 293T or QT6 cells transfected with pCAGGS, pCAGGS-chNHE1, or pCAGGS-huNHE1 were incubated with 8 μg/ml SUJ-Ig or SUA-Ig 48 h after transfection and processed for flow cytometry analysis as described above.
Infection of NHE1-Transfected Cells.
293T or QT6 cells were transfected with pCAGGS, pCAGGS-chNHE1, pCAGGS-huNHE1, pCAGGS-chNHE1AU1, or pCAGGS-huNHE1AU1 as described above. Cells were challenged 48 h later with HIV(EnvJ), HIV(EnvA), or HIV(SARS S) and stained with X-gal 2 days after infection. Cells were subjected to light microscopy and photography, and titers were determined by enumerating blue foci. A fraction of cells was lysed in Brij lysis buffer as above. Lysates were run on SDS/PAGE, and the resulting Western blot was probed with 4E9 or a mouse-anti-AU1 antibody (Covance, Richmond, CA). These experiments were repeated three times with similar results. One representative experiment is shown (Fig. 3).
Cell–Cell Fusion Assay.
An α-complementation-based fusion assay is described in ref. 20. Briefly, 293T cells transfected with pCMV-α and pCB6-EnvJ or pCB6-EnvA were mixed at a 1:1 ratio with 293T cells transfected with pCMV-ω and pCAGGS-chNHE1 or pCB6-Tva 48 h after transfection. Cells were allowed to attach and then were incubated in DMEM adjusted to pH 5 or 7 for 10 min at 37°C. Cells were washed and incubated in normal growth media for 6 h and subjected to X-gal staining, light microscopy, and photography. β-galactosidase activity was measured by a Galacto-Light Plus kit (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank Kathleen Conklin (University of Minnesota, Minneapolis) for providing pBSR54FL, John Young (The Salk Institute for Biological Studies, La Jolla, CA) for providing pCB6-SUA-Ig, and Gilles Bertoia (University of Pennsylvania) for constructing pCB6-EnvJ and performing the EnvJ pH entry assays. This work was supported by National Institutes of Health Grants R01 AI43455, R21 AI059172, and R01 CA76256.
Abbreviations
- ALV
avian leukosis virus
- Env
envelope protein
- NHE1
Na+/H+ exchanger 1
- chNHE1
chicken NHE1
- huNHE1
human NHE1
- SU
Env surface subunit
- TM
transmembrane
- ECL
extracellular loop
- SARS S
severe acute respiratory syndrome coronavirus S.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ256198).
References
- 1.Payne L. N., Howes K., Gillespie A. M., Smith L. M. J. Gen. Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 2.Venugopal K. Res. Vet. Sci. 1999;67:113–119. doi: 10.1053/rvsc.1998.0283. [DOI] [PubMed] [Google Scholar]
- 3.Venugopal K., Smith L. M., Howes K., Payne L. N. J. Gen. Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]
- 4.Chesters P. M., Howes K., Petherbridge L., Evans S., Payne L. N., Venugopal K. J. Gen. Virol. 2002;83:2553–2561. doi: 10.1099/0022-1317-83-10-2553. [DOI] [PubMed] [Google Scholar]
- 5.Silva R. F., Fadly A. M., Hunt H. D. Virology. 2000;272:106–111. doi: 10.1006/viro.2000.0352. [DOI] [PubMed] [Google Scholar]
- 6.Mothes W., Boerger A. L., Narayan S., Cunningham J. M., Young J. A. Cell. 2000;103:679–689. doi: 10.1016/s0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 7.Bates P., Young J. A., Varmus H. E. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 8.Brojatsch J., Naughton J., Rolls M. M., Zingler K., Young J. A. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 9.Elleder D., Stepanets V., Melder D. C., Senigl F., Geryk J., Pajer P., Plachy J., Hejnar J., Svoboda J., Federspiel M. J. J. Virol. 2005;79:10408–10419. doi: 10.1128/JVI.79.16.10408-10419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adkins H. B., Brojatsch J., Naughton J., Rolls M. M., Pesola J. M., Young J. A. Proc. Natl. Acad. Sci. USA. 1997;94:11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crittenden L. B., Stone H. A., Reamer R. H., Okazaki W. J. Virol. 1967;1:898–904. doi: 10.1128/jvi.1.5.898-904.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne L. N., Biggs P. M. Virology. 1964;24:610–616. doi: 10.1016/0042-6822(64)90215-6. [DOI] [PubMed] [Google Scholar]
- 13.Vogt P. K., Ishizaki R. Virology. 1965;26:664–672. doi: 10.1016/0042-6822(65)90329-6. [DOI] [PubMed] [Google Scholar]
- 14.Payne L. N., Brown S. R., Bumstead N., Howes K., Frazier J. A., Thouless M. E. J. Gen. Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 15.Orlowski J., Grinstein S. Pflügers Arch. 2004;447:549–565. doi: 10.1007/s00424-003-1110-3. [DOI] [PubMed] [Google Scholar]
- 16.Gupta A., Edwards J. C., Hruska K. A. Bone. 1996;18:87–95. doi: 10.1016/8756-3282(95)00455-6. [DOI] [PubMed] [Google Scholar]
- 17.Bhartur S. G., Ballarin L. J., Musch M. W., Bookstein C., Chang E. B., Rao M. C. Am. J. Physiol. 1999;276:R838–R846. doi: 10.1152/ajpregu.1999.276.3.R838. [DOI] [PubMed] [Google Scholar]
- 18.Bhartur S. G., Bookstein C., Musch M. W., Boxer R., Chang E. B., Rao M. C. Comp. Biochem. Physiol. A Physiol. 1997;118:883–889. doi: 10.1016/s0300-9629(97)00219-3. [DOI] [PubMed] [Google Scholar]
- 19.Counillon L., Pouyssegur J., Reithmeier R. A. Biochemistry. 1994;33:10463–10469. doi: 10.1021/bi00200a030. [DOI] [PubMed] [Google Scholar]
- 20.Holland A. U., Munk C., Lucero G. R., Nguyen L. D., Landau N. R. Virology. 2004;319:343–352. doi: 10.1016/j.virol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Matsuyama S., Delos S. E., White J. M. J. Virol. 2004;78:8201–8209. doi: 10.1128/JVI.78.15.8201-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. Nature. 2005;435:1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 23.Wakabayashi S., Pang T., Su X., Shigekawa M. J. Biol. Chem. 2000;275:7942–7949. doi: 10.1074/jbc.275.11.7942. [DOI] [PubMed] [Google Scholar]
- 24.Benson S. J., Ruis B. L., Fadly A. M., Conklin K. F. J. Virol. 1998;72:10157–10164. doi: 10.1128/jvi.72.12.10157-10164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai J., Payne L. N., Skinner M. A. J. Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai J., Howes K., Payne L. N., Skinner M. A. J. Gen. Virol. 1995;76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 27.Cui Z., Du Y., Zhang Z., Silva R. F. Avian Dis. 2003;47:1321–1330. doi: 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- 28.Hunt H. D., Lee L. F., Foster D., Silva R. F., Fadly A. M. Virology. 1999;264:205–210. doi: 10.1006/viro.1999.9993. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B., Jin S., Jin J., Li F., Montelaro R. C. Proc. Natl. Acad. Sci. USA. 2005;102:9918–9923. doi: 10.1073/pnas.0501560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbaugh J., Miller A. D., Eiden M. V. Microbiol. Mol. Biol. Rev. 2001;65:371–389. doi: 10.1128/MMBR.65.3.371-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elleder D., Melder D. C., Trejbalova K., Svoboda J., Federspiel M. J. J. Virol. 2004;78:13489–13500. doi: 10.1128/JVI.78.24.13489-13500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adkins H. B., Brojatsch J., Young J. A. J. Virol. 2000;74:3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stedman N. L., Brown T. P., Brown C. C. Vet. Pathol. 2001;38:649–656. doi: 10.1354/vp.38-6-649. [DOI] [PubMed] [Google Scholar]
- 34.Arshad S. S., Smith L. M., Howes K., Russell P. H., Venugopal K., Payne L. N. Avian Pathol. 1999;28:163–169. doi: 10.1080/03079459994894. [DOI] [PubMed] [Google Scholar]
- 35.Stedman N. L., Brown T. P. Vet. Pathol. 2002;39:161–164. doi: 10.1354/vp.39-1-161. [DOI] [PubMed] [Google Scholar]
- 36.Karmazyn M., Gan X. T., Humphreys R. A., Yoshida H., Kusumoto K. Circ. Res. 1999;85:777–786. doi: 10.1161/01.res.85.9.777. [DOI] [PubMed] [Google Scholar]
- 37.Netter R. C., Amberg S. M., Balliet J. W., Biscone M. J., Vermeulen A., Earp L. J., White J. M., Bates P. J. Virol. 2004;78:13430–13439. doi: 10.1128/JVI.78.24.13430-13439.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melikyan G. B., Barnard R. J., Abrahamyan L. G., Mothes W., Young J. A. Proc. Natl. Acad. Sci. USA. 2005;102:8728–8733. doi: 10.1073/pnas.0501864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Putney L. K., Barber D. L. BMC Genomics. 2004;5:46. doi: 10.1186/1471-2164-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilbert J. M., Bates P., Varmus H. E., White J. M. J. Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons G., Reeves J. D., Rennekamp A. J., Amberg S. M., Piefer A. J., Bates P. Proc. Natl. Acad. Sci. USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marzi A., Gramberg T., Simmons G., Moller P., Rennekamp A. J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., et al. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.