Abstract
Diffusion within the extracellular space (ECS) of the brain is necessary for chemical signaling and for neurons and glia to access nutrients and therapeutics; however, the width of the ECS in living tissue remains unknown. We used integrative optical imaging to show that dextrans and water-soluble quantum dots with Stokes–Einstein diameters as large as 35 nm diffuse within the ECS of adult rat neocortex in vivo. Modeling the ECS as fluid-filled “pores” predicts a normal width of 38–64 nm, at least 2-fold greater than estimates from EM of fixed tissue. ECS width falls below 10 nm after terminal ischemia, a likely explanation for the small ECS visualized in electron micrographs. Our results will improve modeling of neurotransmitter spread after spillover and ectopic release and establish size limits for diffusion of drug delivery vectors such as viruses, liposomes, and nanoparticles in brain ECS.
Keywords: drug delivery, integrative optical imaging, nanoparticles, restricted diffusion, somatosensory cortex
The extracellular space (ECS) separates brain cells and normally occupies ≈20% of total brain volume in vivo (1, 2). EM of fixed, adult brain tissue suggests these spaces are only 10–20 nm wide (3, 4), but the ECS in this material has likely contracted after water and electrolyte changes associated with terminal ischemia (4, 5). Because no direct method exists for measuring ECS dimensions in the living state, the width in vivo is unknown. This parameter is important because the ECS is a mandatory route for drugs and drug delivery vectors (nanoparticles, liposomes, and viruses). Furthermore, interstitial space dimensions will affect the diffusion radius of released neurotransmitter, which in turn can shape the relative efficiency of spillover or ectopic release (6–8). Most computational models of neurotransmission assume an ECS width of 20 nm (9–11), although it is known that varying this parameter can significantly affect results (9, 12).
Analyzing how different substances diffuse through brain ECS reveals structural insights (1, 2, 13–20), but such measurements have yet to be used to estimate brain ECS dimensions. One strategy would be to measure the diffusion of a sequence of probe substances of different sizes: as the size approaches the width of the ECS, diffusion should become increasingly restricted. This concept was first applied in a biological setting by Pappenheimer et al. (21), where equivalent “pore” sizes were estimated from transcapillary diffusion measurements based on restricted diffusion (RD) theory. Subsequently, this method has been applied to many different systems, including human cervical mucus (22), cytoplasm (23), and concentrated agarose gels (24), but it has yet to be applied in the brain. Here we adapted the method of integrative optical imaging (IOI), originally described for diffusion measurements in brain slices (15), to determine effective diffusion coefficients (D*) of fluorescent probes spanning a large size range in the rat neocortex in vivo. We first determined the diffusion behavior of two Texas red-labeled polysaccharides: 3,000 Mr dextran (TR-dex3; Stokes–Einstein hydrodynamic diameter, dH = 3 nm) and 70,000 Mr dextran (TR-dex70; dH = 14 nm), for which there already exist IOI-derived data in brain slices (2, 15, 18, 19). Although dextrans of similar size have been used to estimate the dimensions of the glomerular filtration barrier (25) and diffusion paths within cytoplasm (23), use of dextrans with dH larger than ≈20 nm is complicated by conformational flexibility and polydispersity (23). We therefore used a quantum dot conjugate [polyethylene glycol-coated quantum dots with a cadmium selenide core tuned to emit at 655 nm (QD655); overall dH = 35 nm] to extend our measurements and explicitly test whether a substance larger than 20 nm could diffuse through brain ECS in vivo. Our results provide the first direct estimate of ECS width in the living mammalian brain.
Results
Free Diffusion Coefficients (D) and Stokes–Einstein Diameters.
D values were determined by IOI in dilute (0.3%) agarose (Fig. 1A), an essentially “free” medium (15). Fig. 2 A and B shows representative raw and background-subtracted IOI image sequences depicting QD655 diffusion in agarose. Fig. 2C shows typical Gaussian-shaped fluorescence intensity distributions, superimposed with theoretical fits (Eq. 4), along one of four axes through background-subtracted image sequences for QD655, TR-dex70, and TR-dex3. As expected, the time required for the diffusion curves to flatten and broaden is related to probe size (i.e., QD655 > TR-dex70 > TR-dex3). The hydrodynamic diameter (dH) of each probe was estimated from the Stokes–Einstein equation [dH = (kT)/3πηD, where k is Boltzmann’s constant, T is the absolute temperature, and η is the viscosity of water (6.9152 × 10−4 Pa·s at T = 310 K); see ref. 19] by using mean D values (Table 1).
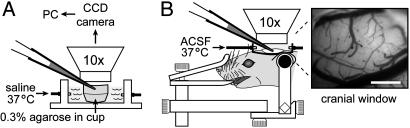
Fig. 1.
Experimental setup for IOI diffusion measurements. Successive images of fluorescent probe diffusion were captured by a cooled charge-coupled device (CCD) camera through a microscope with a ×10 water-immersion objective after pressure ejection from a micropipette into either dilute agarose (A) or somatosensory cortex (B) accessed through an open cranial window. (Scale bar: 500 μm.)
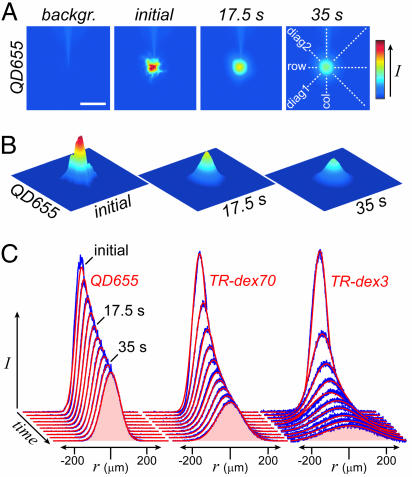
Fig. 2.
Free diffusion measurements. (A) Raw images obtained before (backgr.) and after ejection of QD655 into agarose. (Scale bar: 200 μm.) (B) Final images for analysis, shown in 3D representation, were obtained after subtraction of background fluorescence. (C) Fluorescence intensity profiles were extracted from each image along four axes (depicted in A at far right) and fit to the diffusion equation (see Materials and Methods). Representative data profiles (blue) superimposed with theoretical fits (red) are shown for QD655, TR-dex70, and TR-dex3 at 4.4-s intervals along the row axis. Fitting yielded the following values for D: 1.7 × 10−7 cm2·s−1, QD655; 4.3 × 10−7 cm2·s−1, TR-dex70; 2.3 × 10−6 cm2·s−1, TR-dex3.
Table 1.
Diffusion parameters for dextrans and quantum dots in dilute agarose and normoxic neocortex in vivo
| Molecule | D, 10−7 cm2·s−1 (n) | D*, 10−7 cm2·s−1 (n) | dH, nm |
|---|---|---|---|
| TR-dex3 | 22.2 ± 0.16 (35) | 5.36 ± 0.15 (38) | 2.95 ± 0.02 |
| TR-dex70 | 4.67 ± 0.061 (17) | 0.648 ± 0.044 (13) | 14.1 ± 0.2 |
| QD655 | 1.86 ± 0.049 (45) | 0.0167 ± 0.00068 (6) | 35.4 ± 0.9 |
Values were determined at 37 ± 0.5°C and are reported as mean ± SEM (n independent measurements).
Diffusion in Normoxic Neocortex in Vivo.
D* values were determined 200 μm below the pial surface (cortical layer II) in normoxic somatosensory cortex after the creation of an open cranial window in anesthetized rats (Fig. 1B). Representative image sequences after pressure ejection of TR-dex3 or TR-dex70 into agarose and brain are shown in Fig. 3A and C, respectively, along with fluorescence intensity distributions for TR-dex3 (Fig. 3B) and TR-dex70 (Fig. 3 D and E). The diffusion of each probe was clearly hindered in cortex relative to agarose. Longer times were necessary to visualize dispersion of TR-dex70 in cortex as compared with its diffusion in agarose or TR-dex3 diffusion in either medium. Image sequences (Fig. 4A) and intensity distributions (Fig. 4B) for QD655 revealed an even greater hindrance to diffusion in cortex relative to agarose. Slow diffusion of QD655 in cortex necessitated much longer observation times (typically ≥30 min) compared with TR-dex3 and TR-dex70, but values of D*, although low, were clearly >0 (Fig. 4C). Mean values of D and D* for each probe (Table 1) were used to calculate the tortuosity [λ = (D/D*)1/2], a measure of the hindrance experienced by substances diffusing through the ECS relative to a free medium (1, 2). Results for λ measured in normoxic neocortex are summarized in Fig. 5 together with published in vivo data (14) for the small tetramethylammonium (TMA) cation (74 Mr; dH = 0.51 nm; ref. 2) measured by using real-time iontophoresis and ion-selective microelectrodes (13). It is evident that λ increases markedly with increasing probe diameter in normoxic brain, consistent with RD theory.
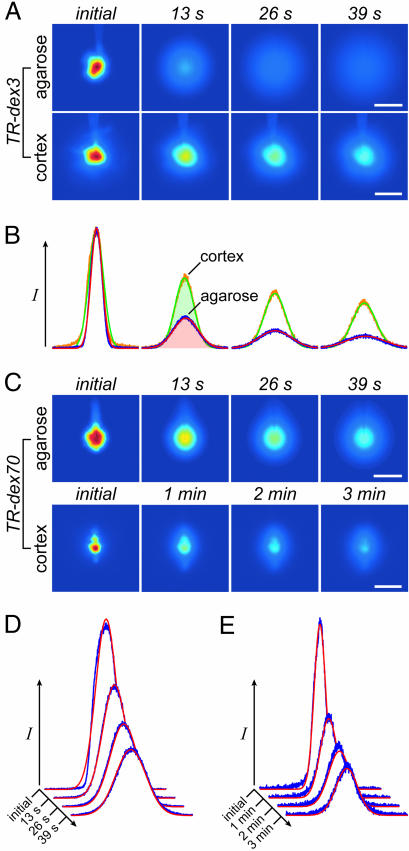
Fig. 3.
Dextran diffusion in normoxic neocortex. (A) Representative images after TR-dex3 ejection into agarose or cortex. (B) Fluorescence intensity profiles and theoretical fits for the images in A yielding D = 2.3 × 10−6 cm2·s−1 and D* = 4.5 × 10−7 cm2·s−1. (C) Representative images after TR-dex70 ejection. (D) Fluorescence intensity profiles and theoretical fits for agarose images in C yielding D = 4.7 × 10−7 cm2·s−1. (E) Profiles and fits for cortex images in C yielding D* = 5.7 × 10−8 cm2·s−1. (Scale bars: 200 μm.)
Fig. 4.
Quantum dot diffusion in normoxic neocortex. (A) Representative images after QD655 ejection into agarose or cortex. (Scale bars: 100 μm.) (B) Fluorescence intensity profiles and theoretical fits for the images in A yielding D = 1.9 × 10−7 cm2·s−1 and D* = 1.6 × 10−9 cm2·s−1. (C) Linear regression of cortex data in A and B; γi2 = 4D* (ti + t0), so regression of γi2/4 upon ti returns a slope of D* (see Materials and Methods).
Fig. 5.
Summary of in vivo λ (mean ± SEM) measured for extracellular probes in normoxic somatosensory cortex. TMA*, λ for TMA in normoxic rat cerebral cortex from Cserr et al. (14).
Diffusion in Neocortex After Terminal Ischemia.
Terminal ischemia follows cardiac arrest and rapidly subjects the brain to energy depletion, cessation of active transport, dissipation of transmembrane ion gradients, and the movement of water from the extra- to intracellular compartment (26). The result is a rapid reduction in ECS to ≈5% total brain volume (16, 20, 26). If ECS width decreases after terminal ischemia, RD theory would predict a reduction in D* that would reflect ECS narrowing. We therefore measured the diffusion of the smallest dextran, TR-dex3, after induction of terminal ischemia. Image sequences (Fig. 6A) and fluorescence intensity distributions (Fig. 6B) for TR-dex3 before and after terminal ischemia showed a marked slowing of diffusion 1 min after induction; D* decreased to just 5% of its normoxic value (terminal ischemia D* = 0.284 ± 0.058 × 10−7 cm2·s−1, n = 5; P < 0.001, Student’s t test) with a >4-fold increase in λ (Fig. 6C).
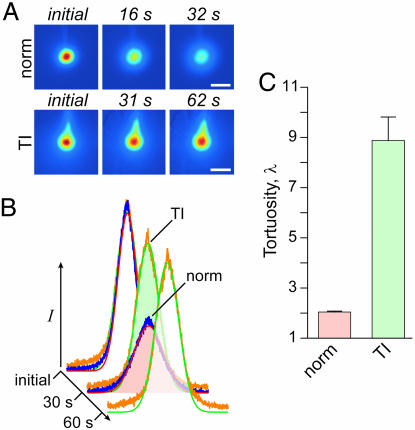
Fig. 6.
TR-dex3 diffusion is reduced in neocortex after terminal ischemia (TI). (A) Representative images after TR-dex3 ejection into normoxic (norm) cortex or 1 min after induction of TI. (Scale bars: 200 μm.) (B) Fluorescence intensity profiles and theoretical fits for the images in A yielding D* (norm) = 5.7 × 10−7 cm2·s−1 and D* (TI) = 3.4 × 10−8 cm2·s−1. (C) Values of λ (mean ± SEM) for TR-dex3 under the two conditions.
Fitting RD Models to the Data Predicts in Vivo ECS Width.
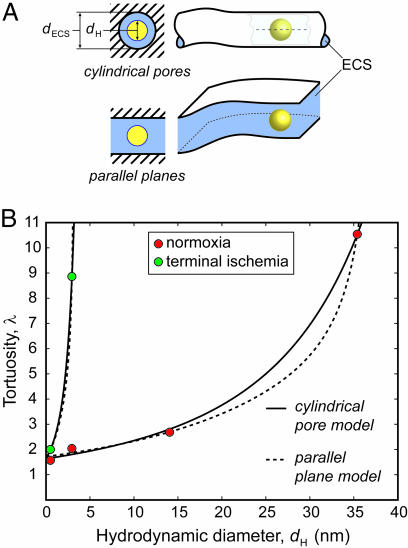
Our results show that much of the rat neocortical ECS under normoxic conditions in vivo must have a width >35 nm, the hydrodynamic diameter for QD655. In theory, a limit for ECS width might be established by injecting increasingly larger probes until diffusion ceased, if not for an obvious problem: as D* approaches 0, it will become increasingly difficult, in a practical time frame, to distinguish between very slow diffusion and no movement at all. Consequently, we estimated ECS width by applying RD theory to our existing data in normoxic brain. At present, the factors responsible for λ ≈ 1.6 for small probes such as TMA are thought to involve some combination of (i) an increased diffusion path length as molecules go around the obstructions imposed by cells (2, 27), (ii) transient retention in dead-space microdomains (18), and (iii) extracellular matrix interactions (20, 28). These factors will be dominant for small probes that explore long-range structure by relatively unrestricted diffusion. At the other extreme studied here, where larger probes generate values of λ significantly greater than 1.6, probe diffusion will be affected mainly by its local environment so that the pore structure of the ECS will dominate. In this regime, two factors will account for RD through pores relative to bulk solution (21, 29): (i) steric hindrance arising from the pore’s limited cross-sectional area and (ii) drag from the pore walls. For simplicity, we modeled the ECS as an isoporous environment using either cylindrical or parallel planar pores (Fig. 7A). Cylindrical pores in the ECS may arise from meshwork-forming extracellular matrix components, particularly hyaluronan and chondroitin sulfate proteoglycans (30, 31). Alternatively, ECS channels might simply be bounded by parallel planes formed by surrounding cell membranes (28). Building on more limited theoretical treatments (28, 32), we separated λ into two components:
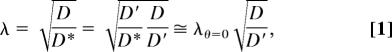 |
where D′ is the interstitial diffusion coefficient. For neutral, inert substances subject to purely steric interactions with pore walls, it can be shown (21, 29) that D′/D will depend on only channel geometry and θ, the ratio of probe diameter to brain ECS width (i.e., θ = dH/dECS), such that D′/D approaches 1 as θ approaches 0. Incorporating appropriate expressions for D′/D (29, 33) into Eq. 1 results in two free parameters: dECS and λθ=0, the λ for a vanishingly small molecule. Analytical expressions for D′/D (valid for 0 ≤ θ < 1) are available for cylindrical pore and parallel plane geometries, assuming axisymmetric diffusion of a sphere that remains on the centerline, a reasonable approximation (29). For cylindrical pore geometry (33)
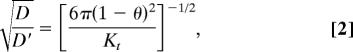 |
where
 |
For parallel plane geometry (29),
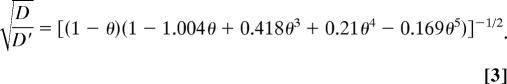 |
Fig. 7.
Model prediction of ECS width (dECS) from experimental λ and dH. (A) Cartoon depiction of models, both of which incorporate a centerline approximation for RD (shown within pore, upper right). (B) Best-fit curves for each model applied to normoxic tissue data, including TMA (14), and for TR-dex3 and TMA (16) after terminal ischemia. Best-fit parameters: cylindrical pore, normoxia: dECS = 63.8 nm, λθ=0 = 1.63, R = 0.97. Parallel plane, normoxia: dECS = 37.7 nm, λθ=0 = 1.72, R = 0.98. Cylindrical pore, ischemia: dECS = 5.7 nm, λθ=0 = 1.64. Parallel plane, ischemia: dECS = 3.2 nm, λθ=0 = 1.69.
Nonlinear least-squares fitting of Eq. 1, substituted with Eq. 2 or Eq. 3, to our results (Fig. 7B) led to independent estimates for dECS and λθ=0. RD theory fit the normoxic tissue data well for both models (correlation coefficient, R > 0.95), yielding dECS = 63.8 nm and λθ=0 = 1.63 for cylindrical pore geometry or dECS = 37.7 nm and λθ=0 = 1.72 for parallel planes. We included TMA data (14) along with our in vivo IOI data for the fitting, but this made little difference to the results (cylindrical pores without TMA: dECS = 64.2 nm and λθ=0 = 1.66; parallel planes without TMA: dECS = 37.8 nm and λθ=0 = 1.77). Model parameters obtained by fitting the TR-dex3 terminal ischemia data along with published TMA measurements under similar conditions (16) suggested a much reduced dECS < 10 nm (Fig. 7B).
Discussion
The major findings of the present study are (i) QD655, a quantum dot bioconjugate with dH = 35 nm, measurably diffuses within rat neocortical ECS in vivo, and (ii) application of RD theory to QD655 and dextran diffusion data indicate that the dECS is 38–64 nm, significantly larger than previous estimates based on EM, where 10- to 20-nm-wide clefts are typically seen (3, 4, 7, 34). Electrophysiological findings in the squid giant axon have suggested the existence of larger periaxonal spaces (≈80 nm) than those visualized in EM material (35), but our study provides the first in vivo evidence for larger spaces in a mammalian species.
Validation of IOI Diffusion Measurements.
The IOI method has already been applied extensively to measure the diffusion of proteins, dextrans, and other substances in agarose and brain slices (15, 17–19, 36–40). D values determined by IOI have generally been in good agreement with values obtained by other methods; in the present study, we obtained D (37°C) = 2.2 × 10−6 and 4.7 × 10−7 cm2·s−1 for TR-dex3 and TR-dex70, respectively. These values are very similar to those reported for D (corrected to 37°C) using dynamic light scattering [2.1 × 10−6 and 4.5 × 10−7 cm2·s−1 for 3,000 and 70,000 Mr dextran, respectively (41)], intrinsic viscosity measurements [5.1 × 10−7 cm2·s−1 for 73,000 Mr dextran (42)] and single-photon fluorescence recovery after photobleaching (FRAP) [4.6–5.9 × 10−7 cm2·s−1 for 71,000 Mr dextran (24, 43)]. Very few studies have used methods other than IOI to determine D* for macromolecules in brain tissue. However, studies in neostriatal brain slices have shown that the diffusion of 70,000 Mr dextran measured by using a multiphoton point source method (44) is remarkably similar to TR-dex70 diffusion measured by IOI in neocortical slices (15), with D* = 0.8 × 10−7 cm2·s−1 in both cases. Recently, a novel single-photon FRAP method for diffusion measurements at the brain surface of mice in vivo failed to reveal any size dependence in values of D*/D for three dextrans covering a range of 4,000–500,000 Mr (45). However, interpretation of this unexpected result is complicated by several factors, including uncertainties about single-photon bleaching distributions in tissue (46), problems with reversible photobleaching (47), and methodological issues that can lead to overestimation of diffusion coefficients (48). Indeed, the value of D* obtained for 70,000 Mr dextran at the cortical surface with this new method is 2.2 × 10−7 cm2·s−1 (49), more than 2-fold greater than D* values obtained in brain slices [≈0.9 × 10−7 cm2·s−1, corrected to 37°C (15, 44)] and more than 3-fold greater than D* measured in the present study (0.65 × 10−7 cm2·s−1). Comparison with in vitro IOI data (15, 19, 36, 39) shows our in vivo λ values for TR-dex3 (λ = 2.04 ± 0.04) and TR-dex70 (λ = 2.68 ± 0.11) to be slightly higher than those determined in neocortical slices (λ = ≈1.8 and 2.25 for TR-dex3 and TR-dex70, respectively). It is not possible to compare the QD655 data with other findings at this time because our study is the first, to our knowledge, to measure the diffusion of any quantum dot in biological tissue.
Comparison with ECS Width Estimates from EM.
Although quantitative EM techniques have often been used to estimate the average width of brain ECS (7, 34, 50), controversy has always existed over how accurately EM preserves the in vivo appearance and size of ECS (4). The smallest widths (≤20 nm) have been estimated from tissue conventionally perfused with aldehyde fixatives and/or osmium tetroxide before dehydration and embedding for EM (4, 7, 34). Although such methods yield excellent cellular morphology and insights into fine ultrastructural details that IOI cannot provide, the distribution of extracellular water is likely compromised because the tissue undergoes terminal ischemia before fixation, leading to ECS contraction (4). Indeed, many early EM studies exhibited a near-complete obliteration of the ECS. Pioneering work by Brightman and others finally established that much of the ECS consists of hydrated channels at least ≈10 nm in width because EM showed that tracers of roughly that size (e.g., ferritin, saccharated iron oxide, and horseradish peroxidase) distributed within brain parenchyma after their introduction into CSF or nearby tissue in living animals (51–53). EM methods using rapid freezing of the superficial ≈10 μm of tissue followed by freeze substitution (50) suggested a larger ECS width (≈40 nm) but poor cellular morphology and the formation of ice crystals complicated interpretation (4). Here we estimated that dECS fell to <10 nm in less than 5 min after terminal ischemia was induced. Although this result is based on only two tracers (TR-dex3 and TMA), its similarity to the ECS gap size routinely visualized by conventional EM provides further evidence that ischemia compromises ECS width in such material (4, 5).
Significance for Nanoparticle Applications in the CNS.
Brain ECS is the conduit through which drugs and drug delivery vectors must diffuse after crossing the blood–brain barrier (54) and consequently forms a critical element of the neurovascular unit (55). Many vectors under investigation, e.g., immunoliposomes and polymer nanoparticles, have sizes >100 nm (55). Our results suggest that these vectors will be too large to transit normal neocortical ECS. We emphasize that, although dECS establishes an upper size limit for transport in brain ECS, diffusion will slow dramatically as the limit is approached. Furthermore, a reduction in dECS with disease or injury, such as that after ischemia, will further restrict interstitial diffusion. Finally, the use of quantum dot bioconjugates for in vivo imaging is a rapidly developing area of nanotechnology (56, 57). Surface modification of quantum dots to make them hydrophilic results in relatively large probes (56), so their diffusion properties in biological tissues will likely dictate their application. Other quantum dot conjugates with different emission spectra and surface chemistry, particularly those similar to or smaller in size than QD655, may be useful for future studies in the CNS.
Significance for CNS Physiology.
It is expected that a larger ECS width will result in decreased lateral diffusion of neurotransmitter as well as a lower extracellular concentration per quantum of transmitter released. It will be important to consider the effect of incorporating our estimate of dECS into models comparing neurotransmitter spillover from the synapse versus ectopic release for the activation of extrasynaptic receptors (6, 7). Simulations have shown ECS width can affect total synaptic current (12) as well as extracellular calcium fluctuations resulting from local influx during neural activity (9).
Our data do not imply that all ECS has the width estimated here, only that a sufficiently large component of the neocortical ECS is well connected by channels of this size so as to allow approximately globular, rigid substances as large as 35 nm, but not greater than 64 nm, to diffuse uniformly and access most of the tissue. The diffusion of large, flexible, linear molecules in hindered media is likely to be different (38) and may be governed by mechanisms other than RD, e.g., reptation (24) or entropic barriers transport (58). Our dECS estimate may be representative of nonneocortical areas where the ECS also occupies ≈20% of the total tissue volume (1, 2), but other areas, e.g., midbrain (17), will likely be different. Variations in ECS width among brain regions will become apparent only after further study.
Our results demonstrate that the behavior of large dextrans and quantum dots diffusing within neocortical ECS may be described by approximating interstitial channels as interconnected fluid phase pores. The available data are insufficient to distinguish between cylindrical and planar pore models at this time; we speculate that the actual ECS microstructure may be a hybrid between the two because of the presence of extracellular matrix components (30, 31). In a simple situation, the ECS width we have estimated using RD theory will equate with the space between cells, but if the extracellular matrix plays a significant role in ECS microstructure the actual membrane separation may exceed dECS. From the practical perspective of a diffusing macromolecule or nanoparticle, however, dECS will be the parameter of importance.
Materials and Methods
Animal Preparation.
Experiments were carried out at the New York University School of Medicine in accordance with National Institutes of Health guidelines and local Institutional Animal Care and Use Committee regulations. Female Sprague–Dawley rats (160–260 g) were anesthetized with urethane (1.5 g·kg−1 i.p.), tracheotomized, and placed in a three-point head holder (Narishige) for preparation of an open cranial window over the left parietal cortex. Atropine sulfate (0.05 mg·kg−1·h−1 s.c.) was administered to diminish bronchial secretions, and body fluids were supplemented with physiological saline as needed (≤5 ml·kg−1·h−1 i.p.). Body temperature was maintained at 37°C, and heart rate/ECG were continuously monitored. A custom fabricated chamber (15-mm inside diameter polypropylene ring fitted with inlet and outlet ports) was affixed to the skull with cyanoacrylate. An ≈3- × 4-mm craniotomy was then performed over the barrel field and trunk region of the primary somatosensory area, and artificial cerebrospinal fluid [composition and osmolality as reported previously (19), except 3 mM KCl] was superfused (2 ml·min−1) via the chamber ports. After careful removal of the dura, animals were transferred to an Olympus BX61WI microscope and clamped to a fixed Gibraltar stage (Burleigh) for imaging experiments after a 1- to 2-h equilibration period. For terminal ischemia measurements, some animals were later administered 1 ml of 1 M KCl intracardially, causing immediate cardiac arrest.
Diffusion Measurements.
We used the IOI method (15, 19), modified for in vivo application. This method employs epifluorescence microscopy with quantitative image analysis to measure fluorescent probe diffusion after a 50- to 200-ms pressure ejection from a micropipette (tip diameters 3–6 μm), approximating a point source (Fig. 1). The projection of the 3D cloud of diffusing probe on the 2D image plane of the camera is described by
and
where Ii is the fluorescence intensity of the ith image at radial distance r from the source in brain and Ei is an expression incorporating the defocused point spread function of the microscope objective (15). A time offset, t0, added to the measured time from the injection, ti, allows the use of a point source description even when a finite initial volume is released (38). Average ejected volumes of 25–50 pl have been reported for IOI (17). Measurements of TMA diffusion by real-time iontophoresis have shown that neither the presence of an IOI micropipette nor the small volumes typically ejected for IOI measurements affect the local ECS volume fraction (19). Eq. 4 is fitted to the data by using a nonlinear algorithm at a succession of times, ti, yielding a sequence of estimates for γi(ti). Linear regression of γi2/4 upon ti gives a slope of D* (or D) using Eq. 5.
TR-dex3 and TR-dex70 (Molecular Probes) were used at concentrations of 1 and 0.1 mM, respectively, in a solution of 154 mM NaCl, and QD655 (Qtracker 655 nontargeted quantum dots surface-coated with methoxy-5,000 Mr polyethylene glycol; Invitrogen) was used as formulated [2 μM in 50 mM borate buffer (pH 8.3)]. QD655 solution tonicity was expected to rapidly equilibrate with interstitial fluid and not affect QD655 diffusion measured over the long observation times used in this study. Measurements of D were performed at 37 ± 0.5°C in either 0.3% agarose made up in 154 mM NaCl (for TR-dex3 and TR-dex70) or 0.2–0.3% agarose made up in PBS (for QD655). No differences were observed between measurements using Isogel agarose (Cambrex Bio Science) or NuSieve GTG agarose (FMC) with TR-dex3 or 0.2–0.3% NuSieve GTG agarose with QD655, so data were pooled for reported D values. All TR-dex70 measurements were performed in Isogel agarose. For determination of D* in brain, artificial cerebrospinal fluid was superfused at 37 ± 0.5°C in the chamber by using an in-line solution heater (Warner Instruments). Mean D* values represent at least four animals per probe for each experimental condition. All data are reported as mean ± SEM.
Acknowledgments
We thank S. Hrabětová, A. Lakkaraju, and L. Tao for comments on an earlier version of the manuscript. This work was supported by National Institutes of Health Grant R01-NS28642 (to C.N.).
Abbreviations
- λ
tortuosity
- D
free diffusion coefficient
- D*
effective diffusion coefficient
- dH
Stokes–Einstein hydrodynamic diameter
- ECS
extracellular space
- IOI
integrative optical imaging
- QD655
polyethylene glycol-coated quantum dots emitting at 655 nm
- RD
restricted diffusion
- TMA
tetramethylammonium
- TR-dex3
Texas red-labeled 3,000 Mr dextran
- TR-dex70
Texas red-labeled 70,000 Mr dextran
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nicholson C., Syková E. Trends Neurosci. 1998;21:207–215. doi: 10.1016/s0166-2236(98)01261-2. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson C. Rep. Prog. Phys. 2001;64:815–884. [Google Scholar]
- 3.Peters A., Palay S. L., Webster H. D. The Fine Structure of the Nervous System. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 4.Van Harreveld A. In: The Structure and Function of Nervous Tissue. Bourne G. H., editor. New York: Academic; 1972. pp. 447–511. [Google Scholar]
- 5.Van Harreveld A., Malhotra S. K. J. Anat. 1967;101:197–207. [PMC free article] [PubMed] [Google Scholar]
- 6.Coggan J. S., Bartol T. M., Esquenazi E., Stiles J. R., Lamont S., Martone M. E., Berg D. K., Ellisman M. H., Sejnowski T. J. Science. 2005;309:446–451. doi: 10.1126/science.1108239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui K., Jahr C. E., Rubio M. E. J. Neurosci. 2005;25:7538–7547. doi: 10.1523/JNEUROSCI.1927-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsui K., Jahr C. E. Neuron. 2003;40:1173–1183. doi: 10.1016/s0896-6273(03)00788-8. [DOI] [PubMed] [Google Scholar]
- 9.Egelman D. M., Montague P. R. Biophys. J. 1999;76:1856–1867. doi: 10.1016/s0006-3495(99)77345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franks K. M., Bartol T. M., Jr, Sejnowski T. J. Biophys. J. 2002;83:2333–2348. doi: 10.1016/S0006-3495(02)75248-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen T. A., DiGregorio D. A., Silver R. A. Neuron. 2004;42:757–771. doi: 10.1016/j.neuron.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Savtchenko L. P., Antropov S. N., Korogod S. M. Biophys. J. 2000;78:1119–1125. doi: 10.1016/S0006-3495(00)76670-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson C., Phillips J. M. J. Physiol. 1981;321:225–257. doi: 10.1113/jphysiol.1981.sp013981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cserr H. F., DePasquale M., Nicholson C., Patlak C. S., Pettigrew K. D., Rice M. E. J. Physiol. 1991;442:277–295. doi: 10.1113/jphysiol.1991.sp018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson C., Tao L. Biophys. J. 1993;65:2277–2290. doi: 10.1016/S0006-3495(93)81324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voříšek I., Syková E. J. Cereb. Blood Flow Metab. 1997;17:191–203. doi: 10.1097/00004647-199702000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Cragg S. J., Nicholson C., Kume-Kick J., Tao L., Rice M. E. J. Neurophysiol. 2001;85:1761–1771. doi: 10.1152/jn.2001.85.4.1761. [DOI] [PubMed] [Google Scholar]
- 18.Hrabětová S., Hrabe J., Nicholson C. J. Neurosci. 2003;23:8351–8359. doi: 10.1523/JNEUROSCI.23-23-08351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorne R. G., Hrabětová S., Nicholson C. J. Neurophysiol. 2004;92:3471–3481. doi: 10.1152/jn.00352.2004. [DOI] [PubMed] [Google Scholar]
- 20.Syková E. Neuroscience. 2004;129:861–876. doi: 10.1016/j.neuroscience.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 21.Pappenheimer J. R., Renkin E. M., Borrero L. M. Am. J. Physiol. 1951;167:13–46. doi: 10.1152/ajplegacy.1951.167.1.13. [DOI] [PubMed] [Google Scholar]
- 22.Saltzman W. M., Radomsky M. L., Whaley K. J., Cone R. A. Biophys. J. 1994;66:508–515. doi: 10.1016/s0006-3495(94)80802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luby-Phelps K., Taylor D. L., Lanni F. J. Cell Biol. 1986;102:2015–2022. doi: 10.1083/jcb.102.6.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pluen A., Netti P. A., Jain R. K., Berk D. A. Biophys. J. 1999;77:542–552. doi: 10.1016/S0006-3495(99)76911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deen W. M., Bridges C. R., Brenner B. M., Myers B. D. Am. J. Physiol. 1985;249:F374–F389. doi: 10.1152/ajprenal.1985.249.3.F374. [DOI] [PubMed] [Google Scholar]
- 26.Hansen A. J. Physiol. Rev. 1985;65:101–148. doi: 10.1152/physrev.1985.65.1.101. [DOI] [PubMed] [Google Scholar]
- 27.Tao L., Nicholson C. J. Theor. Biol. 2004;229:59–68. doi: 10.1016/j.jtbi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Rusakov D. A., Kullmann D. M. Proc. Natl. Acad. Sci. USA. 1998;95:8975–8980. doi: 10.1073/pnas.95.15.8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deen W. M. AIChE J. 1987;33:1409–1425. [Google Scholar]
- 30.Scott J. E., Cummings C., Brass A., Chen Y. Biochem. J. 1991;274:699–705. doi: 10.1042/bj2740699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi Y. Cell. Mol. Life Sci. 2000;57:276–289. doi: 10.1007/PL00000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.el-Kareh A. W., Braunstein S. L., Secomb T. W. Biophys. J. 1993;64:1638–1646. doi: 10.1016/S0006-3495(93)81532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bungay P. M., Brenner H. Int. J. Multiphase Flow. 1973;1:25–56. [Google Scholar]
- 34.Rusakov D. A., Kullmann D. M. J. Neurosci. 1998;18:3158–3170. doi: 10.1523/JNEUROSCI.18-09-03158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Astion M. L., Coles J. A., Orkand R. K., Abbott N. J. Biophys. J. 1988;53:281–285. doi: 10.1016/S0006-3495(88)83090-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao L., Nicholson C. Neuroscience. 1996;75:839–847. doi: 10.1016/0306-4522(96)00303-x. [DOI] [PubMed] [Google Scholar]
- 37.Tao L. J. Neurophysiol. 1999;81:2501–2507. doi: 10.1152/jn.1999.81.5.2501. [DOI] [PubMed] [Google Scholar]
- 38.Prokopová-Kubinová S., Vargová L., Tao L., Ulbrich K., Scaron;ubr V., Syková E., Nicholson C. Biophys. J. 2001;80:542–548. doi: 10.1016/S0006-3495(01)76036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hrabětová S. Hippocampus. 2005;15:441–450. doi: 10.1002/hipo.20068. [DOI] [PubMed] [Google Scholar]
- 40.Thorne R. G., Hrabětová S., Nicholson C. Nat. Mater. 2005;4:713. doi: 10.1038/nmat1489. [DOI] [PubMed] [Google Scholar]
- 41.Shao J., Baltus R. E. AIChE J. 2000;46:1149–1156. [Google Scholar]
- 42.Armstrong J. K., Wenby R. B., Meiselman H. J., Fisher T. C. Biophys. J. 2004;87:4259–4270. doi: 10.1529/biophysj.104.047746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arrio-Dupont M., Cribier S., Foucault G., Devaux P. F., d’Albis A. Biophys. J. 1996;70:2327–2332. doi: 10.1016/S0006-3495(96)79798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stroh M., Zipfel W. R., Williams R. M., Webb W. W., Saltzman W. M. Biophys. J. 2003;85:581–588. doi: 10.1016/S0006-3495(03)74502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binder D. K., Papadopoulos M. C., Haggie P. M., Verkman A. S. J. Neurosci. 2004;24:8049–8056. doi: 10.1523/JNEUROSCI.2294-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown E. B., Wu E. S., Zipfel W., Webb W. W. Biophys. J. 1999;77:2837–2849. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seksek O., Biwersi J., Verkman A. S. J. Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luby-Phelps K. Int. Rev. Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 49.Papadopoulos M. C., Kim J. K., Verkman A. S. Biophys. J. 2005;89:3660–3668. doi: 10.1529/biophysj.105.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trubatch J., Loud A. V., Van Harreveld A. Neuroscience. 1977;2:963–974. [Google Scholar]
- 51.Brightman M. W. Am. J. Anat. 1965;117:193–219. doi: 10.1002/aja.1001170204. [DOI] [PubMed] [Google Scholar]
- 52.Pappas G. D., Purpura D. P. Nature. 1966;210:1391–1392. doi: 10.1038/2101391a0. [DOI] [PubMed] [Google Scholar]
- 53.Brightman M. W., Reese T. S. J. Cell Biol. 1969;40:648–677. doi: 10.1083/jcb.40.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pardridge W. M. Neuron. 2002;36:555–558. doi: 10.1016/s0896-6273(02)01054-1. [DOI] [PubMed] [Google Scholar]
- 55.Begley D. J. Pharmacol. Ther. 2004;104:29–45. doi: 10.1016/j.pharmthera.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Michalet X., Pinaud F. F., Bentolila L. A., Tsay J. M., Doose S., Li J. J., Sundaresan G., Wu A. M., Gambhir S. S., Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larson D. R., Zipfel W. R., Williams R. M., Clark S. W., Bruchez M. P., Wise F. W., Webb W. W. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 58.Nykypanchuk D., Strey H. H., Hoagland D. A. Science. 2002;297:987–990. doi: 10.1126/science.1073277. [DOI] [PubMed] [Google Scholar]