Abstract
Protozoan parasites belonging to the family Trypanosomatidae are characterized by an unusual pathway for the production of mRNAs via polycistronic transcription and trans-splicing of a 5′ capped mini-exon which is linked to the 3′ cleavage and polyadenylation of the upstream transcript. However, little is known of the mechanism of protein synthesis in these organisms, despite their importance as agents of a number of human diseases. Here we have investigated the role of two Trypanosoma brucei homologues of the translation initiation factor eIF4A (in the light of subsequent experiments these were named as TbEIF4AI and TbEIF4AIII). eIF4A, a DEAD-box RNA helicase, is a subunit of the translation initiation complex eIF4F which binds to the cap structure of eukaryotic mRNA and recruits the small ribosomal subunit. TbEIF4AI is a very abundant predominantly cytoplasmic protein (over 1 × 105 molecules/cell) and depletion to ∼10% of normal levels through RNA interference dramatically reduces protein synthesis one cell cycle following double-stranded RNA induction and stops cell proliferation. In contrast, TbEIF4AIII is a nuclear, moderately expressed protein (∼1–2 × 104 molecules/cell), and its depletion stops cellular proliferation after approximately four cell cycles. Ectopic expression of a dominant negative mutant of TbEIF4AI, but not of TbEIF4AIII, induced a slow growth phenotype in transfected cells. Overall, our results suggest that only TbEIF4AI is involved in protein synthesis while the properties and sequence of TbEIF4AIII indicate that it may be the orthologue of eIF4AIII, a component of the exon junction complex in mammalian cells.
INTRODUCTION
The flagellate protozoan parasites belonging to the family Trypanosomatidae include a number of important pathogens responsible for diseases of worldwide impact such as the Sleeping Sickness (Trypanosoma brucei), Chagas' Disease (Trypanosoma cruzi) and the various forms of Leishmaniasis (Leishmania sp.) (www.who.int/tdr). These organisms are unusual in a number of processes necessary for mRNA synthesis and maturation; transcription is polycistronic and monocistronic mRNAs arise after trans-splicing of a capped short exon on to the 5′ end and cleavage and polyadenylation at the 3′ end [reviewed in (1,2)]. As a result of trans-splicing, the 5′ ends of mature trypanosomatid mRNAs all share the same 39 nt leader sequence with a modified cap 4 structure (3). To date, little is known about how these mRNAs are translated, if major differences exist within the process of protein synthesis when compared with other eukaryotes and whether the common leader sequence influences how the mRNAs are recruited for translation.
In eukaryotes, protein synthesis is a complex process which requires a myriad of different macromolecules including RNAs and proteins. The critical initiation step requires a number of translation initiation factors (eIFs) whose activity can be highly regulated [for reviews see (4–7)]. Paramount within these factors is the heterotrimeric eIF4F complex, which is required for the recruitment of the small ribosomal subunit to the 5′ end of the mRNA. eIF4F is composed of the RNA helicase eIF4A, the cap-binding protein eIF4E and the large scaffolding protein eIF4G which mediates interactions between eIF4F and other translation factors as well as the small ribosomal subunit [reviewed in (8)].
eIF4A is the prototype member of the DEAD-box family of RNA helicases which includes several proteins mainly involved in RNA metabolism. These proteins are classified within the superfamily II of a much larger group of related RNA and DNA helicases (9). The RNA helicases couple the hydrolysis of ATP to various activities relevant for RNA function such as rearrangement of inter- or intra-molecular RNA structures, dissociation of RNA–protein complexes and RNA unwinding. The DEAD-box family members are characterized by nine sequence motifs (I, Ia, Ib, II, III, IV, V and VI and the Q motif), as well as several individual amino acids, conserved among the various proteins assigned to this family [for reviews see (10,11)]. Structurally, eIF4A assumes a ‘dumbbell’ shape with two globular domains connected by a flexible linker (12). Comparison with the structure of related RNA and DNA helicases and the Methanococcus jannaschii DEAD-box protein, similar in size to eIF4A (13), have confirmed the basic overall structure of the core helicase domains. The various conserved motifs are positioned in the interface between the two domains and have been implicated in RNA binding and ATP binding and hydrolysis. However, little is known about the molecular basis for RNA specificity and helicase function [reviewed in (11,14)]. In translation initiation, eIF4A binds to the central region of eIF4G, via the eIF4G HEAT domain (15) and, in mammals at least, also to the eIF4G C-terminus (16,17). eIF4A seems to be responsible for melting secondary structures along the mRNA 5′-untranslated region (5′-UTR), facilitating the binding of the small ribosomal subunit and the scanning of the leader region to locate the initiation codon (18,19) [reviewed in (4,6)].
In mammals three different isoforms of eIF4A have been described. Both eIF4AI and II (90% identity between the two proteins) are able to reconstitute the eIF4F subunit and presumably have similar roles in translation (20,21). In contrast, eIF4AIII, only 66% identical to mammalian eIF4AI, is functionally distinct. While eIF4AIII exhibits RNA-dependent ATPase activity and ATP-dependent RNA helicase activity, it does not support binding of the small ribosomal subunit to the mRNA, and inhibits translation in vitro (22). eIF4AIII localizes to the nucleus (23) and recent reports indicate that it may act as an anchoring factor for the exon junction complex (EJC), and is essential for nonsense-mediated decay (NMD) in mammals (24–30).
The mechanisms of translation initiation are virtually unknown in trypanosomatids. A Leishmania eIF4A homologue (called LeiF) was first described in Leishmania braziliensis and Leishmania major as a 45.3 kDa antigen, expressed in both insect and mammalian stages of the parasite life cycle, but its role in translation was not investigated (31,32). Recently, our group has identified multiple L.major homologues for the three eIF4F subunits, all of which are conserved in T.brucei (33). We characterized two putative L.major eIF4A homologues, LmEIF4A1 (LeiF) and LmEIF4A2, with 59 and 52% identities to human eIF4AI, respectively. When assayed with isoform specific antibodies these two factors differ significantly in abundance in L.major promastigotes. LmEIF4A1 is very abundant with over 105 molecules/cell whilst LmEIF4A2 is either absent or present at levels below 104 molecules/cell. Furthermore, only LmEIF4A1 was found to bind specifically to the HEAT domain of one of the Leishmania eIF4G homologues (33).
In this paper we take advantage of the genetic tools available for the study of gene function in T.brucei to extend this analysis of the two trypanosomatid eIF4A homologues. Initially, the mRNA and protein levels of the two T.brucei eIF4A orthologues were analysed during the life cycle. Their intracellular localization was identified through overexpression of enhanced yellow fluorescent protein (EYFP) fusions and their role for parasite viability investigated through RNA interference and overexpression of dominant negative mutants. Our results show that the T.brucei orthologue of LmEIF4A1 (named as TbEIF4AI) is the functional homologue of the eIF4A present in eIF4F. As for the orthologue of LmEIF4A2, it seems to be the functional homologue of the nuclear eIF4AIII present in higher eukaryotes and has been named here as TbEIF4AIII.
MATERIALS AND METHODS
Sequence analysis and molecular modeling
BLAST searches were carried out with the T.brucei genome sequences available at the Gene DB website of the Sanger Institute Pathogen Sequencing Unit (www.genedb.org). Further sequence searches, Clustal W alignments and molecular modeling were done as described previously (33).
PCR and cloning methods
The TbEIF4AI coding sequence was amplified from T.brucei Lister 427 genomic DNA (5′primer, AAG CTT CCG CCA CCA TGG CCC AAC AAG GAA AG; and 3′primer, GGA TCC AGA ACC CTC ACC AAG GTA GGC AGC; added restriction sites used in cloning are underlined) resulting in the entire open reading frame (ORF) flanked by sites for the enzymes HindIII and BamHI. The same strategy was used for the amplification of the TbEIF4AIII sequence (5′primer, AAG CTT CCG CCA CCA TGA CAG CAA CCG CAA GG; and 3′primer, GGA TCC AGA ACC GAA CTG TTC ACC GAC GTT TG). The amplified fragments were then cloned into the vector pGEM-T Easy (Promega) and sequenced. In order to express N-terminal His-tagged fusion proteins both fragments were then recovered by digestion with HindIII and BamHI and subcloned into the same sites of a modified pET15b vector. To generate the TbEIF4AI-EYFP and TbEIF4AIII-EYFP constructs, the two eIF4A fragments were cloned into the HindIII and BamHI sites of p2215, a modified form of pLEW82 (34). To make p2215, the EYFP ORF (Clontech) was obtained as a BamHI/BglII fragment and inserted into the BamHI site of pLEW82. On expression, the resultant fusion protein had the sequence: eIF4A C-terminal residue-GSGSGGG-EYFP. For the RNAi experiments the same two eIF4A DNA fragments were also subcloned into the HindIII/BamHI sites of the transfection vector p2T7-177 (35). Dominant negative mutants were made by altering the sequence of the DEAD box of the helicase (motif II in Figure 1) to DQAD (11,36). Tetracycline-inducible expression of wild type and dominant negative forms of eIF4A was performed using p2280, a derivative of pLEW100 made by introducing a BamHI/BglII DNA segment encoding three tandem myc epitope tags to its BamHI site. The two HindIII/BamHI T.brucei eIF4A fragments were cloned into the same sites of p2280 resulting in the expression of fusion proteins with the myc epitope tags on their C-terminus giving the sequence eIF4A-GSGSGPREQKLISEEDLPREQKLISEEDLPREQKLISEEDLPR.
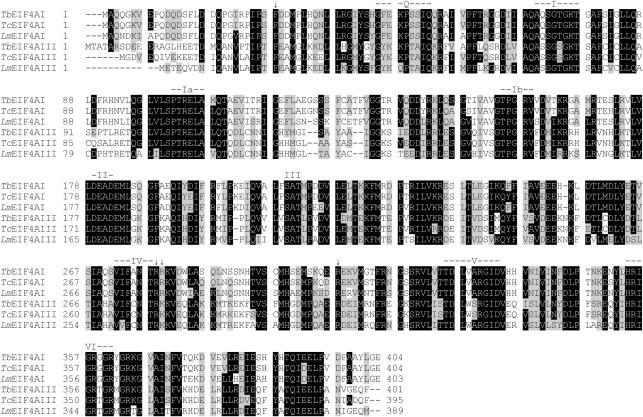
Figure 1.
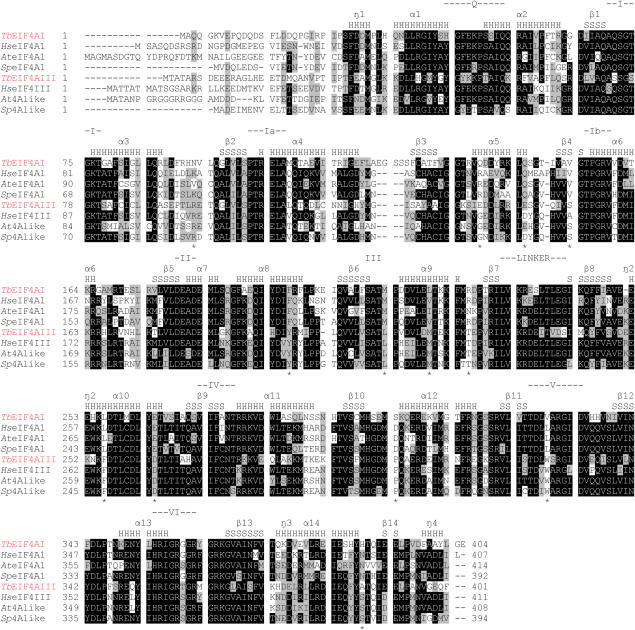
Sequence alignment comparing the T.brucei, T.cruzi and L.major eIF4A homologues. Sequences were aligned with the Clustal W program, from the Centre for Molecular and Biomolecular Informatics (http://www.cmbi.kun.nl/bioinf/tools/clustalw.shtml). Amino acids identical in >60% of the sequences are highlighted in dark gray, while amino acids defined as similar, based on the BLOSUM 62 Matrix, on >60% of the sequences, are shown in pale gray. When necessary, gaps were inserted within the various sequences (dashes) to allow better alignment. The nine motifs typical of DEAD-box RNA helicases (10,11) are highlighted. The single arrows indicate other individual amino acids which seems to be relevant for eIF4A function or RNA binding (12,42). Relevant GenBank accession numbers: LmEIF4AI, AAC24684/AAC24685; LmEIF4AIII, CAJ05468; TbEIF4AI, EAN76544; TbEIF4AIII, EAN79829; TcEIF4AI, EAN98527; TcEIF4AIII, EAN88971.
Parasite growth, transfection and RNAi
Procyclic form T.brucei Lister 427 cells were used throughout. RNAi and ectopic expression of eIF4A were performed using T.brucei Lister 427 29-13, containing integrated copies of pLEW 29 and pLEW13 (34). Procyclic T.brucei forms were propagated in SDM-79 medium at 27°C, supplemented with 10% feotal calf serum (FCS). For the 29–13 cell line, cultures were also supplemented with G418 (15 μg/ml) and hygromycin (25 μg/ml). Parasite growth was monitored microscopically every 24 h. Mid-log phase cultures (106–107 cells/ml) were then used for transfection and total protein extract production. Bloodstream forms (Lister 427) were cultivated in HMI-9 medium (37) at 37°C, 5% CO2, supplemented with 10% FCS. Cultures grown to mid-log phase cultures (105–106 cells/ml) were also harvested for the production of total protein extract.
Plasmids were linearized with NotI prior to electroporation and stable DNA integration was selected using phleomycin (2.5 μg/ml). For the RNAi experiments 1 μg/ml of tetracycline was added to mid-log phase cultures of transfected cells.
RNA analysis
RNA extraction and Northern blots were performed using standard methods (38,39). DNA fragments containing complete ORFs were used as probes for TbEIF4As and EP procyclin. A genomic repeat containing both α- and β-tubulin genes was used to detect tubulin mRNA.
Recombinant protein expression, antibody production and western blots
His-tagged TbEIF4AI and TbEIF4AIII were expressed in Escherichia coli BL21 Star (DE3) using pET15b derived plasmids. The recombinant polypeptides were insoluble after lysing the cells using a French Press. The polypeptides were purified by preparative SDS–PAGE and the bands corresponding to the recombinant proteins were then excised and sent for the production of polyclonal serum (CovalAb). Prior to their use, both antibodies were first affinity purified as described elsewhere (40) with their respective recombinant proteins. Cross-reacting antibodies were eliminated by previous incubation of the anti-TbEIF4AI antisera with TbEIF4AIII recombinant protein and vice versa. To estimate the levels of the eIF4A proteins, first the recombinant proteins were quantified by serial dilutions in SDS–PAGE by comparison with known concentrations of BSA (data not shown). After quantification they were then used in western blots with the respective antisera and compared with serial dilutions of total protein extract from both procyclic and bloodstream forms of T.brucei. The endogenous protein levels were then estimated by the densitometric analysis of the western blot results as described elsewhere (33).
Fluorescence microscopy
For the indirect immunofluorescence assay, wild-type procyclic cells grown to mid-log phase (5 × 106/ml) were harvested, washed with phosphate-buffered saline (PBS)/10 mM glucose and adsorbed to polylysine coated slides. The cells were then fixed in 100% methanol at −20°C/15 min. Antibody detection of TbEIF4AI and III followed standard procedures. DNA was stained using Hoechst 33258. For the analysis of the cells expressing TbEIF4AI-EYFP and TbEIF4AIII-EYFP, aliquots of 5 × 106 cells were harvested, washed with PBS/10 mM glucose and fixed in 0.1% formaldehyde for 5 min. In this case, DNA was stained using Hoechst 33342.
Metabolic labelling
To measure the rate of protein synthesis, [35S]methionine (10 μCi/ml) was added to mid-log cultures which were incubated for 1 h prior to the determination of trichloroacetic acid precipitable incorporation into protein. Parallel incubations in the presence of 50 μg/ml cycloheximide were used to estimate incorporation of radiolabel by processes other than cytoplasmic protein synthesis. For metabolic labeling, cultures were washed twice with methionine-free RPMI 1640 medium and then resuspended at 1 × 107 cells/ml in methionine-free RPMI 1640 containing 50 μCi/ml [35S]methionine and incubated for 1 h at 28°C prior to harvesting and analysis by SDS–PAGE and autoradiography.
RESULTS
Identification of the T.brucei eIF4A homologues
The T.brucei homologues of eIF4A were identified in searches of the genome sequence using human eIF4AI as well as the two Leishmania eIF4A sequences. At the amino acid level, the two T.brucei proteins, TbEIF4AI and TbEIF4AIII, are very similar to their L.major orthologues with identities of 88% for the eIF4AI and 82% for the eIF4AIII pair (to avoid confusion and in view of the data presented below the L.major eIF4A homologues, previously called LmEIF4A1 and 2, have also been renamed to LmEIF4AI and LmEIF4AIII and this nomenclature will be used when needed). The assignment as eIF4A homologues reflects the fact that the two sequences are the closest matches in the two parasite protein databases to human eIF4AI and both share identities of over 50% with the human protein. The third nearest eIF4A homologue in both T. brucei and L. major databases has been assigned to another group of RNA helicases, Dhh1 (33), with an identity of only ∼40% to human eIF4AI. In order to analyse the conservation of the putative eIF4A homologues within an additional member of the family Trypanosomatidae, we performed similar searches using the T.cruzi genome database. Again, orthologues to both proteins could be found in T.cruzi with the third nearest match to human eIF4AI being Dhh1.
Figure 1 shows a sequence alignment comparing the two eIF4A sequences from T.brucei with those from T.cruzi and L.major. Highlighted in the figure are the various conserved motifs typical of eIF4A and related proteins which have been shown to be required for different aspects of the RNA helicase activity. Motifs I, II, VI and the recently identified Q motif (41) have been implicated in ATP binding and hydrolysis; motif III may link nucleotide hydrolysis to helicase function; motifs Ia, Ib, IV and V may be involved in RNA binding [reviewed in (11,14)]. Several conserved arginine residues, which have also been implicated as important for eIF4A/helicase function in yeast eIF4A (12), as well as a conserved N-terminal phenylalanine residue are also shown (42). Overall the alignment confirms the close similarity between the various homologues. In general the N-terminal half of the protein is less conserved than the C-terminal half but only in the very N-terminus are significant differences in the sequences observed. A few conserved differences between the three eIF4AI and three eIF4AIII homologues with potential significance for their function can be identified, such as the replacement of a conserved F46 E47 doublet within the Q motif of eIF4AI by YK in eIF4AIII proteins. Other individual substitutions conserved between the eIF4AI and eIF4AIII homologues can be seen within motifs Q (S50T, S51A), I (Q71S), IV (A275C), V (V328W) and VI (G359T). So far however the functional significance of these substitutions is unknown.
Expression of TbEIF4A mRNAs in T.brucei bloodstream and procyclic forms
To begin the functional characterization of T.brucei eIF4A homologues and assay their expression at the mRNA level, the two genes were amplified, cloned and used as probes in northern blots of RNA from T.brucei procyclic and bloodstream forms (Figure 2A). The membranes were also probed for the constitutively expressed tubulin, to confirm that equal amounts of mRNA were loaded in each lane, and for the procyclic-specific EP procyclin mRNA to verify the stage specificity of both sets of mRNAs (43).
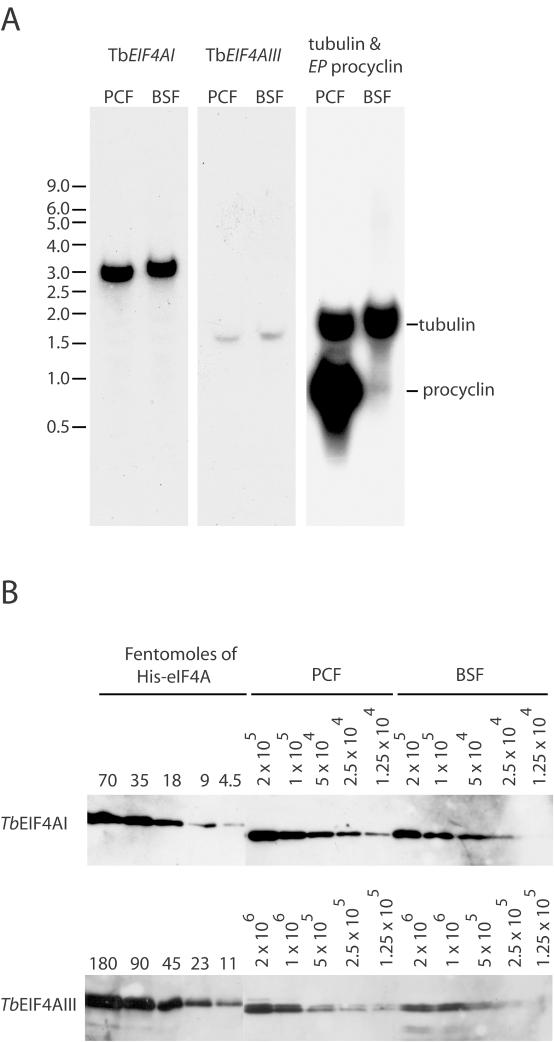
Figure 2.
Expression analysis of TbEIF4AI and III. (A) Total RNA from both procyclic (PCF) and bloodstream (BSF) T.brucei forms was separated on denaturing gels and used in northern blot assays to detect the expression of TbEIF4AI and III. One of the blots was overprobed with tubulin (ubiquitously expressed) and EP procyclin (expressed in procyclics only) as controls. The migration of RNA size markers is indicated on the left in kilobases. (B) Quantification of TbEIF4AI and TbEIF4AIII in the procyclic and bloodstream forms of T.brucei. Recombinant His-tagged TbEIF4AI and III were quantified, diluted to defined concentrations (in fmol) and ran on SDS–PAGE gels with whole parasite extract obtained from known number of cells from both procyclic and bloodstream forms (1.25 × 104–2 × 105 for TbEIF4AI and 1.25 × 105–2 × 106 for TbEIF4AIII). The proteins samples were then transferred to Immobilon-P membranes followed by incubation with the affinity purified isoform specific antisera and goat anti-rabbit IgG conjugated with peroxidase, and detection by ECL. The values obtained for the abundance of the two proteins in fentomoles/105 or 106 cells were then converted in number of molecules/cell.
The two T.brucei eIF4A mRNAs were readily detected and found to be at constant levels throughout the parasite life cycle. However, according to the northern blot, the TbEIF4AI mRNA produces a much stronger signal than TbEIF4AIII. Since both probes used were of similar specific activity, and the exposures times for the films were similar as well, it seems that the TbEIF4AIII mRNA is far less abundant than TbEIF4AI. Remarkably, although the ORFs for both proteins are similar, 1215 versus 1206 bp for TbEIF4AI and TbEIF4AIII, respectively, their mRNAs differ significantly, with the TbEIF4AI message, at ∼3 kb, being nearly twice the length of TbEIF4AIII (∼1.6 kb), probably reflecting a considerable difference in the length of the 3′-UTR.
TbEIF4AI is about 10-fold more abundant than TbEIF4AIII in both procyclic and bloodstream forms
Recombinant His-TbEIF4AI and III were expressed in E.coli, purified from inclusion bodies and used to produce antisera. Affinity purification and depletion was used to produce specific antibodies for each protein (see below, Figure 6A). The antibodies were then used in western blots to analyse the expression of both proteins as well as to estimate their intracellular levels. TbEIF4AI is very abundant (Figure 2B) and although the quantification is only approximate, its levels were estimated at ∼2–5 × 105 and 0.8–1.5 × 105 molecules/cell in procyclic and bloodstream forms, respectively, this difference being a reflection of the relative volumes of the two cell types. These levels are compatible with what has been observed with the L.major orthologue (33) as well as yeast eIF4A (44). In contrast, TbEIF4AIII levels were estimated at ∼2 × 104 and 1 × 104 molecules/cell in procyclic and bloodstream forms, respectively (Figure 2B). These data indicate that TbEIF4AI is present at levels at least 10-fold higher than TbEIF4AIII, a difference which is reminiscent of the situation with the L.major orthologues (33). Since there are estimated to be ∼50 000 mRNAs per procyclic cell (Supplementary Data), TbEIF4AI is in excess relative to mRNA, in contrast to TbEIF4AIII. Overall we conclude that both proteins are expressed constitutively and that only the TbEIF4AI levels are compatible with a role in translation.
Subcellular localization of TbEIF4AI and III
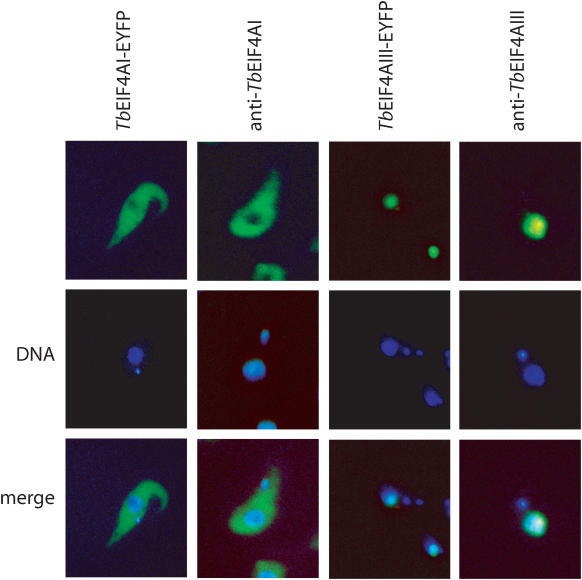
To determine the subcellular localization of the T.brucei eIF4A homologues we used two different experimental approaches. First the TbEIF4AI and TbEIF4AIII ORFs were cloned into the vector p2215 and the construct integrated into the non-transcribed spacer of a ribosomal RNA gene locus in the procyclic cell line Lister 427 29-13. This resulted in a tetracycline-inducible transgene encoding the eIF4A fused at the C-terminus to EYFP. Expression of both constructs was first verified by western blotting and similar levels of expression were observed for both TbEIF4AI and III-EYFP fusion proteins (data not shown). The fluorescent proteins were visualized by microscopy (Figure 3) and strikingly, the two proteins localize differentially within cells. TbEIF4AI-EYFP is found predominantly in the cytoplasm, whilst TbEIF4AIII-EYFP is only found in the nucleus. These results were confirmed for the endogenous proteins through indirect immunofluorescence using isoform specific antibodies: again TbEIF4AI was mainly found in the cytoplasm whilst TbEIF4AIII was only detected in the nucleus (Figure 3).
Figure 3.
Subcellular localization of TbEIF4AI and III in T.brucei procyclic forms. Subcellular localization of the TbEIF4AI and III/EYFP fusion proteins in transfected T.brucei cells was examined with a fluorescence microscope. The localization of native TbEIF4AI and III was also confirmed in wild-type procyclic cells (WT 427) by indirect immunofluorescence using the TbEIF4AI or TbEIF4AIII specific antibodies followed by incubation with the fluorescein-conjugated secondary antibody. Where indicated, the cells were counterstained to locate the nuclear and kinetoplast DNA. Note lack of TbEIF4AIII staining of the kinetoplast.
RNAi of TbEIF4AI and TbEIF4AIII
The function of the two eIF4A homologues was then investigated by knock down of expression through RNA interference. First, both ORFs were subcloned into the vector p2T7-177 vector (35) and the constructs integrated into the procyclic cell line Lister 427 29-13 resulting in cell lines with tetracycline-inducible expression of double-stranded RNA.
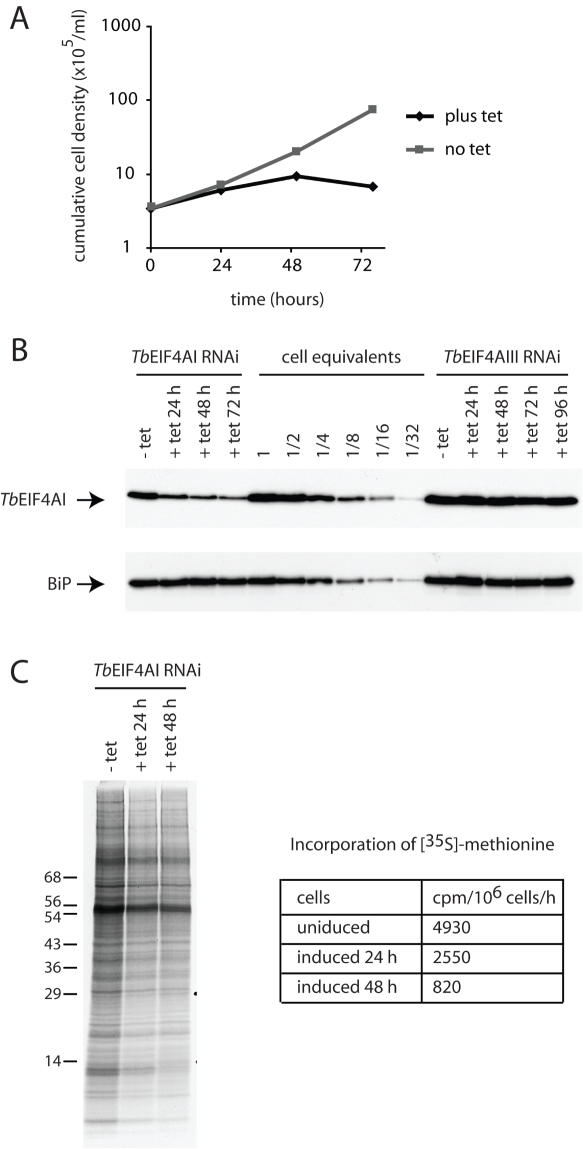
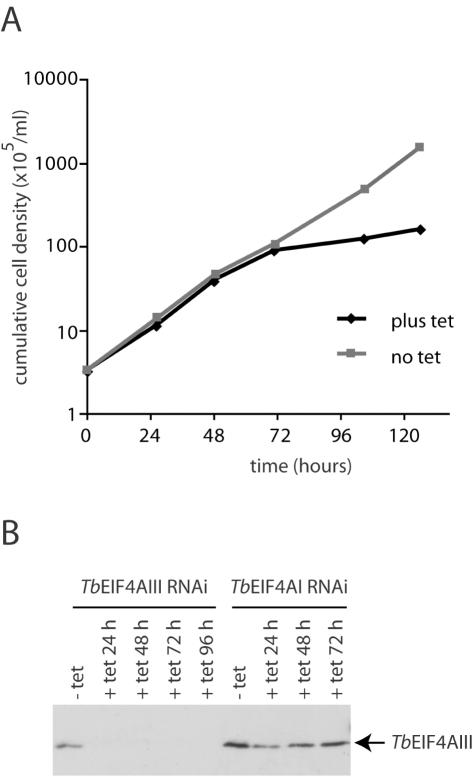
Cell proliferation was reduced within 24 h and ceased around 48 h after induction of TbEIF4AI RNAi (Figure 4A) and the cell density increased by ∼3-fold during this time. Western blotting over a time course after addition of tetracycline showed that the level of the protein decreased to <10% of the starting level but expression was not completely ablated (Figure 4B). Protein synthesis after induction of TbEIF4AI RNAi was monitored in two ways: (i) metabolic labelling to identify any alterations in the complement of polypeptides synthesized, and (ii) the rate of total protein synthesis was measured. There were no substantial changes in the profile of proteins synthesized although a small number of polypeptides appear to be relatively less affected by TbEIF4AI depletion. The overall rate of protein synthesis had halved by ∼22 h, the time at which cell proliferation ceased and was reduced to <20% of the uninduced control by 48 h as shown in Figure 4C.
Figure 4.
RNAi of TbEIF4AI. Procyclic T.brucei cells were transfected with the p2T7-177 derived plasmid containing the TbEIF4AI gene. Transfected cells were selected after growth in the presence of phleomycin and RNA interference induced after tetracycline addition. At regular intervals, cellular growth was monitored by counting the number of viable cells, expression of TbEIF4AI assayed and total protein synthesis investigated by [35S]methionine incorporation. (A) Cell density of transfected cultures with and without tetracycline addition. (B) Western blot analysis of the time course. Note the various dilutions of total cell extract for comparison (1–1/32 cell equivalent—1 cell equivalent equals to 106 cells and was used in the various RNAi lanes). TbEIF4AI was detected with the affinity purified antisera and anti-BiP was used as a loading control. The same blot was probed with both antibodies. Equivalent extracts of cells transfected with the p2T7-177/TbEIF4AIII construct (see also Figure 5) were also used in the blot to monitor for TbEIF4AI levels. (C) [35S]methionine incorporation profile in transfected cells grown without tetracycline or 24 and 48 h after its addition. Total protein synthesis was estimated after RNAi for TbEIF4AI by incubating aliquots of the cells in the presence of [35S]methionine for 1 h followed by TCA precipitation, quantitation of the incorporated radioactivity or SDS–PAGE followed by autoradiography of the selected samples.
The phenotype of cells after RNAi ablation of TbEIF4AIII was different. These cells only showed a dramatic reduction in the rate of proliferation 3 days after induction of RNAi, during which time the cell density increased ∼20-fold (Figure 5A). Levels of TbEIF4AIII fell dramatically during the first 24 h of RNAi (Figure 5B), and the protein was only just detectable in extracts derived from cells at the 48 h time point. It is possible that, owing to its low abundance even in wild-type cells, residuals levels of TbEIF4AIII persist longer than 48 h in the cells after RNAi although they are not detected by the western blotting assay. These residual levels would be responsible for the delayed onset of the growth phenotype. The western blotting results also confirm that lack of TbEIF4AIII is not involved in the phenotype induced by the depletion of TbEIF4AI since no reduction in levels of TbEIF4AIII was observed in cells submitted to the TbEIF4AI RNAi procedure (Figure 5B). Likewise the TbEIF4AIII RNAi does not lead to any reduction in the levels of TbEIF4AI (Figure 4B). These results are compatible with TbEIF4AIII being required only at very low levels so that many cell cycles are required after addition of tetracycline to impair cell growth. In contrast, the levels of TbEIF4AI, despite its abundance in wild-type cells, are much more sensitive to RNAi mediated depletion, consistent with a role in overall protein synthesis.
Figure 5.
RNAi of TbEIF4AIII. Procyclic T.brucei cells were transfected with the p2T7-177/ TbEIF4AIII construct as described for Figure 4, monitored for cellular growth and assayed for expression of TbEIF4AIII. (A) Cell density of transfected cultures at different time points with and without tetracycline addition. (B) Western blot analysis of the time course for both the TbEIF4AIII and TbEIF4AI RNAi experiments using the TbEIF4AIII antibodies. Samples from the same experiment assayed in Figure 4B were assayed for TbEIF4AIII expression.
Expression of dominant negative mutants of TbEIF4AI and III in transfected procyclic cells
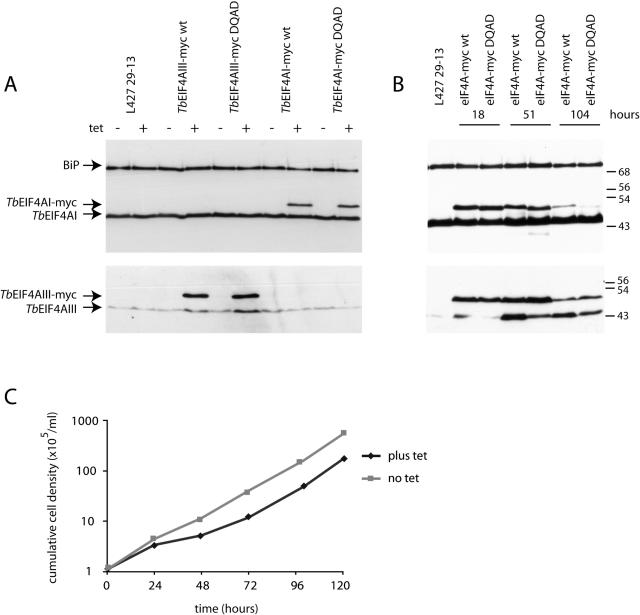
The helicase activity of eIF4A is essential for protein synthesis and viability and some mutations that abolish its activity can act as dominant negative mutants. Wild-type T.brucei helicases and equivalent DEAD-box mutant transgenes, in which the glutamic acid residue in the DEAD motif II (Figure 1) was substituted with a glutamine (DEAD→DQAD), were expressed using a tetracycline-inducible promoter. This mutation induces a dominant negative phenotype in mammalian eIF4AI, resulting in potent inhibition of protein synthesis and is widely used to abrogate the function of DEAD-box proteins (11,36). The transgenes encoded a C-terminal triple myc tag to distinguish the expression of the transgene from the endogenous protein. Wild-type and mutant versions of the two proteins were then expressed in procyclic Lister 427 29-13 cells and analysed by western blotting (Figure 6). The tetracycline regulation of expression was effective and the wild type and mutant proteins were expressed at similar levels (Figure 6A). The level of expression relative to the endogenous protein varied; the expression from the TbEIF4AI transgenes was lower than expression from the endogenous gene whereas expression from the TbEIF4AIII transgenes was several fold higher than the endogenous protein. All the myc-tagged transgenes localized correctly (data not shown).
Figure 6.
Expression of myc-tagged dominant negative mutants of TbEIF4AI and III in procyclic cells. (A) Western blot analysis of the expression of the various TbEIF4AI and III/myc fusions in transfected cells in the absence or after exposure to tetracycline for 18 h. In each case the expression was detected using antibodies specific to each of the eIF4A homologues. The TbEIF4AI western blot was simultaneously probed with anti-BiP as a loading control. (B) Time course expression of the different versions of TbEIF4A-myc after tetracycline addition to the culture. The TbEIF4AI western blot was simultaneously probed with anti-BiP as a loading control. (C) Effect of the expression of the dominant negative form of TbEIF4AI-myc on the growth of the transfected cells in culture.
Cell growth and transgene expression was monitored over a time course (Figure 6B). Expression from the TbEIF4AI transgene reduced over the time course and was barely detectable by 104 h. We have observed this diminution of expression over time with other, but not all, transgenes expressed from vectors derived from pLEW100 and are unsure of the cause. The only transgene that had any effect on growth was the mutant form of TbEIF4AI (Figure 6C), all others grew at the same rates as the control cultures without tetracycline (data not shown). At 18–51 h after the addition of tetracycline, the expression levels of the TbEIF4AI transgenes were readily detectable and the mutant, but not the wild type, produced a slowing of growth. As the expression of the transgene reduced, the culture returned to the same rate of growth as the no tetracycline control. In contrast, the significantly overexpressed TbEIF4AIII mutant transgene had no effect on growth of the culture. Interestingly, the expression of the TbEIF4AIII transgenes, but not the TbEIF4AI transgenes, resulted in increased levels of the endogenous protein. Overall, the results are compatible with the RNA helicase activity of TbEIF4AI being strictly required for growth. As for TbEIF4AIII, the lack of a slow growth phenotype when the dominant negative mutant is expressed suggests that either its RNA helicase activity is not required for the protein function or it is not affected by the DEAD→DQAD mutation. Either option strongly indicates that TbEIF4AIII is not active in translation.
Mapping of isoform specific amino acids
The results described above for TbEIF4AIII are reminiscent of what is known of mammalian eIF4AIII (Discussion). Human eIF4AIII, identified previously as a negative regulator of translation (22), has been shown to be a component of the EJC, with roles in mRNA export, cytoplasmic RNA localization and NMD (24–27). Pairwise sequence comparisons between TbEIF4AI and III (or their orthologues in T.cruzi and L.major) and the functionally divergent human eIF4AI/eIF4AIII do not show a clear match between either of the parasite homologues with the two human sequences. Indeed, the overall identity between human eIF4AI/eIF4AIII (66%) is greater than that between either protein and the two trypanosomatid eIF4As (∼55–60%).
The kinetoplastid eIF4AI and eIF4AIII sequences were then aligned with putative eIF4AI and eIF4AIII homologues from the major lines of eukaryotic evolution (Figure 7). The homologues from Arabidopsis thaliana and Schizosaccharomyces pombe were identified using BLAST searches of non-redundant sequence databases using the human eIF4AI or eIF4AIII sequences as queries. The alignment in Figure 7 does not show any continuous sequence of amino acids that distinguish between all putative eIF4AI or eIF4AIII homologues. However, at various positions, interspersed within the sequences common to both sets of proteins, individual amino acids can be identified which are conserved and unique either to the eIF4AI or eIF4AIII proteins. Table 1 lists 13 positions where a clear difference could be found between the two sets of sequences. Several, but not all, of these amino acid substitutions are also shared by an eIF4AIII-related protein from Saccharomyces cerevisae, Fal1p, a nucleolar protein shown to be required for 40S ribosomal subunit formation (45). Fal1p, however, does not seem to be involved in EJC formation since a search in S.cerevisae for similar EJC constituents, conserved in other fungi and in plants, such as Magoh or Y14, did not produce any clear homologues.
Figure 7.
Sequence alignment comparing TbEIF4AI and III with the putative eIF4AI and eIF4AIII from selected organisms. (A) Sequences were aligned as described in Figure 1 and the various DEAD-box motifs are shown as indicated previously. The predicted secondary structural elements derived from the modelling shown in Figure 8 and from Ref. (46) are indicated numbered α1–α13/η1–η4 (alpha-helices—H) and β1–β14 (beta-strands—S). Asterisk indicates amino acids which distinguish between the eIF4AI and eIF4AIII homologues. Further relevant GenBank accession numbers: human (Hs) eIF4AI, AAX43035; human eIF4AIII (HseIF4A3), P38919; S.pombe (Sp) eIF4A1, CAA56772; S.pombe eIF4A-like protein (Sp4Alike), CAA92238; A.thaliana (At) eIF4A1, NP_177417; A.thaliana eIF4A-like protein (At4Alike), NP_188610.
Table 1.
Summary of the amino acid substitutions identified between the putative eIF4AI/eIF4AIII homologues from the main lineages of eukaryotic evolution
| Position: TbEIFAI | Substitution: 4AI > 4AIII | Secondary structure | Domain | Overall position in predicted tertiary structure |
|---|---|---|---|---|
| 93 | K/N/V > R | Loop | N-terminal | Near α5/exposed |
| 139 | Q/R > E/G | α 5 | N-terminal | Exposed |
| 146 | Q/A > E/D | α 5 | N-terminal | Exposed |
| 153 | V > S/Aa | β4 | N-terminal | Buried |
| 197 | F > Y | α8 | N-terminal | Partially exposed |
| 213 | M > L | Loop | N-terminal | Interface/next to Motif III/partially buried |
| 220 | L/V/I > M | α9 | N-terminal | Buried |
| 226 | R/N > T | Loop | C-terminal | Exposed |
| 256 | L > Fa | α 10 | C-terminal | Partially buried |
| 264 | E > Da | α 10 | C-terminal | Exposed |
| 303 | S/D > Pa | Loop | C-terminal | Exposed |
| 328 | V/L > Wa | Loop | C-terminal | Interface/Motif V/Buried |
| 388 | H/N > A/Sa | Loop | C-terminal | Exposed |
aThese substitutions but not the others are present in the S. cerevisae nucleolar protein Fal1p [may be related to the eIF4AIII proteins (45)].
The various amino acid substitutions listed in Table 1 (TbEIF4A I numbering), indicated by a star in the alignment in Figure 7, discriminate between all putative eIF4AI and eIF4AIII homologues compared, including the two trypanosomatid proteins. These substitutions are located in the two globular domains present in eIF4A and related DEAD-box helicases (11,14). Both the N- and the C-terminal domains have been shown to participate in the binding to RNA and ATP required for the helicase/ATPase activities, but few roles have been postulated for them regarding specific protein functions. The alignment results clearly show that candidate eIF4AIII homologues are present throughout the various eukaryotic lineages, although it has only been functionally characterized in metazoans. The unique substitutions are also indicative of amino acids involved in specific aspects of eIF4AI/III function in general (see below).
Molecular modelling of TbEIF4AI and TbEIF4AIII
To understand the functional implications of the observed amino acids substitutions to eIF4A function, not only in trypanosomatids but also in eukaryotes in general, we modeled the structures of both TbEIF4AI and III based on the solved structure of either the yeast S.cerevisae eIF4A (12) or the related DEAD-box protein Dhh1p (46). The structure of yeast eIF4A is in an open conformation with the two globular domains positioned apart and non-interacting. In contrast, Dhh1p is in a closed conformation with the two domains facing each other. Most of the conserved motifs in Dhh1p are positioned in close spatial proximity facing the cleft between the two domains. Both sets of models were validated as described (33) and found to have self-consistency in terms of sequence–structure compatibility and to be of good overall quality. For our analysis we favored the closed conformation structure since the two domains need to interact in order to fully form the ATP- and RNA-binding sites (12,46,47).
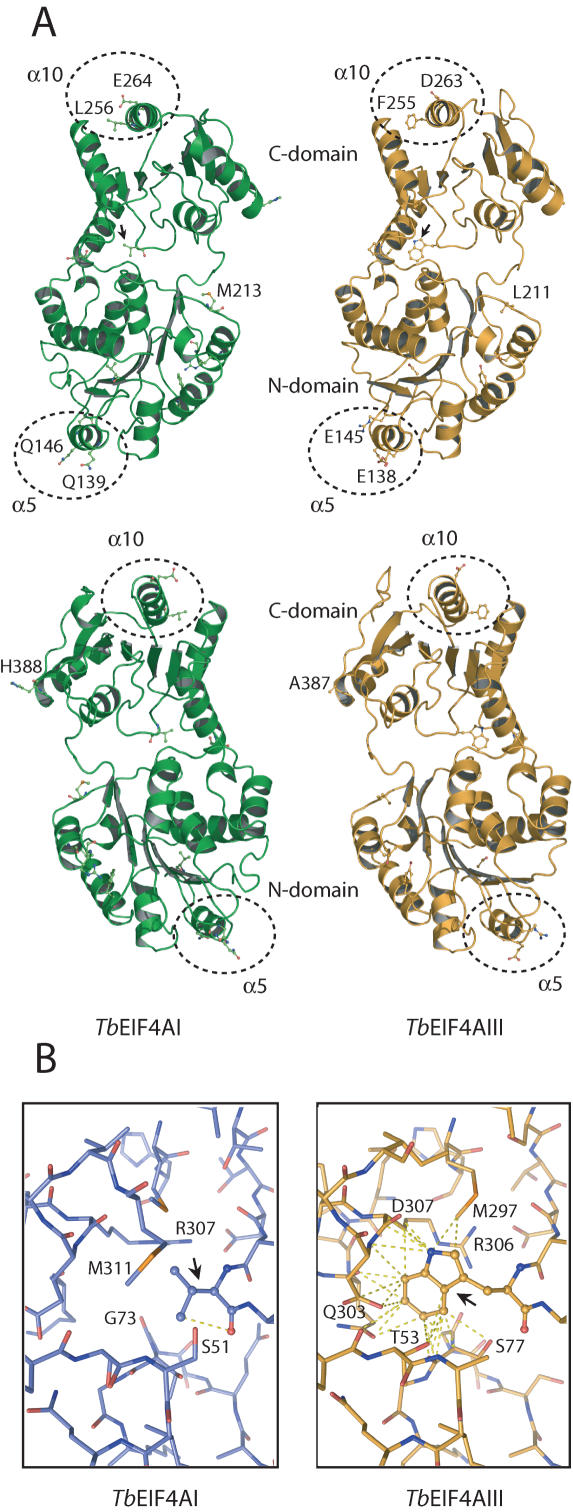
Figure 8A shows the ribbon drawing for the predicted structures of TbEIF4AI and TbEIF4AIII. Highlighted in the figure are several of the diagnostic amino acid substitutions identified in the eIF4A alignment (Figure 7). Of special interest is the V/L328W substitution in motif V, in the C-terminal domain. Motif V lies in a loop positioned in the interface between the two domains and, in Dhh1p, several amino acids in this motif are seen to make direct interactions with specific amino acids in motifs I and Q, positioned in the N-terminal domain (46). In the models shown here both the V and W residues in TbEIF4AI and III, respectively, are protruding from the main polypeptide backbone in the direction of a cleft in the proteins' N-terminal domain. To investigate the likelihood of either amino acid interacting with neighboring chains, atoms in these chains were first identified which are positioned within a radius of 4 Å from the two residues. These are the only ones capable of forming non-covalent interactions to atoms in either amino acid and the full set of potential interactions are shown in Figure 8B as dotted lines. The substantially larger W residue in TbEIF4AIII is capable of making a number of interactions with neighboring amino acids in both the N- and C-terminal domains, as well as with the polypeptide backbone. In contrast, the V residue in TbEIF4AI is very limited in the number of interactions it can establish. It is possible then that the presence of the W residue in TbEIF4AIII, and other eIF4AIII homologues, can enhance the interaction between the helicase's two domains as compared to the V/L residue in the eIF4AI and even Dhh1p proteins.
Figure 8.
Molecular modelling of TbEIF4AI and III highlighting the position of amino acids unique to the eIF4AI or eIF4AIII homologues. Diagrams were created with the program PyMol (http://www.pymol.org). (A) Ribbon diagrams of the overall structure of both TbEIF4AI and III viewed as in (46) (upper panel) or rotated 180° about its long axis (lower panel). The structure is in a closed conformation where the two, N- and C-terminal, domains are facing each other. The arrows indicate the position of the L328W substitution which lies in the loop containing Motif V and is positioned in the interface between the two domains. The dotted circles delimit the two helices discussed in the text, α5 and α10. The H/N388A/S and M213L substitutions are also indicated (their numbering differ however from the eIF4AI/eIF4AIII sequences—for instance, H388 in TbEIF4AI is equivalent to A387 in TbEIF4AIII and so on). (B) Balls and sticks representation showing the neighbourhood of the L328W substitution in both TbEIF4AI and III. The dotted lines indicate the atoms in the neighbouring amino acid chains which are positioned within a radius of 4 Å from the atoms in either the L or W residues. In both (A and B), the relevant amino acids are listed.
Other potentially interesting eIF4AIII-like substitutions map in helices 5 (Q/R139E/G, Q/A146D/E) and 10 (L256F, E264D), on the N- and C-terminal domains, respectively. These helices are largely exposed to the solvent on the external side of the proteins (Table 1) and thus the amino acids involved could mediate eIF4A binding to functional partners. Indeed, recent evidence strongly supports such a hypothesis. First, the binding surface for eIF4GII has been mapped to the C-terminal domain of eIF4AI (47). A double mutation in human eIF4AI which prevents binding to eIF4GII maps to helix 10 and targets the same glutamate residue (E264) found to be unique to the eIF4AI sequences. Likewise, the two substitutions in helix 5 are included within a proposed eIF4AIII specific motif (motif C) which has just been found to constitute part of the binding site for the EJC component MNL51 (30). Moreover, a further unique substitution identified in Figure 7, which lies in an exposed loop near the C-terminal end of the eIF4A proteins (H/N388A/S—also shown in Figure 8), lies within another proposed motif (motif H) found to be required for eIF4AIII to bind spliced mRNA and to rescue NMD in eIF4AIII depleted cells (30). In summary, we have identified several individual amino acids conserved in either eIF4AI or eIF4AIII sequences which may play significant roles in these proteins' functions not only in trypanosomatids but also in eukaryotes in general.
DISCUSSION
The results presented here provide strong support that only one of the two eIF4A homologues identified in trypanosomatids is involved in the initiation of translation. The abundance of the TbEIF4AI protein, its constitutive expression during the parasite life cycle as well as its cytoplasmic localization, the effect of RNAi depletion and the dominant negative phenotype of the DEAD→DQAD mutation are all compatible with what is expected of this protein. In contrast, TbEIF4AIII does not seem to play an obvious role in protein synthesis. The nuclear localization of TbEIF4AIII, its low abundance, longer response to the RNAi induced phenotype and lack of inhibition by the dominant negative mutant all indicate an essential role in RNA metabolism in the nucleus unrelated to eIF4A function in translation. These results are also compatible with what is known of the L.major orthologues; LmEIF4AI binds strongly to at least two eIF4G homologues whereas LmEIF4AIII has a reduced binding activity [(33) and C. R. S. Reis, unpublished data]. Mammalian eIF4AIII localizes mainly to the nucleus (23), is present in levels ∼10-fold lower than eIF4AI in HeLa cells and does not function in protein synthesis (22). An unusual feature of human eIF4AIII is that the DEAD→DQAD mutation has no effect on its activity in EJC formation and NMD (30). Thus, TbEIF4AIII behaves similarly to human eIF4AIII in several important aspects and, coupled with the sequence analysis data, our results are consistent with it being an eIF4AIII orthologue with functions possibly conserved along most major lines of eukaryotic organisms.
As part of the EJC, eIF4AIII binds directly to the core proteins Magoh, Y14 and MLN51 (25–30) and also to other proteins required for EJC function such as the TAP and Aly/REF proteins involved in nuclear mRNA export (25). Magoh homologues have been clearly identified in the three trypanosomatid genomes finished to date, T.brucei (GenBank, AAZ12053), T.cruzi (EAN97132) and L.major (CAJ06870) and possible TAP homologues can also be found. We have also tried to identify candidate Y14, MLN51 or Aly/REF homologues but so far without success. However, both Y14 and Aly/REF are small RNA-binding proteins with single RRMs, a category which includes many proteins with unassigned functions in those three genomes (48). It may be possible that, due to the degree of evolutionary distance between trypanosomatids and animals, homologues to these two proteins cannot be clearly identified by sequence analysis alone. As for MLN51 it is poorly conserved outside the metazoans so it is unlikely also for homologues to be identified in trypanosomatids only by sequence analysis. Nevertheless the strong conservation of the Magoh sequences between the human and parasite homologues (over 50% identity) is an indication that the EJC may be present throughout the major groups of eukaryotes and that eIF4AIII-like proteins may be active within this complex.
In a very recent study eight eIF4AIII specific motifs (named A to H) were identified in an alignment comparing various eIF4AIII homologues with the human eIF4AI and II proteins. Selected amino acids in some of these motifs, as well as in the canonical eIF4A motifs I, Ia and VI, were then mutated in recombinant or in vivo overexpressed eIF4AIII to investigate their requirement for eIF4AIII function (30). In the alignment provided here, which includes both T.brucei eIF4A homologues, as well as eIF4AI sequences from divergent organisms, no continuous set of amino acids were found to be typical of either eIF4AI or eIF4AIII proteins. However, unique amino acid substitutions were identified which distinguish eIF4AIII-like proteins from eIF4AI homologues in all sequences investigated so far. Several of these substitutions not only coincide with some of the proposed eIF4AIII specific motifs (motifs C, E and H), but also are included in two of those motifs found to be involved in specific eIF4AIII functions such as binding to the EJC partner MLN51 (motif C) and requirements for binding to spliced mRNA and for NMD (motif H) (30). However some of the unique eIF4AI/eIF4AIII substitutions identified here do not coincide with the remaining proposed motifs. These might be involved in mediating other aspects of eIF4A function and should be considered as targets for further investigation.
Very few protein coding genes in trypanosomatids contains a cis-intron (49,50). However, every cytoplasmic mRNA is trans-spliced to form the mature 5′ end of the mRNA and this splice site is possibly the location of EJC binding. The function of the EJC in these organisms remains obscure specially considering that the splice site is always to the 5′ side of the ORF. The EJC-mediated mechanism of NMD seems to be restricted to mammalian cells [reviewed in (51,52)] and indeed there is strong evidence that NMD does not occur in trypanosomatids (38). In mammals, both the EJC and the nuclear cap-binding complex (CBC, composed of two subunits CBP20 and CBP80) bind to precursor mRNAs in the nucleus, prior to or during the splicing event, and remain bound to the mRNAs until they are transported to the cytoplasm and/or translated for the first time (53,54). In T.brucei a novel CBC has been described which consists of a CBP20 subunit (also present in yeast and humans) plus four other polypeptides, one of which is importin-α (known to associate with CBC in other eukaryotes) and three novel proteins only present in trypanosomatids. The parasite CBC has been implicated in the early steps of mRNA maturation, prior to the trans-splicing event whereas the polycistronic precursor mRNA is cleaved into mature monocistronic units (55). At this stage it still remains to be determined whether TbEIF4AIII and other components of the putative EJC are also necessary for mRNA processing, export from the nucleus or even translation in trypanosomatids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank M. Moore for sharing unpublished data and J. Bangs for the gift of the BiP antibody. O. Thiemann and D. Souza allowed us access to the computing facilities necessary for the modelling work. The work in Cambridge was funded by the Wellcome Trust and by a Nuffield Foundation summer studentship to N. Marinsek. The Brazilian scientists were supported by grants/studentships from CNPq and CAPES. Funding for the laboratory in Recife came in part from a grant from FACEPE/CNPq/CT INFRA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Campbell D.A., Thomas S., Sturm N.R. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Liang X.H., Haritan A., Uliel S., Michaeli S. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell. 2003;2:830–840. doi: 10.1128/EC.2.5.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry K.L., Watkins K.P., Agabian N. Trypanosome mRNAs have unusual ‘cap 4’ structures acquired by addition of a spliced leader. Proc. Natl Acad. Sci. USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershey J.W.B., Merrick W.C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control Of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- 5.Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preiss T., Hentze M.W. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 7.Sonenberg N., Dever T.E. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 8.Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya A.E., Koonin E.V. Helicases-amino-acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 10.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 11.Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 12.Caruthers J.M., Johnson E.R., McKay D.B. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Story R.M., Li H., Abelson J.N. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc. Natl Acad. Sci. USA. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 15.Marcotrigiano J., Lomakin I.B., Sonenberg N., Pestova T.V., Hellen C.U., Burley S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 16.Imataka H., Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H.S., Cho M.H., Zakowicz H., Hegamyer G., Sonenberg N., Colburn N.H. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol. Cell. Biol. 2004;24:3894–3906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen P.J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoder-Hill J., Pause A., Sonenberg N., Merrick W.C. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J. Biol. Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
- 22.Li Q., Imataka H., Morino S., Rogers G.W., Jr, Richter-Cook N.J., Merrick W.C., Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holzmann K., Gerner C., Poltl A., Schafer R., Obrist P., Ensinger C., Grimm R., Sauermann G. A human common nuclear matrix protein homologous to eukaryotic translation initiation factor 4A. Biochem. Biophys. Res. Commun. 2000;267:339–344. doi: 10.1006/bbrc.1999.1973. [DOI] [PubMed] [Google Scholar]
- 24.Ferraiuolo M.A., Lee C.S., Ler L.W., Hsu J.L., Costa-Mattioli M., Luo M.J., Reed R., Sonenberg N. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl Acad. Sci. USA. 2004;101:4118–4123. doi: 10.1073/pnas.0400933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan C.C., Dostie J., Diem M.D., Feng W., Mann M., Rappsilber J., Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palacios I.M., Gatfield D., St J.D., Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 27.Shibuya T., Tange T.O., Sonenberg N., Moore M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nature Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 28.Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le H.H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nature Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 29.Tange T.O., Shibuya T., Jurica M.S., Moore M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibuya T., Tange T.O., Stroupe M.E., Moore M.J. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12:360–374. doi: 10.1261/rna.2190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skeiky Y.A., Guderian J.A., Benson D.R., Bacelar O., Carvalho E.M., Kubin M., Badaro R., Trinchieri G., Reed S.G. A recombinant Leishmania antigen that stimulates human peripheral blood mononuclear cells to express a Th1-type cytokine profile and to produce interleukin 12. J. Exp. Med. 1995;181:1527–1537. doi: 10.1084/jem.181.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skeiky Y.A., Kennedy M., Kaufman D., Borges M.M., Guderian J.A., Scholler J.K., Ovendale P.J., Picha K.S., Morrissey P.J., Grabstein K.H., et al. LeIF: a recombinant Leishmania protein that induces an IL-12-mediated Th1 cytokine profile. J. Immunol. 1998;161:6171–6179. [PubMed] [Google Scholar]
- 33.Dhalia R., Reis C.R., Freire E.R., Rocha P.O., Katz R., Muniz J.R., Standart N., de Melo Neto O.P. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol. Biochem. Parasitol. 2005;140:23–41. doi: 10.1016/j.molbiopara.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 35.Wickstead B., Ersfeld K., Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol. Biochem. Parasitol. 2002;125:211–216. doi: 10.1016/s0166-6851(02)00238-4. [DOI] [PubMed] [Google Scholar]
- 36.Pause A., Methot N., Svitkin Y., Merrick W.C., Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirumi H., Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 38.Webb H., Burns R., Ellis L., Kimblin N., Carrington M. Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short-lived protein factor. Nucleic Acids Res. 2005;33:1503–1512. doi: 10.1093/nar/gki298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrington M., Roditi I., Williams R.O. The structure and transcription of an element interspersed between tandem arrays of mini-exon donor RNA genes in Trypanosoma brucei. Nucleic Acids Res. 1987;15:10179–10198. doi: 10.1093/nar/15.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minshall N., Thom G., Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordin O., Tanner N.K., Doere M., Linder P., Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 43.Roditi I., Carrington M., Turner M. Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature. 1987;325:272–274. doi: 10.1038/325272a0. [DOI] [PubMed] [Google Scholar]
- 44.von der Haar T., McCarthy J.E. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 45.Kressler D., de la,C.J., Rojo M., Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Z., Coller J., Parker R., Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberer M., Marintchev A., Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Gaudenzi J., Frasch A.C., Clayton C. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot. Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mair G., Shi H., Li H., Djikeng A., Aviles H.O., Bishop J.R., Falcone F.H., Gavrilescu C., Montgomery J.L., Santori M.I., et al. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000;6:163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conti E., Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Lejeune F., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Lejeune F., Ishigaki Y., Li X., Maquat L.E. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 55.Li H., Tschudi C. Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol. Cell Biol. 2005;25:2216–2226. doi: 10.1128/MCB.25.6.2216-2226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.