Abstract
The biosynthetic pathway of l-tartaric acid, the form most commonly encountered in nature, and its catabolic ties to vitamin C, remain a challenge to plant scientists. Vitamin C and l-tartaric acid are plant-derived metabolites with intrinsic human value. In contrast to most fruits during development, grapes accumulate l-tartaric acid, which remains within the berry throughout ripening. Berry taste and the organoleptic properties and aging potential of wines are intimately linked to levels of l-tartaric acid present in the fruit, and those added during vinification. Elucidation of the reactions relating l-tartaric acid to vitamin C catabolism in the Vitaceae showed that they proceed via the oxidation of l-idonic acid, the proposed rate-limiting step in the pathway. Here we report the use of transcript and metabolite profiling to identify candidate cDNAs from genes expressed at developmental times and in tissues appropriate for l-tartaric acid biosynthesis in grape berries. Enzymological analyses of one candidate confirmed its activity in the proposed rate-limiting step of the direct pathway from vitamin C to tartaric acid in higher plants. Surveying organic acid content in Vitis and related genera, we have identified a non-tartrate-forming species in which this gene is deleted. This species accumulates in excess of three times the levels of vitamin C than comparably ripe berries of tartrate-accumulating species, suggesting that modulation of tartaric acid biosynthesis may provide a rational basis for the production of grapes rich in vitamin C.
Keywords: idonate dehydrogenase, transcriptional profiling, organic acid, biosynthesis, Vitis vinifera
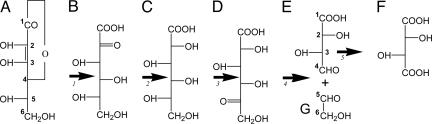
L-Tartaric acid (TA) (1) is the primary nonfermentable soluble acid in grapes and the principal acid in wine (2), contributing important aspects to the taste, mouthfeel, and aging potential of the vinified product. Moreover, exogenous TA is used as a flavorant and antioxidant for a range of foods and beverages, including grape juice and wine. Many plants make TA (3), yet in contrast to the fruit acids malic and citric, pioneering work in the 1950s showed that the metabolic origin of TA lay outside the oxidative metabolism of sugars (4). Uniquely for a fruit acid, TA biosynthesis begins with l-ascorbic acid (AA, vitamin C) (5). A key step in TA formation is the cleavage of a six-carbon intermediate between either position C2/C3 or C4/C5, depending on the plant species (6). The former reaction yields oxalic acid (OxA) (7) and l-threonate (8), which in plants of the Geraniaceae is then converted to TA in the leaves (7), whereas the latter “direct pathway” yields TA and a two-carbon compound, possibly glycoaldehyde, and is the pathway proposed to predominate in the Vitaceae (5). It was recently shown using intact berries of Vitis vinifera that OxA and TA are formed by AA catabolism in these tissues, suggesting that, in the grapevine, both pathways are functional (9). In grape berries, TA biosynthesis is limited to the period of development from postanthesis until the onset of ripening or “veraison” (10, 11).
Radioisotope tracer studies reveal that catabolism of AA to TA in the direct pathway (5, 9, 10, 12–18) proceeds via the conversion of AA to 2-keto l-idonic acid, with successive reduction to l-idonic acid (12, 14) and oxidation to 5-keto d-gluconic acid (10, 12, 14). In the penultimate step, 5-keto d-gluconic acid is cleaved between carbons 4 and 5 to yield the four-carbon l-threo-tetruronate (13), which is oxidized to form TA. It is proposed that a glycoaldehyde is formed from the remaining two-carbon fragment (15) (Fig. 1). Kinetic analyses using 14C-labeled intermediates suggest that oxidation of idonate to 5-keto d-gluconic acid (Fig. 1, step 3) is the rate limiting step in this pathway (7, 12, 17).
Fig. 1.
Proposed pathway for TA formation via AA in the grapevine (7). Compounds identified by letters are: A, AA; B, 2-keto l-gulonic acid; C, l-idonic acid; D, 5-keto d-gluconic acid; E, l-threo-tetruronate (13); F, TA; G, glycoaldehyde (15). Italicized numbers denote specific reactions referred to in text.
Despite having established the nature of chemical intermediates in TA biosynthesis, none of the enzymes responsible for the proposed reactions have been identified. Moreover, attempts to identify the corresponding enzyme activities in crude cell extracts have been unsuccessful. Cleavage of 5-keto d-gluconic acid (Fig. 1, step 4) was proposed to be nonenzymic because of its close kinetic correlation with respect to 14C conversion into TA (14), although this work was focused more closely on examining 14C flux rather than enzymatic activities. This hypothesis was subsequently refuted when 18O tracer experiments suggested the cleavage mechanism to involve a putative hydrolase (18). More recently (19), it has been shown that, in Gluconobacter suboxidans, 5-keto d-gluconic acid can be used as the substrate for a dedicated transketolase. These authors speculated that further oxidation of the resulting four-carbon semialdehyde by a succinate semialdehyde dehydrogenase homolog could result in formation of TA. The paucity of specific biochemical and molecular data pertaining to the mechanism and regulation of TA biosynthesis in grapevines is surprising, given the multibillion dollar value of the crop and the close ties between TA and AA metabolism.
The biosynthetic pathway for AA has only recently been solved (20), and a renaissance of interest has occurred with various ways of increasing vitamin C accumulation proposed (21), such as overexpressing a key enzyme in a different pathway (22) or by manipulating enzymes involved in its recycling (23). TA biosynthesis represents an unusual fate for AA, and its accumulation to high levels in ripe berries (≈7 mg·g−1 fresh weight, ref. 24) suggests a highly active metabolic pathway that may compete for AA with those redox-associated functions more usually linked with in vivo AA pools. Here we combine transcript profiling of EST data and metabolite profiling of organic acids to identify the gene and characterize the enzyme for the proposed rate-limiting step in TA biosynthesis from vitamin C. We also identify a naturally occurring grape species that lacks this gene. The species, Ampelopsis aconitifolia, not only lacks detectable TA but hyperaccumulates vitamin C (AA) compared to related TA-forming species. These results are consistent with the proposed in vivo function of this gene and suggest modulation of TA biosynthesis as a means to increase vitamin C in plants.
Results
Identification of Candidate Genes by EST Transcriptional Profiling.
Preliminary experimental work to identify and isolate enzymatic activity capable of promoting reactions within the pathway of TA formation was unsuccessful (data not shown). In particular, soluble protein extracts prepared from immature berries did not contain measurable levels of enzymes capable of cleaving 5-keto d-gluconate to a four-carbon intermediate, and similar results were found when other intermediates in the pathway were tested. Therefore, we adopted a molecular approach to identify candidate genes for tartrate biosynthetic enzymes. The search for genes and enzymes for this pathway is complicated by the absence of TA formation in the primary plant model, Arabidopsis thaliana, and the absence of simple molecular-genetic tools in the grapevine. In lieu of genetic analysis, we interrogated transcript data compiled from an extensive EST collection that relates the expression of thousands of Vitis genes to many tissues and developmental stages of the grapevine. Transcripts that were differentially expressed in TA-accumulating tissues, including preveraison berries and young leaves, and which were represented by at least six ESTs per contig (see Materials and Methods), formed a set of 87 candidates for further analysis (Table 2, which is published as supporting information on the PNAS web site).
Initial gene annotations obtained from tblastx and interpro scan (25) analyses of the 87 candidates were suggestive of a variety of metabolic processes. Candidate selection was further refined by searching conceptually translated candidate sequences for similarity to domains associated with oxido-reductase activity, the reaction chemistry proposed for at least three of the conversions from AA to TA (Fig. 1). A subset of four candidate genes was selected based on these combined expression and protein homology criteria and further tested for likely involvement with TA biosynthesis using a combination of molecular and biochemical analyses.
Characterization of the Natural Variation in Tartaric Acid: Family Vitaceae.
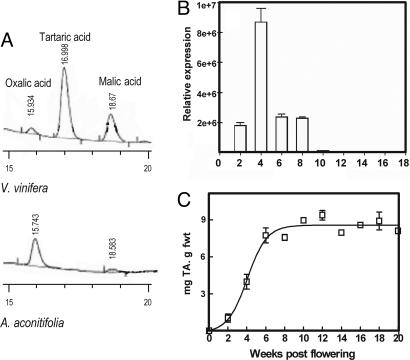
In model plants such as Arabidopsis, the function of candidate genes can often be tested based on overexpression or RNAi phenotypes. However, grapevines remain recalcitrant to facile transformation, and none of the model plants accumulate tartaric acid in their fruit; instead, we sought to correlate the expression of candidate genes with TA metabolism. Toward this end, we characterized the organic acid profiles in berries collected from 28 species of the family Vitaceae, representing four genera with natural distributions across four continents. Among the organic acids surveyed, TA and OxA are metabolic breakdown products of vitamin C (9), whereas the biosynthesis of malic and succinic acids is linked to primary sugar metabolism. All acids measured are important to grape and wine chemistry. As shown in Fig. 5, which is published as supporting information on the PNAS web site, a wide range of acid accumulation was detected across the species surveyed. In particular, tartaric acid levels in berries ranged from a high of 15 mg per g fresh weight, to three of the 28 species with <1.5 mg per g fresh weight. Only one species, A. aconitifolia, contained no detectable tartaric acid (Fig. 2A). Berries of A. aconitifolia also contained high levels of OxA (≈1.8 mg per gram fresh berry weight, Fig. 5), whereas malic acid levels were among the lowest recorded across the 28 species (≈2 mg per gram fresh berry weight). The accumulation of tartaric acid was further assayed in young leaves of this species, and in agreement with data obtained from berries, tartaric acid could not be detected (data not shown). The identification of a species of grapevine in which tartaric acid accumulation was not detected suggested its possible utility for the verification of candidate tartaric acid biosynthetic genes.
Fig. 2.
Identification of a candidate TA biosynthetic gene. (A) HPLC of organic acids from V. vinifera (Upper) and A. aconitifolia (Lower). (B) Quantitative RT-PCR analysis of candidate gene mRNA levels measured every 2 weeks during berry development in berries of V. vinifera relative to expression of ubiquitin with mRNA levels standardized to the same concentration. (C) Accumulation of TA during development of berries from V. vinifera. Bars show standard error, n = 3.
Candidate Gene Verification by PCR.
RT-PCR assays were used to compare the transcript abundance of TA-biosynthesis candidate genes between TA-accumulating species and the non-TA accumulating A. aconitifolia. Transcripts of one candidate gene, contig 1029130, were absent from berries of A. aconitifolia, but present in the tissues and at the times when TA biosynthesis occurs in V. vinifera (Fig. 2B), Vitis californica, and Ampelopsis brevipedunculata (data not shown). Transcript accumulation of the remaining candidate genes was not correlated with TA biosynthesis. PCR analysis of genomic DNA with 1029130-specific primers efficiently amplified fragments from species of Vitis, Parthenocissus, and Ampelopsis, but not from A. aconitifolia (data not shown). The potential for a mismatch or point mutation causing the lack of amplification from A. aconitifolia was addressed by designing four sets of primers, covering the entire ORF. None of these primer pairs nor those spanning the full-length gene resulted in amplification from genomic DNA of A. aconitifolia, although all worked efficiently on DNA from other grape genera, as well as related species of Ampelopsis (data not shown). Quantitative RT-PCR, measuring changes in transcript accumulation every 2 weeks during berry development, revealed that 1029130 was strongly transcribed in the preveraison V. vinifera berry, with a distinct peak of transcription 2–6 weeks after flowering (Fig. 2B). This pattern of transcription is consistent with an exponential increase in TA accumulation during the early stage of V. vinifera grape berry development (Fig. 2C).
In viticultural practice, management of canopy structure and leaf area provides a means to modulate incident light on berry clusters and thereby impact berry metabolism. We measured the effect of light on organic acid accumulation and 1029130 transcript levels by shading individual clusters during early development. The results reveal strong reductions to TA pools in shaded clusters compared to untreated controls (Fig. 6A, which is published as supporting information on the PNAS web site) and corresponding decreases in 1021930 transcript (Fig. 6B). By contrast, levels of malate and oxalate were unaffected by the shading treatment (Fig. 6C).
Protein Studies and Enzymology.
The deduced 366-aa, 40.1-kDa protein encoded within contig 1029130 shares 77% amino acid sequence identity with putative NAD+-linked sorbitol dehydrogenases from a number of plant species. Homology analysis revealed global similarity to COG1063, a family of zinc-dependent dehydrogenase proteins that includes an Escherichia coli protein (P39346), which catalyses the reversible oxidation of l-idonate to 5-keto d-gluconic acid within the gluconic acid metabolism pathway (26). This reaction occurs also within the TA biosynthetic pathway (Fig. 1, step 3); thus, the identification of a homologous gene expressed in coincidence with TA accumulation was of great interest. Although these two proteins exhibit only 31% sequence identity at the amino acid level, functional classification by means of svmprot (http://jing.cz3.nus.sg/cgi-bin/svmprot.cgi) indicated an 87% likelihood that the encoded protein belonged to the class of oxidoreductase enzymes acting on the CH-OH group of donors (27). Further evidence that the protein encoded within contig 1029130 was active in the conversion of l-idonate to 5-keto d-gluconate was obtained from in vitro assay of its activity, as described below.
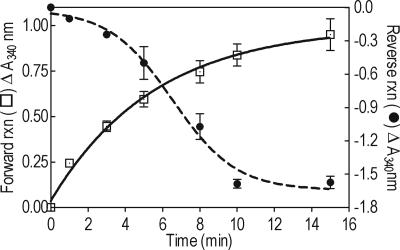
Overexpression of the candidate gene in E. coli and purification of the soluble recombinant protein yielded a ≈40-kDa product when examined by SDS/PAGE (data not shown). The ability of the purified recombinant protein to catalyze the interconversion of l-idonate and 5-keto d-gluconate was assayed by spectrophotometry in a simple NAD+-coupled reaction. The purified protein showed high catalytic activity for l-idonate and, in the reverse reaction with NADH as the cofactor, for 5-keto d-gluconate as substrates (Fig. 3). No activity was observed if NADH was used as the cofactor with l-idonate, or if NAD+ was used as the cofactor with 5-keto d-gluconate; furthermore, no activity was observed for either forward or reverse reactions with NADP+ or NADPH, respectively, as the cofactor (Fig. 7, which is published as supporting information on the PNAS web site). d-gluconic acid and 2-keto l-gulonic acid (Fig. 1-2) (intermediates proposed to be involved in TA biosynthetic pathways) and AA (Fig. 1-1) were also tested as substrates with NAD+ as cofactor; each displayed very low rates of NADH formation (Fig. 7). In additional experiments, HPLC was used to separate the stopped reaction mixes; a single peak, formed in the reverse reaction (used because there was no coelution with compounds in the reaction mixture), cochromatographed with authentic l-idonate and was absent in all controls (Fig. 8, which is published as supporting information on the PNAS web site). LC-mass spectrometry (MS) was used to confirm that the mass of the reaction product matched that of authentic l-idonate (data not shown). As stated above, the recombinant protein, which we have named l-idonate dehydrogenase (l-IdnDH), is most similar in amino acid sequence to sorbitol dehydrogenase; when sorbitol was tested as a substrate, NAD+ reduction occurred but at a very low rate (Fig. 7). We suggest that l-IdnDH belongs to the sorbitol dehydrogenase family of dehydrogenases, with principal activity against l-idonate, representing a characterized enzyme associated with TA biosynthesis in higher plants. Kinetic analysis of the purified recombinant enzyme suggested a Km for l-idonate in the forwards reaction of 2.2 mM, and for 5-keto d-gluconate in the reverse reaction of 12.5 mM (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 3.
The activity of recombinant V. vinifera l-IdnDH. Forward reaction l-idonate to 5-keto d-gluconate as indicated by an open square. Reverse reaction, 5-keto d-gluconate to l-idonate as indicated by a filled circle. Reactions were followed by oxidation and reduction of NADH and NAD, respectively, and measured as change in absorbance at 340 nm. Bars show standard error, n = 3.
In Vivo Studies of Enzymatic Activity.
Earlier evidence from radioisotope tracer studies in grape berries showed that 14C radiolabel from both l-idonate and 5-keto d-gluconate was incorporated into TA (10, 12). To investigate and confirm the involvement of l-idonate and 5-keto d-gluconate in TA biosynthesis, we fed 100 mM concentrations of these intermediates to slices of grape berries from V. vinifera and A. aconitifolia; this resulted in a drop in the pH of the feeding solutions from pH 7.0 to 5.5 and 6.2 when 100 mM l-idonate and 5-keto d-gluconate were added, respectively. In both species, uptake of the applied intermediates into the washed berry slices was confirmed by HPLC (data not shown). Berry slices from V. vinifera were able to take up both substrates and rapidly accumulate TA; however, no formation of TA was detected in berry slices prepared from A. aconitifolia when incubated with either intermediate (Table 1). The product formed in V. vinifera was confirmed as TA by HPLC and LC-MS (data not shown). In berry slices of V. vinifera, 10 mg more TA per g of berry slice was formed after 3 h of incubation with 5-keto d-gluconate compared to the control without substrate addition (H2O).
Table 1.
Change in tartaric acid concentration (mg·g−1) in V. vinifera berry slices incubated with 100 mM l-idonate, 5-keto d-gluconate, or water
| Time, h | Control (H2O) | l-Idonate | 5-Keto d-gluconate |
|---|---|---|---|
| 0 | 0.0 | 0.0 | 0.0 |
| 1 | 0.0 ± 0.8 | 3.4 ± 1.3 | 1.9 ± 1.3 |
| 2 | 0.2 ± 0.4 | 4.0 ± 0.2 | 4.1 ± 0.3 |
| 3 | −3.9 ± 0.6 | 6.2 ± 2.2 | 11.1 ± 5.6 |
Substrate addition to A. aconitifolia did not stimulate the formation of tartaric acid.
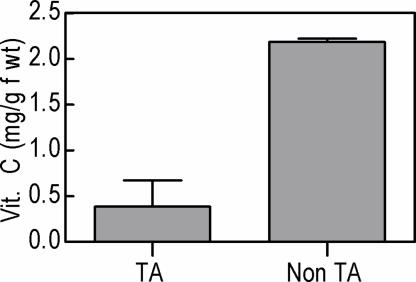
Assessing the total organic acid profile of berries from the TA accumulating and non accumulating species revealed that in A. aconitifolia, where TA accumulation does not occur and which lacks the gene encoding l-IdnDH, AA accumulates to three times the concentration observed in the TA accumulating species V. vinifera (Fig. 4). No corresponding accumulation of l-idonate was observed in berries of A. aconitifolia (data not shown).
Fig. 4.
Vitamin C measured in ripe berries of TA-accumulating V. vinifera and non-TA-accumulating A. aconitifolia. Five berries were sampled per replicate. Bars show standard error, n = 3.
Discussion
Despite their structural similarity, markedly different biochemical processes form TA and l-malic acids, which accumulate to high levels in grape berries during ripening. TA accumulation occurs in a very small number of plant species, the most significant of these being the cultivated grapevine V. vinifera. TA levels within grape berries confer organoleptic properties important in the palatability of table grapes, but more importantly, provide the basis for the control of pH that is crucial during the production of wine. We have demonstrated enzymatic activity for a key component (l-IdnDH) of the direct TA biosynthetic pathway from AA in the grapevine V. vinifera. A combination of transcript and metabolite analyses in Vitaceous species was used to identify and correlate candidate gene expression with TA formation.
Preliminary biochemical investigations to identify TA-biosynthetic-specific enzyme activity in soluble protein extracts prepared from immature grape berries were unsuccessful. It is suggested that, despite the use of extraction buffer systems designed to maximize the recovery of enzymatic activity from these tissues (28), the combination of high organic acid and high phenolic conditions was refractory to the recovery of a measurable amount of soluble protein. Earlier work (data not shown) to isolate glucosyltransferase enzymes from similar tissues was likewise unsuccessful, with only one-tenth to one-fortieth of the yield of protein observed in extractions from mature berries compared to leaves (28).
Although the ease of transformation in model plant systems such as Arabidopsis provides researchers with reverse genetic tools to link genes to phenotypes, many nonmodel systems, including the grapevine, display recalcitrance to facile transformation. Consequently, despite their undisputed economic and cultural importance, the biology of these species remains largely unexplored. Here, we combined transcript and metabolic profiling to target a key gene in the biosynthesis of TA in grapevines, hitherto a pathway whose occurrence was postulated but for which no enzymological or molecular evidence existed. Transcript profiling has been used widely in combination with metabolite analysis (29) in the search for glucosyltransferase activities in M. truncatulata (30), and during fruit development in tomato (31). Recent work provided a method for the interrogation of transcript data from a large collection of sequenced and catalogued grapevine cDNAs (32); we have used this resource to refine the search for candidate genes expressed in tissues known to be active in TA biosynthesis. Likely involvement in TA biosynthetic reactions was further tested by examining the domain architecture of deduced protein sequences for evidence of oxidoreductase-type reaction domains. A number of potential candidates were identified by this combined approach. To further test each of these candidate genes and in the absence of a simple transgenic assay system in grapevines, we surveyed the organic acid profiles of 28 members of the family Vitaceae. Considerable differences in the accumulation of tartaric, oxalic, malic, and succinic acids were observed, suggesting that the full metabolomic analysis of grape berry composition will reveal numerous metabolite differences within this plant family. Crucial to the present investigation, one species, A. aconitifolia, was identified in which TA could not be detected. PCR analysis of berry messenger RNA and genomic DNA indicated that one candidate gene was absent from the A. aconitifolia genome.
Enzymological evidence confirmed the capacity of the encoded enzyme to complete a key step in TA biosynthesis, the conversion of l-idonate to 5-keto d-gluconic acid, providing evidence for a biochemical component of this pathway. The product of the “reverse reaction,” l-idonate, was identified by HPLC-mass spectrometry, confirming the specificity of the enzyme for its postulated substrate. The lack of measurable activity with other substrates further supported the specific role proposed for the enzyme. An l-IdnDH also occurs in the gluconate II pathway of E. coli, where gluconate is used as an energy source in the human gut (26). We suggest also the reduction of 5-keto d-gluconate to l-idonate in Fusarium sp. no. 125 reported in the early 1960s may have been catalysed by a similar enzyme (33, 34). Interestingly, the identity between E. coli l-IdnDH and grapevine l-IdnDH is only 31% at the amino acid level, although both share a zinc-binding alcohol dehydrogenase backbone. The grapevine protein is more similar to sequences encoding NAD+-linked 2-iditol dehydrogenases from a number of plants including Arabidopsis thaliana, Malus domestica (apple), Eriobotrya japonica (loquat), and Lycopersicon esculentum (tomato). In addition to the classical C-terminal Rossman fold characteristic of NAD(P)+ dehydrogenases, these enzymes contain a predicted ketose reductase domain associated with sorbitol dehydrogenase activity. The divergence of paralogous genes due to small changes in amino acid sequence has been reported to alter the specificity and catalytic activity of enzymes (35, 36); the origin of the unusual substrate specificity possessed by l-IdnDH may be explained by a similar series of events. The absence of the l-IdnDH paralog, specifically from A. aconitifolia but not from related TA-forming species, is consistent with a gene deletion in A. aconitifolia as an underlying cause of a lack of TA accumulation in this species.
Berry slices prepared from immature V. vinifera tissue exhibited a high rate of TA biosynthesis when fed l-idonate or 5-keto d-gluconic acid; moreover, this activity was absent from berries of the nontartrate accumulating species A. aconitifolia. l-idonate might be expected to accumulate in berries of this species, but this was not the case. Instead, we observed that berries of A. aconitifolia possess significantly higher levels of ascorbate, the biosynthetic precursor of tartrate (12) and an abundant organic acid in many fruit. If l-idonate or 5-keto gluconic acid act to feedback inhibit the conversion of ascorbic acid to 2-keto l-gulonic acid, then this could explain why ascorbic acid accumulates in A. aconitifolia. This finding suggests the intriguing possibility that in planta deletion of components of the TA biosynthetic pathway may lead to increased amounts of AA (vitamin C) within the developing berry, lending additional support to the conclusion that l-IdnDH is a key component of the pathway converting vitamin C to TA.
If the lack of TA in A. aconitifolia were solely due to the absence of the l-IdnDH gene, then exogenously added 5-keto d-gluconate would likely restore TA formation to berries of A. aconitifolia. However, our results showed that the addition of 5-keto d-gluconate to berry slices of A. aconitifolia did not have this effect (Table 1), suggesting that additional factors contribute to the lack of TA accumulation in this species. Further investigation of candidate genes encoding additional TA-biosynthetic steps, including the characterization of gene knock-out and knock-down phenotypes produced by RNA interference technology, may serve to answer these questions.
The present work provides biochemical and molecular evidence for the proposed rate-limiting step in formation of TA in the grapevine and demonstrates the utility of combining molecular, metabolic, and biochemical approaches to resolve otherwise intractable biosynthetic phenomena. Biochemical data support the proposed function of l-IdnDH in the conversion of l-idonate to 5-keto d-gluconate in vitro and, by virtue of the identification of a grapevine species lacking the gene encoding this enzyme, in vivo also. Furthermore, evidence provided from the examination of precursor uptake experiments allows a glimpse into the intricacies of the role of AA as a biosynthetic precursor, and suggests a rational basis to design grapes, already the world's most important fruit crop, containing elevated levels of the vital human micronutrient vitamin C.
Materials and Methods
Chemicals.
All chemicals and reagents used were of analytical grade or higher. Authentic samples of organic acids and their salts were obtained from Sigma, Fluka, Riedel den Haan, and BDH as applicable. Authentic sodium l-idonate was obtained as a gift from Kazumi Saito (Kyoto University, Kyoto, Japan).
In Silico Identification of Preveraison Expressed Transcripts.
Differentially expressed transcripts were predicted by comparing EST frequencies across 56 independent cDNA libraries, either using a simple Boolean logic for unigenes with at least six ESTs, or based on false discovery rate calculations as described (32). Preliminary candidate identification was achieved by using tblastx (37).
Motif Searching Among Candidate Gene Products.
The structures of intermediates in the conversion of ascorbate to tartrate revealed the likely involvement of dehydrogenases, which are members of the oxidoreductase family of proteins. The deduced protein sequences of the candidate transcripts were analyzed to identify putative oxidoreductase domains and motifs via pfam, ncbi, brenda, and interpro protein domain blast interfaces (25).
Plant Material.
Berry and foliar samples were from vines grown in Winters, northern California, and Adelaide, South Australia. Samples from 28 species of Vitaceae were used in experiments. Sampling occurred in the 2003 and 2004 growing seasons and was from a minimum of two vines per species. Multiple bunches of berries were collected, and three five-berry subsamples were selected from a random 50-berry sample for HPLC analysis. Foliar samples were collected from these same plants for isolation of RNA and DNA. For experiments comparing light and shaded fruit for TA concentration and l-IdnDH transcripts, field-grown grapes of Vitis vinifera cv. Cabernet Sauvignon were used. Bunches in a zone of highly exposed berry clusters were enclosed in boxes to eliminate sunlight (<95% of ambient). Air flow was maintained around bunches to avoid temperature differences between shaded and nonshaded berries (38).
Characterization of Natural Variation in Tartaric Acid: Family Vitaceae.
HPLC was used to quantify organic acid content in berry samples essentially as described (9). Samples were prepared as described above. Ascorbic acid determination was essentially as described (9) but using fresh plant material (not frozen or stored) and measuring absorbance at 244 nm instead of 210 nm. Peak identity and quantification were determined by comparison with a set of authentic standards. AA (quantified at 244 nm) and TA (210 nm) were confirmed by LC-MS. For Fig. 4, fresh berries at the same physiological developmental stage from A. aconitifolia (0.9 g) and V. vinifera (2.1 g) were measured for organic acid using the HPLC system as above. Peaks were integrated, and the data were adjusted to calculate the concentration as mg of AA per 100 g of berry fresh weight.
PCR Validation of Gene Expression.
Total RNA from V. vinifera, V. californica, Ampelopsis brevipedunculata, and A. aconitifolia was extracted from washed grape berries and leaves as described (39). Genomic DNA was extracted by using a genomic DNA extraction kit (MoBio) according to manufacturer's instructions, from V. vinifera, V. californica, A. aconitifolia, A. brevipedunculata, and Parthenocissus tricuspifolia. For candidate genes, internal 18- to 22-bp primers were designed with annealing temperatures of 55°C. cDNA synthesis used Superscript III reverse transcriptase (Invitrogen). Full-length l-IdnDH was amplified by using primers GGGCATATGATGGGGAAAGGAGGCAACTCTG (forward) and CCGGATCCTTAGAGATTAAACATGACCTTG (reverse). Ubiquitin and actin sequences were used for amplification controls. PCRs were composed of 1.5 pmol of each primer, 0.5 μl of Taq DNA polymerase, 0.5 μl of dNTPs, 1 μl of MgCl (25 mM), 1 μl (500 ng·μl−1) of template cDNA, 2 μl of 10× (Mg2+-free) buffer, made to 20 μl with deionized H2O (dH2O). A 3-min heating at 95°C was followed by 32 cycles of 95°C (30 s) denaturation, 55°C (30 s) annealing temp, 1 min per kilobase product size extension times, and a final extension of 5 min. Quantitative RT-PCR was conducted by using 10 μl of Bio-Rad real-time PCR reagent, 1 μl of template cDNA (single-strand cDNA was made from mRNA standardized at a concentration of 500 ng·μl−1), 8.4 μl of dH2O, and 0.6 μl (1.5 pmol) of F/R primer [internal primers were as follows: AAGTTTGCCTTGTGGGTTTG (forward) and AAGGCTTCCTCCACATCCTT (reverse)]. The standard curve for l-IdnDH expression was generated by using l-IdnDH cDNA template over a series of concentrations. Relative expression is based on a correction factor, which was calculated based on the expression of the ubiquitin control gene in all templates.
Molecular Cloning and Recombinant Protein Formation of l-IdnDH.
Full-length l-IdnDH was amplified from V. vinifera RNA obtained from berries sampled 4 weeks after anthesis and cloned via pTOPO2.1 (Invitrogen) into pET14b (Novagen) for prokaryotic expression. The sequence verified construct was transformed into E. coli BL21 (DE3) pLysS (Novagen) for induction. Transformants (50 ml in a 250-ml Erlenmeyer flask) were grown in Terrific Broth supplemented with ampicillin (100 μg·ml−1) and chloramphenicol (34 μg·ml−1). Once the culture reached an optical density of 0.9 at 600 nm, the expression of l-IdnDH was induced by addition of 0.4 mM isopropyl β-d-thiogalactoside. Cells were harvested by centrifugation 3 h after induction and resuspended in sonication buffer (4 ml/g wet cell paste; Clontech) containing 10 mM 2-mercaptoethanol, followed by three freeze–thaw cycles in liquid nitrogen. Protease mixture (Roche) was added, and cells were disrupted by reciprocation through a 20-gauge needle followed by centrifugation at 13,500 × g for 10 min at 4°C and then divided into soluble and insoluble fractions. Final purification of soluble material was achieved by using Talon resin (Clontech).
l-IdnDH Enzyme Assay.
Assays (150 μl) contained 15 μl of purified protein extract (≈0.09 mg/ml) preequilibrated to 30°C in 100 mM Tris·HCl, pH 8/330 μM NAD+ or NADH, in a glass cuvette zeroed at A340 nm before addition of substrate to a final concentration of 50 mM. The forward and reverse reactions (l-idonate to 5-keto d-gluconic acid and 5-keto d-gluconic acid to l-idonate, respectively) were followed by change in absorbance at A340 nm. HIS-TAG purified protein was used in all assays; therefore, untransformed E. coli reactions are not presented in the data. Substrates tested included l-idonate (supplied as the monosodium salt), 5-keto d-gluconic acid (supplied as the monopotassium salt), 2-keto l-gulonic acid, AA, d-sorbitol, and d-gluconic acid. Reactions were stopped with 50 μl of 1 M HCl and centrifuged at 13,500 × g for 5 min; the supernatant (20 μl) was loaded onto an HPLC column (Prevail OA 4.6 × 250 mm; Alltech Associates). The mobile phase was 25 mM KH2PO4 adjusted to pH 2.0 with phosphoric acid, at a flow rate of 0.5 ml/min. Detection of the organic acids including l-idonate and 5-keto d-gluconic acid was by UV absorbance at 210 nm.
Characterizing the Catalytic Activity of l-IdnDH.
Km and Vmax values of the recombinant l-IdnDH were determined for the forward and reverse reactions. Conditions of the reaction were as described above, using ≈0.09 mg/ml purified protein and varying concentrations of substrate. The recombinant protein used in kinetic assays was prepared fresh and frozen once at −80°C before use. A Bio-Rad benchtop spectrophotometer was programmed to measure kinetics of enzyme activity, set at A340 nm, measuring ΔA340 nm every 5 s for 100 s. The Km and Vmax calculations were determined from double reciprocal Lineweaver–Burke graphs. Freezing and thawing stability of the protein was tested because activity was not constant over time. The variation of activity with pH was studied for the forward and reverse reactions. Oxidation and reduction reactions were measured between pH 5.4 and 8.5 using the following buffers: Mes, pH 5.4–6.2; Pipes, pH 6.2–6.6; Tris, pH 6.5–8.5. Standard assays were carried out as described, with the buffer concentration fixed at 100 mM throughout.
Precursor Feeding Experiments.
The substrates of the forward and reverse reactions catalyzed by l-IdnDH, sodium l-idonate, and potassium 5-keto d-gluconate (100 mM in deionized H2O), were added to slices (≈1 mm thick) prepared from berries of A. aconitifolia and V. vinifera. Measurements of TA content in berry tissues after incubation with the substrates (n = 3 per time point) were made hourly from 0 to 3 h via HPLC and confirmed by LC-MS.
Supplementary Material
Acknowledgments
We thank Alberto Iandolino, Francisco Goes Da Silva, Brent Kaiser, and Steve Tyerman for helpful discussions. Kazumi Saito is gratefully acknowledged for the gift of sodium l-idonate. Simon Robinson, Susan Wheeler, Renata Ristic, and Alberto Iandolino are thanked for RNA and grape samples. This work was supported by the Australian Government's Cooperative Research Centre Program and conducted by the Cooperative Research Centre for Viticulture. Financial support was also provided by the California Department of Food and Agriculture.
Abbreviations
- TA
l-tartaric acid
- AA
l-ascorbic acid
- OxA
oxalic acid
- l-IdnDH
l-idonate dehydrogenase.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ124868).
References
- 1.Pasteur L. Compt. Rend. 1860;51:298. [Google Scholar]
- 2.Hale C. R. Nature. 1962;195:917–918. [Google Scholar]
- 3.Stafford H. A. Am. J. Bot. 1959;46:347–352. [Google Scholar]
- 4.Loewus F. A., Stafford H. A. Plant Physiol. 1958;33:155–156. doi: 10.1104/pp.33.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito K., Kasai Z. Phytochemistry. 1969;8:2177–2182. [Google Scholar]
- 6.Banhegyi G., Loewus F. A. In: Vitamin C. Functions and Biochemistry in Animals and Plants. Asard H., May J. M., Smirnoff N., editors. London: BIOS. Sci. Publ.; 2004. pp. 33–48. [Google Scholar]
- 7.Loewus F. A. Phytochemistry. 1999;52:193–210. [Google Scholar]
- 8.Green M. A., Fry S. C. Nature. 2005;433:83–87. doi: 10.1038/nature03172. [DOI] [PubMed] [Google Scholar]
- 9.DeBolt S., Hardie J., Tyerman S. D., Ford C. M. Aust. J. Grape Wine Res. 2004;10:134–142. [Google Scholar]
- 10.Saito K., Kasai Z. Plant Cell Physiol. 1982;23:499–508. [Google Scholar]
- 11.Coombe B. G., McCarthy M. G. Aust. J. Grape Wine Res. 2000;6:131–135. [Google Scholar]
- 12.Malipiero U., Ruffner H. P., Rast D. M. J. Plant Physiol. 1987;129:33–40. [Google Scholar]
- 13.Saito K. Phytochemistry. 1992;31:1219–1222. [Google Scholar]
- 14.Saito K., Kasai Z. Plant Physiol. 1984;76:170–174. doi: 10.1104/pp.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito K., Loewus F. A. Plant Cell Physiol. 1979;20:1481–1488. [Google Scholar]
- 16.Saito K., Loewus F. A. Plant Cell Physiol. 1989;30:905–910. [Google Scholar]
- 17.Saito K., Morita S. I., Kasai Z. Plant Cell Physiol. 1984;25:1223–1232. [Google Scholar]
- 18.Saito K., Ohmoto J., Kuriha N. Phytochemistry. 1997;44:805–809. [Google Scholar]
- 19.Salusjarvi T., Povelainen M., Hvorslev H., Eneyskaya E. V., Kulminskaya A. A., Shabalin K. A., Neustroev K. N., Kalkkinen N., Miasnikov A. N. Appl. Microbiol. Biotechnol. 2004;65:306–314. doi: 10.1007/s00253-004-1594-6. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler G. L., Jones M. A., Smirnoff N. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- 21.Smirnoff N., Running J. A., Gatzck S. In: Vitamin C: Functions and Biochemistry in Animals and Plants. Asard H., May J. M., Smirnoff N., editors. London: BIOS. Sci. Publ.; 2004. pp. 7–29. [Google Scholar]
- 22.Agius F., Gonzalez-Lamothe R., Caballero J. L., Munoz-Blanco J., Botella M. A., Valpuesta V. Nat. Biotechnol. 2003;21:177–181. doi: 10.1038/nbt777. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z., Young T. E., Ling J., Chang S.-C., Gallie D. R. Proc. Natl. Acad. Sci. USA. 2003;100:3525–3530. doi: 10.1073/pnas.0635176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iland P. G., Coombe B. G. Am. J. Enol. Vit. 1988;39:71–76. [Google Scholar]
- 25.Mulder N. J., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bausch C., Peekhaus N., Utz C., Blais T., Murray E., Lowary T., Conway T. J. Bacteriol. 1998;180:3704–3710. doi: 10.1128/jb.180.14.3704-3710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai C. Z., Han L. Y., Chen X., Chen Y. Z. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford C. M., Høj P. B. Aus. J. Grape Wine Res. 1998;4:48–58. [Google Scholar]
- 29.Suzuki H., Achnine L., Xu R., Matsuda S. P. T., Dixon R. A. Plant J. 2002;32:1033–1048. doi: 10.1046/j.1365-313x.2002.01497.x. [DOI] [PubMed] [Google Scholar]
- 30.Achnine L., Huhman D. V., Farag M. A., Sumner L. W., Blount J. W., Dixon R. A. Plant J. 2005;41:875–887. doi: 10.1111/j.1365-313X.2005.02344.x. [DOI] [PubMed] [Google Scholar]
- 31.Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G. B., Tanksley S. D., Giovannoni J. J. Plant Cell. 2005;17:2954–2965. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goes da Silva F., Iandolino A., Al-Kayal F., Bohlmann M. C., Cushman M. A., Lim H., Ergul A., Figueroa R., Kabuloglu E. K., Osborne C., et al. Plant Physiol. 2005;139:574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takagi Y. Agric. Biol. Chem. 1962;26:717–718. [Google Scholar]
- 34.Takagi Y. Agric. Biol. Chem. 1962;26:719–720. [Google Scholar]
- 35.Broun P., Shanklin J., Whittle E., Somerville C. Science. 1997;282:1315–1317. doi: 10.1126/science.282.5392.1315. [DOI] [PubMed] [Google Scholar]
- 36.Henikoff S., Greene E. A., Pietrokovski S., Bork P., Attwood T. K., Hood L. Science. 1997;278:609–614. doi: 10.1126/science.278.5338.609. [DOI] [PubMed] [Google Scholar]
- 37.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Downey M. O., Harvey J. S., Robinson S. P. Aust. J. Grape Wine Res. 2004;10:55–74. [Google Scholar]
- 39.Iandolino A. B., Goes da Silva F., A., Lim H., Choi Williams L. E., Cook D. R. Plant Mol. Bio. Rep. 2004;22:269–278. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.