Abstract
Rational engineering of complicated metabolic networks involved in the production of biologically active plant compounds has been greatly impeded by our poor understanding of the regulatory and metabolic pathways underlying the biosynthesis of these compounds. Whereas comprehensive genome-wide functional genomics approaches can be successfully applied to analyze a select number of model plants, these holistic approaches are not yet available for the study of nonmodel plants that include most, if not all, medicinal plants. We report here a comprehensive profiling analysis of the Madagascar periwinkle (Catharanthus roseus), a source of the anticancer drugs vinblastine and vincristine. Genome-wide transcript profiling by cDNA-amplified fragment-length polymorphism combined with metabolic profiling of elicited C. roseus cell cultures yielded a collection of known and previously undescribed transcript tags and metabolites associated with terpenoid indole alkaloids. Previously undescribed gene-to-gene and gene-to-metabolite networks were drawn up by searching for correlations between the expression profiles of 417 gene tags and the accumulation profiles of 178 metabolite peaks. These networks revealed that the different branches of terpenoid indole alkaloid biosynthesis and various other metabolic pathways are subject to differing hormonal regulation. These networks also served to identify a select number of genes and metabolites likely to be involved in the biosynthesis of terpenoid indole alkaloids. This study provides the basis for a better understanding of periwinkle secondary metabolism and increases the practical potential of metabolic engineering of this important medicinal plant.
Keywords: cell culture, periwinkle, profiling, secondary metabolism, jasmonate
The medicinal plant Catharanthus roseus L. G. Don is of enormous pharmaceutical interest because it contains >120 terpenoid indole alkaloids (TIAs), some of which exhibit strong pharmacological activities (1). Ajmalicine, an antihypertensive alkaloid, and vinblastine and vincristine, antineoplastic bisindole alkaloids, are already in clinical use. The latter two anticancer compounds are produced only in very low amounts in C. roseus plants (2) and, despite significant efforts, cell cultures are not yet a valid alternative for production. Although undifferentiated Catharanthus cells can produce fairly high levels of several monomeric alkaloids (e.g., ajmalicine and serpentine), vindoline, the compound that, together with catharanthine, is one of the building blocks for the in vivo formation of bisindole alkaloids (Fig. 1), is not synthesized.
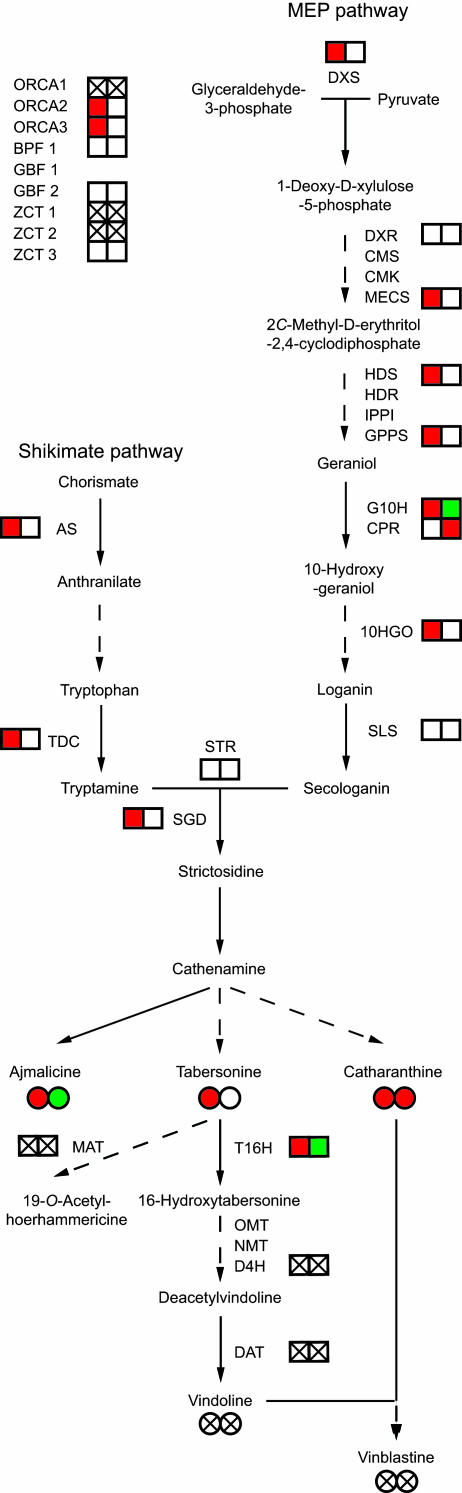
Fig. 1.
Biosynthesis of C. roseus TIAs. Metabolites are given as full names in lowercase and enzymes as abbreviations in capitals. Full and dashed arrows mark single and multiple conversion steps between intermediates, respectively. In the upper left corner, transcription factors binding to promoters of TIA biosynthetic genes are indicated. In addition, a graphical snapshot shows the relative transcript (boxes) and metabolite (circles) accumulation levels in samples harvested 12 h after elicitation. The left and right boxes or circles reflect the influence of jasmonate and auxin, respectively. Red, induced accumulation compared with control without the phytohormone; green, repressed accumulation compared with control without the phytohormone; white, no effect of the phytohormone on accumulation; crossed, no transcript or metabolite accumulation detected. Enzymes and transcription factors listed: ORCA, octadecanoid-responsive Catharanthus AP2-domain; BPF, box P-binding factor; GBF, G-box binding factor; ZCT, zinc finger Catharanthus transcription factor; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; CMS, 4-diphosphocytidyl-2C-methyl-d-erythrol 4-phosphate synthase; CMK, 4-diphosphocytidyl-2C-methyl-d-erythrol kinase; MECS, 2C-methyl-d-erythrol-2,4-cyclodiphosphate synthase; HDS, GCPE, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; HDR, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; IPPI, isopentenylpyrophosphate isomerase; G10H, geraniol 10-hydroxylase; CPR, cytochrome P450 reductase; 10HGO, 10-hydroxygeraniol oxidoreductase; SLS, secologanin synthase; STR, strictosidine synthase; SGD, strictosidine β-d-glucosidase; AS, anthranilate synthase; TDC, tryptophan decarboxylase; T16H, tabersonine 16-hydroxylase; OMT, O-methyltransferase; NMT, N-methyltransferase; D4H, desacetoxyvindoline 4-hydroxylase; DAT, deacetylvindoline 4-O-acetyltransferase; MAT, acetyl-CoA:minovincine-O-acetyltransferase.
All TIAs in C. roseus are derived from the central precursor strictosidine, which is a fusion product of the shikimate pathway-derived tryptamine moiety and the plastidic nonmevalonate pathway-derived secologanin moiety (Fig. 1). Starting from the amino acid tryptophan and the monoterpenoid geraniol, the biosynthesis of bisindole alkaloids in C. roseus involves at least 35 intermediates and 30 enzymes (1, 3). At least seven different (sub)cellular compartments are involved in TIA biosynthesis (1), and extensive transport of intermediates is required, as evidenced from enzyme targeting and the cell type-specific expression of biosynthetic genes. The TIA biosynthetic pathway is under strict developmental and environmental control. For instance, the late steps of vindoline formation are confined to idioblast and laticifer cells, whereas the epidermal layers of immature leaves and stems are the only locations in which the early phases of TIA biosynthesis take place (3). D4H expression and vindoline accumulation are affected both by light and methyl jasmonate (MeJA) in C. roseus seedlings (4), whereas TIA production in C. roseus cell cultures can be induced by various phytohormones and biotic and abiotic elicitors (1). Ultimately, transcription factors are responsible for coordinating the expression of biosynthetic genes in response to these external and internal signals. Members of the plant-specific AP2/ERF transcription factor family have been identified in C. roseus, namely ORCA2 and ORCA3, whose expression is induced by MeJA (5, 6). ORCA proteins control the transcription of genes, such as STR, involved in TIA biosynthesis by binding specifically to a promoter element involved in jasmonate- and elicitor-responsive gene expression. Several other C. roseus transcriptional regulators have been identified, all binding to the STR promoter (7–9), suggesting a considerable degree of complexity in the control of TIA biosynthesis.
Therefore, a deeper understanding of the regulatory system governing TIA metabolism is of particular interest and could eventually make successful metabolic engineering of alkaloid biosynthesis possible. To gain such knowledge, we used the cDNA-amplified fragment length polymorphism (AFLP) technology in combination with targeted and nontargeted liquid chromatography-mass spectrometry to study secondary metabolism in elicited C. roseus cells. By integrating the genome-wide transcript and metabolite profiles, we were able not only to visualize most of the known genes involved in TIA biosynthesis in a single experiment but also to draw previously undescribed gene-to-gene and gene-to-metabolite networks. This study provides insights into the complex regulation not only of TIA metabolism but also of (secondary) metabolism in C. roseus in general.
Results
Metabolite Analysis.
Before initiating functional genomics-driven gene discovery for TIA metabolism in periwinkle cells, we needed to identify the conditions in which differential accumulation of the desired metabolites can be observed (10). In the case of C. roseus, the literature suggests that TIA accumulation is strongly influenced by the complex interaction of phytohormones such as auxins and jasmonates (11). Therefore, it seemed most promising to investigate the combined effects of these two hormones on TIA accumulation in C. roseus cells, applying growth and elicitation conditions similar to those described in ref. 11. TIA-targeted metabolite profiling showed pronounced accumulation of tabersonine and ajmalicine within 4 h and catharanthine within 12 h after MeJA elicitation (Fig. 3, which is published as supporting information on the PNAS web site).
We then performed a large-scale nontargeted liquid chromatography-mass spectrometry analysis on the same samples used for TIA-targeted analysis. Before normalization and peak filtering, the metabolic profile of this nontargeted analysis revealed 3,891 peaks by using the MZmine LC/MS toolbox (12). Peak filtering resulted in a final set of 178 peaks. Using an internal library of masses and retention times, the following TIAs were identified: ajmalicine, tabersonine, catharanthine, yohimbine, cathenamine, secologanine, lochnerinine, 16-methoxy-2,3-dihydro-3-hydroxytabersonine, and desacetoxyvindoline. As anticipated, all of these compounds are synthesized upstream of D4H in the pathway (Fig. 1). The remaining peaks contained metabolites most abundantly in the range of 300–400 m/z, the expected range for monomeric TIA metabolites. Some also displayed retention times similar to the TIAs identified. More detailed analytical investigation would be required for conclusive chemical characterization.
Average linkage hierarchical clustering of the metabolite accumulation profiles revealed the existence of various subclusters, two of which are particularly important: one consisting of 43 metabolites, of which accumulation is stimulated by auxins, and the other with 53 metabolites, of which accumulation is repressed by auxins (Fig. 4, which is published as supporting information on the PNAS web site). The differential auxin-mediated regulation of some of these metabolites might be enforced by or dependent on the presence of MeJA. Most importantly, however, not all monomeric TIAs identified are found within the same auxin-modulated subcluster, indicating divergent regulation of metabolite biosynthesis and accumulation within the TIA pathway. For instance, auxins enhanced the accumulation of tabersonine and catharanthine, the two building blocks for bisindole alkaloid biosynthesis, whereas they repressed ajmalicine, a monomeric TIA not involved in the synthesis of bisindole alkaloids. This result corroborates the findings obtained by TIA-targeted metabolite analysis and underscores the reliability of our C. roseus metabolome data set. The negative effect of auxins on ajmalicine levels has also been observed in ref. 11, but the levels of other monomeric TIAs were not assessed in that study.
Transcript Profiling.
The cDNA-AFLP technique (13) was applied for genome-wide transcript profiling by using the same samples as those used for metabolite analysis. Using 128 BstYI + 1/MseI + 2 primer combinations, the quantitative temporal accumulation patterns of 10,790 transcript tags were determined and analyzed. In total, 561 differentially expressed transcript tags were isolated (hereinafter referred to as CR tags). Direct sequencing of the PCR products gave good-quality sequences for 417 fragments (74%) (Table 1, which is published as supporting information on the PNAS web site). A unique sequence could not be attributed unambiguously to the remaining 26%, indicating that they might not represent unique gene tags. Upon blasting of the 417 nucleotide sequences of the cDNA-AFLP tags with the 236 C. roseus European Molecular Biology Laboratory entries publicly available before this study, <10% gave a (near) perfect match. Thus, the vast majority of the tags identified here are previously undescribed C. roseus sequence information. blast searches with the sequences from the C. roseus cDNA-AFLP tags revealed that 37% of the CR tags displayed no sequence similarity to any known plant genes.
The CR tags could be divided into different subclusters, based either on their expression profiles or their annotation. Average linkage hierarchical clustering analysis of the expression profiles showed that three main forces steer the formation of clusters (Fig. 5, which is published as supporting information on the PNAS web site). Listed in order of impact, these forces are the addition of MeJA (irrespective of auxin presence), growth (independent of the exogenous application of hormones or elicitors), and the presence of auxin (irrespective of MeJA presence). These factors modulate the expression of 42.8%, 33.8%, and 28.5% of the CR tags, respectively. Only for 5.2% of the CR tags was the expression affected both by MeJA and auxin. According to the Functional Catalog of the Munich Information Center for Protein Sequences (http://mips.gsf.de/projects/funcat), the CR tags can be classified into eight broadly defined functional groups. The functional category “Metabolism and Energy” in particular is one of the major groups (Fig. 5) and includes, as anticipated, a large number of TIA genes. The tags that corresponded to genes reported to be associated with TIA biosynthesis, and isolated as differentially expressed by cDNA-AFLP, include tags for genes associated with biosynthesis of the terpenoid moiety, biosynthesis of the indole moiety, biosynthesis of monomer TIAs, and transcription factors regulating TIA biosynthesis (Fig. 6A, which is published as supporting information on the PNAS web site). In all these cases, the tags showed a (near) perfect match with the gene sequences encoding the isoforms reported to catalyze (or regulate) the known enzymatic reactions. Moreover, the cDNA-AFLP set also includes tags with close similarity (>70%) to 10HGO, STR, T16H, and DAT but whose sequence does not perfectly match that of the isoenzyme reported to catalyze the corresponding enzymatic reactions. Because in plant secondary metabolism even close sequence similarity is not sufficient to indicate a correct functional annotation, further analysis will be required to ascertain whether the gene products corresponding to these tags catalyze identical reactions or whether they possibly generate structurally related alkaloids or other types of secondary metabolites. As can be seen in Fig. 6A, the expression of all these genes can be induced by elicitation with MeJA and either stimulated or repressed by auxin. RT-PCR was performed for all of the remaining known but undetected TIA biosynthesis genes and showed that all but two (T16H and TDC) were either not differentially expressed or not transcribed at all (Fig. 6B).
Integrated Transcriptome and Metabolome Analysis.
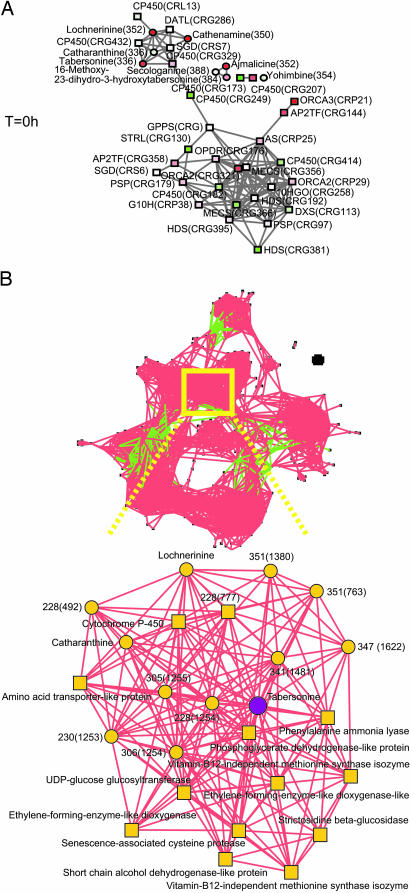
The accumulation profiles of the 178 metabolite peaks retained were combined with the expression profiles of the 417 transcripts for integrated analysis. A principal component analysis with mean scaling was performed first to explore the variability structure of the data (14). The first two principal components (PCs), accounting for 64% of the total variability, revealed clear separations: the auxin-treated cells from the nontreated cells by the first PC and each group across the time domain by the second PC (Fig. 7 and Table 2, which are published as supporting information on the PNAS web site). Correlation network analysis was used to establish gene-to-gene and gene-to-metabolite coregulation patterns (15). The Pearson correlation coefficient between each pair of variables (either gene or metabolite) across the profiles, including all time points and conditions, was calculated. To best visualize the complex networks of secondary metabolism in C. roseus cells, two different subsets of the profiles were analyzed. The correlation network analysis was first performed for a select subset of identified metabolites and genes (Fig. 2A) chosen to include the 9 TIA metabolite peaks identified and the 34 gene tags identical to or with close sequence similarity to genes that encode proteins catalyzing either jasmonate or TIA biosynthesis. Given their importance in TIA biosynthesis, all putative cytochrome P450 (CYP450) and AP2 transcription factors also were included. Variable correlation coefficient cutoff values were applied to draw the edges as part of the exploratory study of correlation structure within the subset. Fig. 2A presents one example with a cutoff of 0.55. With the exception of three tags corresponding to an AP2 factor (CRG20), a lipoxygenase possibly involved in jasmonate biosynthesis (CRG48), and a CYP450 (CRG96), all of the CR tags belonging to the gene classes used in this subset could be integrated into the network. The most striking observations were (i) the presence of a strongly correlated gene-to-gene network that includes practically all of the known early pathway TIA genes and the ORCA transcription factors, (ii) the presence of a gene-to-metabolite cluster comprising a part of the TIA metabolites and a set of CYP450 genes with as-yet-unspecified function, and (iii) some smaller correlation groups (i.e., only gene or metabolite pairs or triplets) containing, for instance, ajmalicine, thus confirming the differential regulation of this particular biosynthetic branch.
Fig. 2.
Gene-to-metabolite networks in elicited C. roseus cells. Metabolites are represented by circles and transcripts by squares. (A) Correlation network for a select subset of identified metabolites and genes consisting of nine TIA metabolite peaks and gene tags identical to or with close sequence similarity to genes, encoding for proteins catalyzing jasmonate and TIA biosynthesis, respectively, and all putative cytochrome P450 and AP2 transcription factors. Correlations between the variables are calculated from the complete profiles across all conditions and time points, and edges are drawn when the linear correlation coefficient is >0.55. Tags corresponding to AP2TF (CRG20), CYP450 (CRG96), and LOX (CRG48) also were included in the gene subset, but these data points have been removed from the figure because no correlation was observed. (B) Correlation network for the complete set of metabolites and genes (Upper), with a zoom-in on the tabersonine node and 24 of its nearest neighbors (Lower). Red lines represent positive correlations (C > 0.8).
An unbiased subset was then visualized, subtracted from the complete network across all transcript and metabolite profiles, and centered on the tabersonine node by using the cutoff for absolute value of correlation coefficient C > 0.8 (Fig. 2B). The gene-to-metabolite network around the tabersonine node consisted of 11 metabolites and 13 genes (with a blast hit) representing the nearest neighbors. Many of the unassigned metabolites included in this cluster displayed masses in the range of 300–400 m/z, the expected range for monomeric TIA metabolites and, thus, might constitute yet-unknown intermediates or side products of TIA metabolism. Accordingly, this network centered on tabersonine includes numerous tags corresponding to enzymes with currently unidentified activity and substrate specificity that are, thus, likely to code for some of the missing links in the biosynthesis of tabersonine or other monomeric TIAs.
Discussion
Generation of C. roseus ESTs by cDNA-AFLP-Based Transcript Profiling.
Large-scale gene discovery programs in medicinal plants such as C. roseus are hampered enormously by the fact that standard transcript profiling methods, such as serial analysis of gene expression or microarray analysis, are not applicable because of the lack of large sequence repertoires. In contrast, the cDNA-AFLP technology can be used to identify genes in nonmodel plant species and acquire quantitative expression profiles at the same time (16). Indeed, in this study of jasmonate-elicited C. roseus cell suspensions, genome-wide cDNA-AFLP transcript profiling allowed us to build a substantial collection of known and previously undescribed genes from this medicinally important plant.
The number of 37% “no-hit” tags in blast searches is higher than the 26% reported in a similar tobacco BY-2 profiling analysis (16). This finding was not unexpected considering the scarcity of gene sequences from either C. roseus or other species belonging to the Apocynaceae family, which can be used for tag sequence extension in blast searches, compared to the abundant EST collections for tobacco, tomato, and other Solanaceae species.
Importantly, using the cDNA-AFLP technique, we were able to monitor in one single experiment all but two of the known genes involved in TIA biosynthesis that were differentially expressed under our experimental conditions. Furthermore, their expression profile was linked to that of multiple previously undescribed genes with unknown activities, some of which could code for missing links in TIA biosynthesis. In comparison, a recently performed systematic 2D PAGE proteomic analysis of TIA-producing C. roseus cell suspension cultures identified 58 proteins, among which only two are known to be involved in TIA biosynthesis (17), thus underscoring the current value of the cDNA-AFLP technology. In addition, based on public European Molecular Biology Laboratory sequences, the size of a tag of a particular gene of interest can be predicted precisely with cDNA-AFLP profiling, provided no small genotype-specific nuclear polymorphisms alter the presence of restriction enzyme sites or the sequence important for AFLP primer specificity around these sites. We used this feature to assess the quality and utility of a larger number of samples of elicited C. roseus cells harvested for a pilot cDNA-AFLP experiment (data not shown) and subsequently selected the most appropriate set of samples for the complete genome-wide cDNA-AFLP analysis (Fig. 5). Based on released gene sequences, we were able to precisely locate, in addition to the differentially regulated TIA biosynthesis tags, tags corresponding to genes encoding enzymes involved in the cytosolic mevalonate pathway or transcription factors (BPF 1, ref. 18; GBF 2, ref. 9) whose corresponding transcript steady-state levels remained constant throughout the different hormone or elicitor treatments (Fig. 6C).
Upon overlay of the TIA gene expression profiles with the TIA metabolite accumulation profiles on the pathway presented in Fig. 1, our findings clearly extend earlier observations concerning the coordinated regulation of TDC and STR by MeJA in Catharanthus cells (3) to the entire TIA pathway (up to 16-hydroxytabersonine). In agreement with ref. 19, later pathway genes such as D4H and DAT were not expressed under our experimental conditions, justifying the fact that vindoline does not accumulate in cell cultures despite the availability of precursor metabolites.
In addition to tags corresponding to TIA genes, cDNA-AFLP analysis also revealed clusters of CR tags involved in other metabolic pathways such as the S-adenosyl methionine (SAM) cycle (Fig. 8, which is published as supporting information on the PNAS web site) and phenolic compound synthesis (Table 1). SAM is an important biological compound involved in many essential biochemical processes, above all acting as the major methyl donor in reactions catalyzed by methyltransferases (20). An example of a specific SAM-dependent enzyme in TIA biosynthesis is the OMT that catalyzes the formation of 16-methoxytabersonine (21). Interestingly, a corresponding gene tag was found in the cDNA-AFLP analysis for all of the enzymes involved in the SAM cycle (Fig. 8), some of which cluster around the tabersonine node (Fig. 2B).
Depicting Gene-to-Gene and Gene-to-Metabolite Networks for TIA Biosynthesis.
Integration of transcriptomics and metabolomics data will be crucial for the study of gene-to-metabolite networks for (secondary) metabolism in plants, both at the regulatory and catalytic levels. We have performed a linear correlation network analysis of transcripts and nontargeted metabolites from elicited periwinkle cells to create previously undescribed gene-to-gene and gene-to-metabolite networks and, thereby, discover previously undescribed genes involved in TIA biosynthesis. In contrast to previous studies, in which long-term effects on metabolism (days after application of nutritional stress; ref. 22) or steady-state situations (transgenic plants; ref. 23) were evaluated, we focused on inducible short-term effects (gene expression and metabolite accumulation within hours after MeJA application). Accordingly, all of the known TIA genes visualized in our cDNA-AFLP analysis are induced early by adding MeJA. It should be noted that the transcripts corresponding to previously released TIA genes are all involved in the early part of the TIA pathway (upstream of cathenamine). With the exception of T16H and D4H, late pathway genes (from cathenamine to deacetylvindoline) have not yet been isolated. In contrast, the TIA metabolites detected all derive from the late part of the pathway (see Fig. 1). Early pathway intermediates (upstream of cathenamine) were not detected in C. roseus cells by using ethanol as an extraction solvent (except for secologanine). This finding could have been due to the use of unspecific extraction conditions (here optimized for monomeric TIA alkaloids), a high turnover rate, or the absence of differential accumulation patterns.
Nevertheless the correlation networks constructed allowed us to identify those genes most likely to be involved in TIA metabolism and, therefore, locate possible candidates coding for some of the missing links in the biosynthesis of monomeric TIAs. Out of the complex set of all metabolites and transcripts identified, a short list can be made of those whose accumulation or expression is closely correlated with the accumulation of specific metabolites, such as tabersonine. For instance, like any plant species, Catharanthus synthesizes a large number of CYP450 enzymes, most of which have unknown activity. Several of these enzymes now have been picked up in our transcriptome study. In our correlation network, the tag CRG329 is directly linked with the metabolite tabersonine (Fig. 2). Furthermore, lochnerinine, a TIA with an epoxy function at positions 6 and 7 similar to lochnericine (24), is linked with tabersonine and two other as-yet-uncharacterized CYP450 transcripts (CRG432 and CRL13) (Fig. 2), suggesting that their respective gene products could encode for proteins similar in function to tabersonine-6,7-epoxidase (25). This hypothesis seems also to be reinforced by phylogenetic analysis of the previously undescribed CR CYP450 genes (Table 3, which is published as supporting information on the PNAS web site). Although the corresponding cDNA-AFLP tags were too short to perform statistically significant phylogenetic tree analysis, we were able to tentatively place them in CYP450 subclasses based on the first blast hits of the tag sequences. This in silico analysis clearly added further weight to the putative function of these enzymes in TIA metabolism; two of the highlighted tags (CRG329 and CRG432) seem to belong to the same CYP450 subclass as one of the CYP450s known to be involved in TIA biosynthesis, i.e., T16H. Likewise, at the regulatory level, the ORCA and two other as-yet-uncharacterized AP2-domain transcription factor-encoding genes (CRG358 and CRG144) are clearly linked with early pathway structural genes, whereas other transcription factor-encoding genes (such as CRG101) did not correlate with any of the TIA genes or metabolites selected (Fig. 2). Members of the AP2/ERF-domain transcription factor family play a central role in the regulation of plant stress responses, and the two previously undescribed members that cluster with the TIA structural genes may control, for instance, a distinct set of target genes as described for ORCA3 (6).
Furthermore, the correlation network analysis could help elucidate the function and pathways of previously undescribed and known compounds. For example, the unknown compounds with m/z values 409 and 305 cluster together with catharanthine, lochnerinine, and tabersonine (Fig. 2), whereas the compounds with m/z value of 321 and 349 (most likely serpentine) cluster with ajmalicine (data not shown), suggesting differential regulation of biosynthesis or accumulation.
In conclusion, we believe that the comprehensive profiling approach described here is an excellent example of the enormous potential of a gene discovery platform based on an open transcript profiling method such as cDNA-AFLP to dissect (secondary) metabolism in nonmodel plant systems. The power of this approach was also successfully demonstrated by Croteau and coworkers (26) in their studies of taxol biosynthesis and by our own study of tobacco nicotine biosynthesis (16, 27). Therefore, the large quantity of previously undescribed Catharanthus ESTs that have been generated will undoubtedly provide us with exciting opportunities to map both biosynthetic pathways and signaling cascades, ultimately leading to the creation of previously undescribed genetic tools to stimulate production of pharmaceutical compounds of high importance.
Materials and Methods
Plant Cell Cultures and Elicitation.
C. roseus cell suspensions were grown as described in ref. 28. MeJA elicitations started on day 6 after inoculating 2 g fresh weight of cells in 25 ml of medium (with or without NAA) in 100-ml Erlenmeyer flasks. MeJA at a final concentration of 50 μM, or an equivalent of DMSO (the MeJA solvent) as a control, was added to the culture. Samples were harvested for metabolite and transcript profiling by vacuum filtration 0, 1, 4, 8, and 12 h after elicitation or 0, 4, and 12 h after the addition of DMSO. The samples were lyophilized and stored at −20°C until extraction.
Extraction and Sample Preparation.
The modified TIA extraction protocol of Whitmer et al. (29) was followed. Briefly, 100 mg of lyophilized cells were spiked with an internal standard (vincamine from Sigma-Aldrich) and extracted with 15 ml of ethanol in an ultrasonic bath for 10 min. After centrifugation at 2,711 × g for 10 min, the solvent was decanted and evaporated to dryness. The dry samples were stored at −20°C until analysis. The samples were redissolved in a 1:1 mixture of acetonitrile (Rathburn Chemicals, Walkerburn, U.K.) and 10 mM ammonium acetate (Merck, Darmstadt, Germany) and adjusted to pH 10.
Liquid Chromatography-Mass Spectrometry Analysis.
After centrifugation, a 25-μl aliquot was loaded onto a reverse-phase C18 column (Xterra MS C18, 4.6 × 150 mm, 5 μm; Waters) at 35°C and eluted after 30 min under isocratic conditions of 10 mM ammonium acetate at pH 10 and acetonitrile (55:45) by applying a flow of 1 ml/min and a split of 0.2 ml/min reaching the mass spectrometer. The separation was performed with an HT-Alliance 2795 system (Waters) and was monitored with a 996 photodiode array detector (200–270 nm; Waters) and a Micromass (Manchester, U.K.) Quattro micro triple quadrupole mass spectrometer (Waters) equipped with an electrospray source. The ion source was operated at 3.20 kV capillary voltage and 45 V cone voltage. Source and desolvation temperatures were 130°C and 290°C, respectively. Desolvation gas flow was 900 liters/h and cone gas flow 30 liters/h. The full-scan mode function was applied to record the protonated molecular ions. Targeted liquid chromatography-mass spectrometry was performed as described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The data processing methods for peak detection, alignment, and normalization were the same as those described in ref. 30.
cDNA-AFLP Analysis and Data Processing.
RNA from C. roseus cells was prepared with Concert Plant RNA Reagent (Invitrogen). Sample preparation and cDNA-AFLP-based transcript profiling were performed as described in ref. 13. For transcript profiling, all 128 possible BstYI + 1/MseI + 2 primer combinations were used. The data were processed essentially as described in ref. 31. For normalization within each primer combination, 25% of the genes with the lowest coefficient of variation value were marked as constitutively expressed. Gene tags displaying expression values with a coefficient of variation >0.6 were considered as differentially expressed and, after visual inspection, were taken for further analysis. To characterize the isolated cDNA-AFLP fragments, the sequences, directly obtained from the reamplified PCR product, were compared against nucleotide and protein sequences in publicly available databases by blast sequence alignments (32). Tag sequences were replaced with longer EST or isolated cDNA sequences, when available, to increase the chance of finding significant homology (see Table 1). The similarity threshold for blast searches was defined at 1 × 10−3. However, because of the small size of some tags, in some cases a lower value was accepted when unambiguous matches were observed (see Table 1).
RT-PCR.
RNA and single-stranded cDNA for RT-PCR analysis were prepared as described for cDNA-AFLP analysis. Expression of gene products reported to be involved in TIA biosynthesis was verified by RT-PCR with gene-specific primer pairs (data not shown). PCR products were visualized on ethidium bromide-stained agarose gels.
Data Analysis of Transcriptional and Metabolic Profiles.
Both metabolic and gene expression data sets were normalized separately with the method based on maximum likelihood estimate of scaling parameters (33) by using the complete data set in parameter estimation calculations (15). To better match the size of the transcriptional data set, the peaks from the metabolic profile data set were filtered based on coefficient of variation and retention time. Peaks eluting within the first 3 min were excluded, because retention is unstable within this period. Furthermore, peaks with too low signal-to-noise ratio, low intensity, low variability, and natural isotope peaks were removed. Only peaks with coefficient of variation >0.7 across all conditions were retained. The principal component analysis (14) was performed by using PLS Toolbox package (Eigenvector Research, Wenatchee, WA) and matlab (Mathworks, Gouda, The Netherlands). tom sawyer visualization 6.0 (Tom Sawyer Software, Oakland, CA) was used for the generation of correlation networks.
Supplementary Material
Acknowledgments
We thank Prof. R. Verpoorte (Leiden University, Leiden, The Netherlands) for generously providing the C. roseus cell line and J. Rikkinen for excellent technical assistance in tissue culture work. This research was funded by the National Technology Agency of Finland (Tekes) “NeoBio” program (to K.-M.O.-C.), Marie Curie Fellowship QLK4-2002-51547 from the European Community “Quality of Life” program (to H.R.), the Academy of Finland SysBio Program (to M.K.), and a Marie Curie International Reintegration Grant from the European Community (to M.O.).
Abbreviations
- AFLP
amplified fragment length polymorphism
- MeJA
methyl jasmonate
- TIA
terpenoid indole alkaloid.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.van der Heijden R., Jacobs D. I., Snoeijer W., Hallard D., Verpoorte R. Curr. Med. Chem. 2004;11:607–628. doi: 10.2174/0929867043455846. [DOI] [PubMed] [Google Scholar]
- 2.Noble R. L. Biochem. Cell Biol. 1990;68:1344–1351. [PubMed] [Google Scholar]
- 3.St-Pierre B., Vazquez-Flota F. A., De Luca V. Plant Cell. 1999;11:887–900. doi: 10.1105/tpc.11.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez-Flota F. A., De Luca V. Phytochemistry. 1998;49:395–402. doi: 10.1016/s0031-9422(98)00176-9. [DOI] [PubMed] [Google Scholar]
- 5.Menke F. L. H., Champion A., Kijne J. W., Memelink J. EMBO J. 1999;18:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Fits L., Memelink J. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- 7.Chatel G., Montiel G., Pré M., Memelink J., Thiersault M., Saint-Pierre B., Doireau P., Gantet P. J. Exp. Bot. 2003;54:2587–2588. doi: 10.1093/jxb/erg275. [DOI] [PubMed] [Google Scholar]
- 8.Pauw B., Hilliou F. A. O., Martin V. S., Chatel G., de Wolf C. J. F., Champion A., Pré M., van Duijn B., Kijne J. W., van der Fits L., et al. J. Biol. Chem. 2004;279:52940–52948. doi: 10.1074/jbc.M404391200. [DOI] [PubMed] [Google Scholar]
- 9.Sibéril Y., Benhamron S., Memelink J., Giglioli-Guivarc'h N., Thiersault M., Boisson B., Doireau P., Gantet P. Plant Mol. Biol. 2001;45:477–488. doi: 10.1023/a:1010650906695. [DOI] [PubMed] [Google Scholar]
- 10.Oksman-Caldentey K. M., Inzé D. Trends Plant Sci. 2004;9:433–440. doi: 10.1016/j.tplants.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Gantet P., Imbault N., Thiersault M., Doireau P. Plant Cell Physiol. 1998;39:220–225. [Google Scholar]
- 12.Katajamaa M., Orešič M. BMC Bioinformatics. 2005;6:179.1–179.12. doi: 10.1186/1471-2105-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breyne P., Dreesen R., Cannoot B., Rombaut D., Vandepoele K., Rombauts S., Vanderhaeghen R., Inzé D., Zabeau M. Mol. Genet. Genomics. 2003;269:173–179. doi: 10.1007/s00438-003-0830-6. [DOI] [PubMed] [Google Scholar]
- 14.Jackson J. E. User's Guide to Principal Components. New York: Wiley; 1991. [Google Scholar]
- 15.Orešič M., Clish C. B., Davidov E. J., Verheij E., Vogels J. T. W. E., Havekes L. M., Neumann E., Adourian A., Naylor S., van der Greef J., et al. Appl. Bioinformatics. 2004;3:205–217. doi: 10.2165/00822942-200403040-00002. [DOI] [PubMed] [Google Scholar]
- 16.Goossens A., Häkkinen S. T., Laakso I., Seppanen-Laakso T., Biondi S., De Sutter V., Lammertyn F., Nuutila A. M., Söderlund H., Zabeau M., et al. Proc. Natl. Acad. Sci. USA. 2003;100:8595–8600. doi: 10.1073/pnas.1032967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs D. I., Gaspari M., van der Greef J., van der Heijden R., Verpoorte R. Planta. 2005;221:690–704. doi: 10.1007/s00425-004-1474-4. [DOI] [PubMed] [Google Scholar]
- 18.van der Fits L., Zhang H., Menke F. L. H., Deneka M., Memelink J. Plant Mol. Biol. 2000;44:675–685. doi: 10.1023/a:1026526522555. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez-Flota F., De Luca V., Carrillo-Pech M., Canto-Flick A., de Lourdes Miranda-Ham M. Mol. Biotechnol. 2002;22:1–8. doi: 10.1385/mb:22:1:001. [DOI] [PubMed] [Google Scholar]
- 20.Fontecave M., Atta M., Mulliez E. Trends Biochem. Sci. 2004;29:243–249. doi: 10.1016/j.tibs.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Schröder G., Unterbusch E., Kaltenbach M., Schmidt J., Strack D., De Luca V., Schröder J. FEBS Lett. 1999;458:97–102. doi: 10.1016/s0014-5793(99)01138-2. [DOI] [PubMed] [Google Scholar]
- 22.Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. Proc. Natl. Acad. Sci. USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tohge T., Nishiyama Y., Hirai M. Y., Yano M., Nakajima J., Awazuhara M., Inoue E., Takahashi H., Goodenowe D. B., Kitayama M., et al. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 24.Laflamme P., St-Pierre B., De Luca V. Plant Physiol. 2001;125:189–198. doi: 10.1104/pp.125.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez S., Compagnon V., Crouch N. P., St-Pierre B., De Luca V. Phytochemistry. 2003;64:401–409. doi: 10.1016/s0031-9422(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 26.Schoendorf A., Rithner C. D., Williams R. M., Croteau R. B. Proc. Natl. Acad. Sci. USA. 2001;98:1501–1506. doi: 10.1073/pnas.98.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Sutter V., Vanderhaeghen R., Tilleman S., Lammertyn F., Vanhoutte I., Karimi M., Inzé D., Goossens A., Hilson P. Plant J. 2005;44:1065–1076. doi: 10.1111/j.1365-313X.2005.02586.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Sayed M., Choi Y. H., Frederich M., Roytrakul S., Verpoorte R. Biotechnol. Lett. 2004;26:793–798. doi: 10.1023/b:bile.0000025879.53632.f2. [DOI] [PubMed] [Google Scholar]
- 29.Whitmer S., van der Heijden R., Verpoorte R. Plant Cell Tissue Organ Cult. 2002;69:85–93. [Google Scholar]
- 30.Orešič M., Rischer H., Oksman-Caldentey K. M. In: Biotechnology in Agriculture and Forestry: Plant Metabolomics. Dixon R., Willmitzer L., Saito K., editors. Heidelberg: Springer; 2006. pp. 277–289. [Google Scholar]
- 31.Vandenabeele S., Van Der Kelen K., Dat J., Gadjev I., Boonefaes T., Morsa S., Rottiers P., Slooten L., Van Montagu M., Zabeau M., et al. Proc. Natl. Acad. Sci. USA. 2003;100:16113–16118. doi: 10.1073/pnas.2136610100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J. H., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartemink A. J., Gifford D. K., Jaakkola T. S., Young R. A. Proc. SPIE Int. Soc. Opt. Eng. 2001;4266:132–140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.