Abstract
We describe two homologues of the mammalian divalent metal transporter (DMT1) for Schistosoma mansoni, a pathogenic intravascular parasite of humans. Schistosomes have a high nutritional and metabolic demand for iron. Nucleotide sequences of the parasite homologues, designated SmDMT1A and -B, are identical in all but the 5′-regions. The predicted amino acid sequences share at least 60% identity with DMT1 (=Nramp2) of humans, mice, and rats, and at least 55% identity with Nramp1 from mice, humans and Caenorhabditis elegans. SmDMT1A is expressed in differentiating eggs, miracidia, cercariae, schistosomula, and adults, whereas SmDMT1B is expressed in all but the miracidium and occurs at lower levels than SmDMT1A in differentiating eggs and cercariae. An iron-responsive element, present at the 3′-untranslated region of many DMT1 molecules, is not present in schistosome mRNAs studied here. A Western blot analysis of adult worm preparations using a homologous rabbit serum raised against a schistosome DMT1 peptide and a heterologous serum raised against mammalian DMT1, revealed a band approximating 115 kDa. By immunofluorescence microscopy, the schistosome DMT1s localize primarily to the tegument. Iron uptake assays demonstrated that SmDMT1s were able to rescue yeast growth in ferrous iron-transport deficient yeast (fet3fet4). The results suggest that schistosomes express molecules for ferrous iron transport in their tegument, suggesting trans-tegumental transport as one means of iron acquisition for these parasites.
Schistosomes are platyhelminth parasites of humans, responsible for significant disease in many tropical countries. Adult schistosomes inhabit the portal blood system of their hosts, where they feed extensively on the hemoglobin from host erythrocytes as the primary protein source. Female schistosomes have been recorded to imbibe up to 330,000 erythrocytes/h, equivalent to a daily intake of 0.88 μl of blood, whereas males imbibe only 39,000 cells/h (1).
Erythrocytes are digested within the gastrodermis by a series of extracellular cytolytic and specific proteolytic enzymes that catabolize hemoglobin to dipeptides or free amino acids for absorption by the gastrodermal syncytium (1-3). Hemoglobin is a rich source of amino acids, but heme, its catabolic by product, is potentially toxic to the parasites and is polymerized into hematin prior to egestion. The heme-bound iron, which accounts for much of the iron present in schistosome tissues, is thus egested from the body of the worms without absorption into somatic tissues.
Iron is a trace element, essential in a wide variety of metabolic activities including immunological resistance, oxidative metabolism, and biomineralization (4, 5). A surplus of iron in organisms has potentially lethal effects (6), because the element can induce the formation of toxic hydroxyl radicals that damage lipid membranes, proteins, and DNA (7-9). Schistosome tissues are iron-rich, and much of this abundance can be attributed to heme and heme polymers present in the gastrodermal lumen as by-products of hemoglobin digestion. In addition to this gut store, female schistosomes express a vitelline cell-specific ferritin molecule, which presumably provides iron for embryogenesis (10).
In mammals, ferrous ions are absorbed across the intestinal brush border by a divalent metal transporter (DMT1),4 a member of the highly conserved Nramp (natural resistance-associated macrophage protein) family (11). Nramp proteins have a wide range of functions in eukaryotes but are primarily involved in trans-membrane transport of divalent metals. DMT1 is an integral membrane transporter containing 10-12 transmembrane domains (7, 11) and can accept a broad range of transition metal ions, among which Fe2+ is transported with high affinity (11, 12).
We hypothesized that because schistosomes have large tissue stores of iron (13) and live in an iron-rich environment, it was probable that the parasites have pathways for iron acquisition from the bloodstream. Moreover, because Nramp proteins are ubiquitous and highly conserved, we reasoned that a DMT1 homologue is present in these parasites. This paper describes the identification and preliminary characterization of two schistosome DMT1 molecules from Schistosoma mansoni.
EXPERIMENTAL PROCEDURES
Parasites—Adult S. mansoni (Puerto Rican strain, both sexes) worms were perfused from BALB/c mice 7-10 weeks after subcutaneous challenge with cercariae and washed thoroughly in phosphate-buffered saline (PBS). Schistosome eggs were isolated from mouse livers (14), and miracidia were hatched in non-chlorinated water from finely diced mouse livers. Cercariae, collected from infected Biomphalaria glabrata snails, were kindly provided by Dr. Fred Lewis (Biomedical Research Institute, Rockville, MD).
Degenerate PCR—Degenerate primers were designed from consensus regions identified by alignment of Caenorhabditis elegans (NM_076730), Mus musculus (L33415), Drosophila melanogaster (AY058543), and Homo sapiens (AF046997) DMT1 nucleotide sequences. A nested PCR strategy was employed to amplify schistosome cDNA. The first reaction used a previously constructed S. mansoni adult λ-ZAP cDNA library as template and degenerate primers P23F 5′-ATGWSCATWGCSTAYYTRGAYCCHGGHAAYAT and P1274R 5′-AGDBCSVRWGCARBCARVCA in conditions specified for use with Amplitaq DNA polymerase (Applied Biosystems, Foster City, CA). The second PCR was performed using 1 μl of PCR product as template and the nested degenerate primers P250F 5′-GCYGWDCCRATRACYTCYKGCAT and P600R 5′-GACTTRACYAARGCDGAATGCA. PCR parameters were as follows: 95 °C of denaturation for 1 min, 50-65 °C of primer annealing for 1 min (melting temperature dependant on primers pairs used), 72 °C of primer extension for 3 min, and cycle for 30 cycles. A final extension of 7 min was performed at 72 °C before storing the samples at 4 °C. PCR products were gel extracted using the QIA-quick® gel extraction and purification kit and cloned into TOPO TA cloning vector pCR4-TOPO (Invitrogen), and the DNA insert was sequenced. Sequences were compared with those in the GenBank (nr) and dbEST databases using the National Centre for Biotechnology Information (NCBI) Blast program. Gene-specific primers were designed from the resultant sequence and full-length SmDMT1-A and -B sequences were obtained using cDNA generated by the GeneRacer Kit (Invitrogen).
Molecular Phylogeny—To demonstrate the similarities among related Nramp sequences from a diversity of organisms, a phylogenetic tree was constructed from inferred amino acid sequences. A variety of vertebrate Nramp1 and Nramp2 sequences were selected for inclusion, as well as sequences from several invertebrates (NCBI accession numbers are listed in Fig. 1B). No full-length Nramp sequences are available from any species of parasitic or free-living platyhelminth, other than S. mansoni. However, the trace archive at NCBI contains data from a whole genome shotgun sequencing project for the free-living triclad planarian Schmidtea mediterranea. This archive was searched, using as the query nucleotide sequences from the most conserved portions of the Nramp genes of S. mansoni. Several exons of the triclad homolog were thus identified. By extending the search in 5′- and 3′-directions, most of the triclad gene was eventually obtained. The entire genomic sequence for the gene was not obtained because of the presence of very long introns.
FIGURE 1.
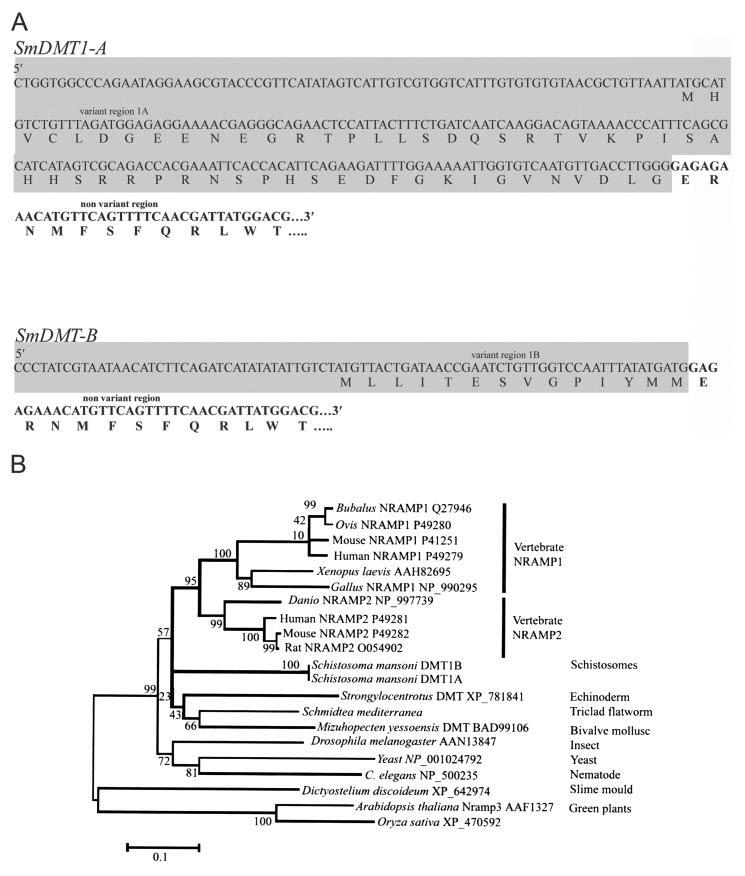
A, identification of two isoforms of the DMT1 mRNAs in S. mansoni. Sequences of the alternative 5′-regions of the S. mansoni DMT1 mRNA are labeled as variant region 1A and variant region 1B, respectively. Each molecule possesses a downstream non-variant region. B, minimum evolution tree inferred from amino acid sequences of NRAMP genes from flatworms and a variety of other organisms (see text for more details of tree construction). Species names, gene name (where assigned), and published accession numbers are shown for each sequence. Bootstrap values (1000 resamplings) are shown to the left of the relevant nodes. The tree is midpoint-rooted.
ClustalW (15) was used to align the sequences. Only sites for which data were available for S. mediterranea (374 sites) were included in the analyses. All sites in which an alignment gap occurred in any sequence were omitted from the analysis. Trees were constructed in MEGAv3.0 (16) using the minimum evolution method. The substitution model for the tree shown in Fig. 1B was the JTT model. Uniform rates among sites were assumed. The dataset was bootstrapped 1000 times, and resulting values shown on branches of the tree. The tree is midpoint-rooted.
Stage-specific Expression of SmDMT1-A and -B—Expression of SmDMT1-A and -B (S. mansoni divalent metal transporter 1 A and B) transcripts in key life cycle stages (male and female adult worms, eggs, cercariae, and miracidia) of the parasite was determined by RT-PCR. Total RNA was extracted from each of the life cycle stages of the parasite using TRIzol reagent (Invitrogen) in accordance with the manufacturer's instructions. Single stranded cDNA was generated by priming 1 μg of total RNA with 500 ng of oligo(dt)12-18 random primers (Boehringer, Mannheim) with Superscript II reverse transcriptase (Invitrogen). Control reactions (without the addition of reverse transcriptase) were included to monitor for contamination with genomic DNA and primers targeting a 291-bp region of the S. mansoni gene encoding triosephosphate isomerase were used to amplify constitutively expressed positive control cDNA (17). Specific forward primers for SmDMT1A (5′-ATGCATGTCTGTTTAGATGGAGAGGAAAACGAGGGCAG′) and SmDMT1B (5′-CCACCCTATCGTAATAACATCTTCAGATCATATATATTG) were used with the same reverse primer (5′-CAAAATAGTAATAAGTACACCAGCCCATAA) to amplify 555-bp and 435-bp target fragments, respectively. PCRs were performed on single stranded cDNA as follows: hot start at 94 °C for 2 min followed by 30 cycles of 94 °C for 1 min; 55 °C for 1 min and 72 °C for 1 min. The products were resolved on a 1.5% agarose gel and stained with ethidium bromide.
Antisera—A rabbit antiserum was raised against a synthetic peptide (CTREGKIPVR) corresponding to a region of the fourth extracellular loop of schistosome DMT1 (see Fig. 2) and synthesized as a KLH-fusion protein (Sigma Genosys). Prior to use, anti-KLH antibodies were removed from the serum by affinity purification. For this, the serum was passed through KLH-loaded HiTrap-NHS columns (Amersham Biosciences) according to the manufacturer's instructions. Sera were tested prior to use for cross-reactivity by PAGE and Western blot analysis (data not shown). A heterologous rabbit antiserum, raised against a 15-amino-acid peptide corresponding to amino acids 230-243 between transmembrane regions 5 and 6 of the rat DMT1 molecule (18) was also used for Western blot analysis and immunolocalization.
FIGURE 2.
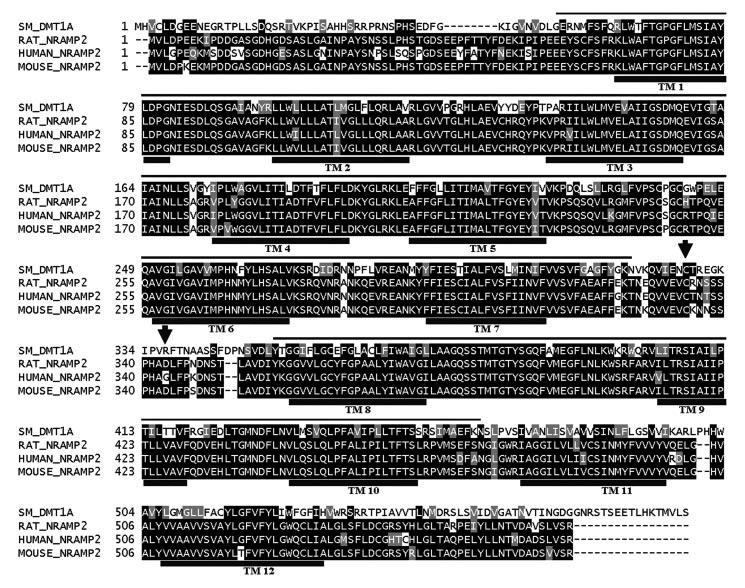
Multiple sequence alignment (ClustalW) of deduced amino acid sequence of S. mansoni DMT1-A with Nramp2 sequences of rat (Rat_Nramp2, GenBank™ accession number O54902), human (Human_Nramp2, P49281), and mouse (Mouse_Nramp2, P49282). Putative transmembrane reg ions are spanned by a black line and labeled TM. Arrows denote the position of the peptide sequence used to generate antisera against SmDMT1. The thin line over alignment indicates the length of the partial Nramp gene from S. mediterranea available for phylogenetic analysis.
A membrane extract of adult S. mansoni worms was prepared using the cellular labeling and immunoprecipitation kit (Roche Applied Science), according to manufacturer's instruction. Whole parasite extract was prepared by solubilization of worms as described previously (19) and denaturation by boiling at 100 °C for 10 min in SDS reducing buffer (Bio-Rad). As a positive control for Western blotting using the heterologous serum, a mouse small intestine extract was added to SDS glycine sample buffer but not denatured.
Adult parasite extracts, mouse intestine, and KLH protein (Sigma) were separated by SDS-PAGE, transferred to nitrocellulose membranes, incubated with primary sera diluted at 1:50,000 for SmDMT1-KLH antiserum, 1:5000 for KLH-depleted serum, and 1:1000 for the heterologous serum. Preimmune rabbit sera at equivalent dilutions were used as negative controls. In addition, KLH-reactive fraction of the SmDMT1 sera was used as the control. After incubation with primary sera, the nitrocellulose membranes were washed in 0.05% Triton X-100, 0.1% Tween 20/phosphate-buffered saline and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2000, Bio-Rad) to detect bound antibodies.
For immunofluorescence, unfixed adult S. mansoni worms were snap-frozen in Cryomatrix (ThermoShandon) for cryostat sectioning. All incubations used 2% skim milk powder in phosphate-buffered saline as incubation buffer. Cryosections were incubated with SmDDNA generated by the GeneRacer Kit MT1 serum (1:20) or with the heterologous antiserum diluted (1:100 in incubation buffer) followed by goat anti-rabbit secondary antibody conjugated to Cy3 fluorophore (Jackson ImmunoResearch Laboratories) (1:500 in incubation buffer). Negative controls were as described for Western blotting (above). Sections were examined and photographed using a Leica DM IRB fluorescence microscope equipped with a Leica DC 500 digital camera.
Functional Expression of Schistosome DMT1 cDNAs in Yeast—Both S. mansoni DMT1 sequences were cloned into the NotI site of pFL61 vector (gift of Dr. D. Day, University of Western Australia). The ferrous iron transport-deficient yeast strain DEY1453 (fet3fet4) (20) was transformed using lithium acetate-mediated transformation and selected for growth on SD media containing 20 mg ml-1 glucose and appropriate autotrophic requirements and supplemented with FeCl3 to promote growth of yeast (21). Complementation experiments were performed following methods of Eide and colleagues (22). As positive control, Arabidopsis thaliana ferrous iron transporter (AtIRT1), a member of the Nramp family (23), known from previous yeast complementation assays to be an efficient Fe2+ transporter (21), was used. As negative control, yeast was transformed with empty pFL61 vector.
RESULTS
Identification of Two Isoforms of SmDMT1—Degenerate primers were designed from conserved regions of DMT1 amino acid sequences from C. elegans, M. musculus, H. sapiens, and D. melanogaster and used to amplify DNA from an adult S. mansoni cDNA library. Complete sequence data were obtained from the amplification of 5′- and 3′-ends of S. mansoni adult RACE cDNA.
SmDMT1-A is a 2249-bp cDNA containing an open reading frame of 583 amino acids with a predicted Mr of 64,130 Da, and SmDMT1-B is a 2084-bp cDNA with a predicted open reading frame of 543 amino acids predicted to encode a 59,730-Da protein. Both cDNAs are identical except for variant N-terminal regions (Fig. 1). The variant region in SmDMT1A consists of 239 bp of unique DNA that is predicted to encode 54 amino acids before the shared sequence begins. The variant region of SmDMT1B is considerably shorter and consists of 81 bp of unique DNA that is predicted to encode 14 amino acids before the shared sequence.
In Silico Analysis of SmDMT1—Sequences were compared with those in the GenBank™ and dbEST databases using the National Centre for Biotechnology Information (NCBI) Blast program. The SmDMT1 amino acid sequence shows homology to the Nramp family of functionally related proteins. The members of this family are divalent cation transporters and are defined by a conserved hydrophobic core of 10-12 transmembrane domains. SmDMT1 (excluding the variant 5′-regions) shows strong identity with Nramp2 molecules from human (P49281) 63%, mouse (P49282) 63%, rat (O54902) 63% and D. melanogaster (P49283) 58% (Fig. 2). Identity is also shared with Nramp1 from human (P49279) 61%, mouse (P41251) 59%, and C. elegans (Q21434) 54% with identity to the yeast SMF1 (P38925) and SMF2 (P38778) proteins and manganese transport proteins from bacteria at 30% (figure not shown). Based on sequence comparison and hydropathy plots, SmDMT1 is predicted to contain 12 transmembrane spanning regions, which is characteristic of this family of proteins.
In mammals, the biosynthetic rates for both transferrin receptor 1 (TfR1) and ferritin are regulated by iron. A highly conserved iron-responsive element (IRE) in the 5′-untranslated portion of the ferritin mRNA and the 3′- UTR of mammalian TfR1/mRNA mediates iron-dependent post-transcriptional control of their expression. The IRE functions by forming a specific stem-loop structure that interacts with a transacting factor in an iron-dependent fashion. This element has been reported in the 3′-UTR of human DMT1 sequences (12, 24) and follows the 9-nucleotide consensus sequence of 5′-CNNNNNCAGUGN-3′ (25). The two schistosome transcripts include a conserved polyadenylation signal (AATAAA) 22 nucleotides upstream of the poly(A)+ tail indicating that the full 3′-UTR has been obtained. Analysis of the 3′-UTR of both SmDMT1 revealed a sequence of 5′-AAAUAAACAGUGN-3′. This shared schistosome sequence was identical in all respects to the IRE consensus sequence except for an adenine that replaces the upstream cytosine, that forms the so-named “C” bulge of the consensus IRE. Because the IRE is absolutely conserved (25), the schistosome sequence is unlikely to function as a regulatory element. In addition, a program designed to analyze sequences for regulatory RNA elements (50), including IRE sequence motifs, also failed to recognize any IREs in the 3′ UTR of SmDMT1.
All sequences chosen for inclusion in the phylogenetic tree (Fig. 1B) fall into the “archetype” Nramp group (16, 26). The flatworm genes clearly fall within this group. The clusters at the top of the tree are vertebrate Nramp1 and Nramp2. Sequences from invertebrates (and plants, yeast, and slime mold) were all very different from each other and from those of vertebrates. Outside the vertebrate clade, almost all bootstrap values were very low indicating poor support for any of the groupings observed (apart from Nramps from the two plant species). An Nramp from the free-living flatworm S. mediterranea fell closest to that of the bivalve mollusk in the tree presented here, but analyses using other models of substitution sometimes provided weak support for a sister group relationship with S. mansoni. Therefore, despite belonging to the same phylum, S. mediterranea and S. mansoni do not resemble one another closely in the sequence of the Nramp gene. Neither has the schistosome sequence converged on the vertebrate sequences, thereby providing little support for a postulate of molecular mimicry by the parasite in the case of Nramp.
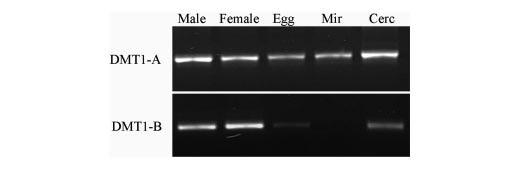
Stage-specific Expression of SmDMT1-A and -B—RT-PCR amplification of the SmDMT1-A and -B mRNA from different life cycle stages of S. mansoni showed that SmDMT1-A is transcribed in all the stages evaluated. SmDMT1-B is transcribed in comparable amounts in adult male and female worms, to a much lesser extent in cercariae and egg, and is not expressed in the miracidial stage (Fig. 3).
FIGURE 3.
RT-PCR analysis of expression patterns of SmDMT1-A and -B in S. mansoni. Lane 1, adult males; lane 2, adult females; lane 3, eggs; lane 4, miracidia; lane 5, cercariae.
PAGE and Western Blotting—Western blot analysis of adult S. mansoni using an antiserum directed against the SmDMT1-KLH fusion peptide recognized a number of bands (Fig. 4A). After KLH-directed antibodies were removed from the serum, the depleted serum recognized a single band of ∼115 kDa (Fig. 4A). An antiserum directed against rat DMT1 also revealed a single diffuse band, representing a protein or aggregate in S. mansoni adult worms at ∼115 kDa in size (Fig. 4B). This latter serum was also used to probe mouse small intestine extract, as was a preimmune serum (Fig. 4B). No bands were identified using the preimmune rabbit serum against mouse small intestine, but the anti-ratDMT1 serum identified a smear spanning 85-90 kDa (Fig. 1A). Mouse DMT1 has been reported to range from 65 to 116 kDa (18, 27-29), and our results are consistent with these observations.
FIGURE 4.
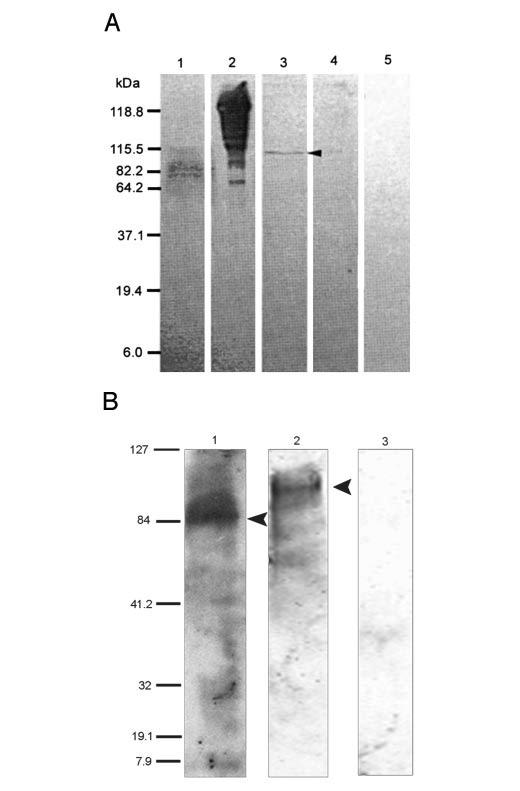
Western blot analysis of S. mansoni DMT1. A, an antiserum directed against a SmDMT1 peptide was used to probe membrane extracts of S. mansoni adults (lanes 1 and 3) and purified KLH (lanes 2 and 4). Lanes 1 and 2 were probed with unpurified SmDMT1-KLH serum, and lanes 2 and 4 were probed with the same serum after KLH-reactivity was depleted. The depleted serum recognized a single band of ∼115 kDa (arrow). Membrane extract probed with normal rabbit serum was used as negative control(lane 5). B, a heterologous rabbit anti-rat DMT1 serum was used to probe mouse small intestine (lane 1) and S. mansoni worm (lane 2) extracts. Arrows highlight the position of bands. Preimmune rabbit serum against mouse small intestine (lane 3) was used as a negative control. The sizes of the molecular weight markers are indicated (kDa).
Immunolocalization—Immunofluorescence labeling of adult worm sections with an anti-SmDMT1-KLH fusion protein, the same antiserum from which KLH immunoreactivity had been depleted (anti-SmDMT1), and a heterologous serum raised against rat-DMT1 gave strong positive labeling for the distal cytoplasm of the tegument (Fig. 5, A, B, D, and E). None of these reagents displayed positive immunoreactivity for the gastrodermis of the parasite. The anti-SmDMT1-KLH serum gave a strong positive reaction over the tegument and parenchyma and labeled internal ducts of the parasites (Fig. 5E). Many cross-reactive molecules occur in the parenchyma and excretory ducts of schistosomes but, significantly, not the tegument of adult worms (30,31). The KLH-depleted anti-SmDMT1 serum retained positive immunoreactivity for the tegument (Fig. 5D). No reaction was detected in sections labeled with preimmune sera (Fig. 5, C and F) or when primary antiserum was omitted (data not shown).
FIGURE 5.
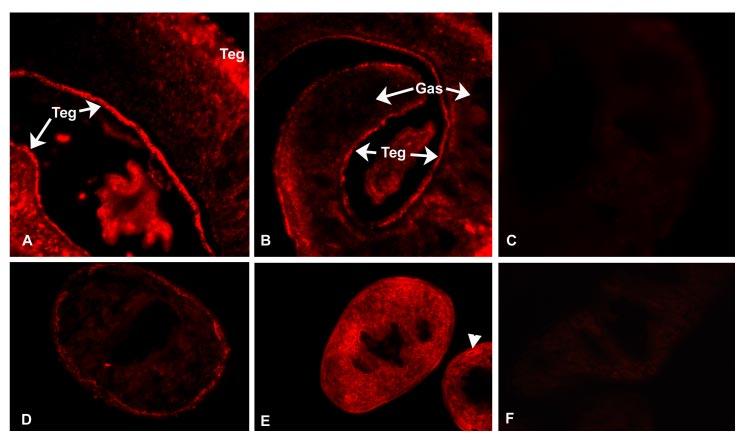
Immunolocalization of DMT1 in S. mansoni adult worms using primary rabbit sera and secondary sera conjugated to Cy3. A and B, cryostat sections labeled with rabbit anti-rat DMT1 serum shows strong tegumental (Teg) labeling. No labeling was observed over the gastrodermis (Gas). C, no reaction was detected in sections labeled with corresponding preimmune serum. D, tegument reactivity in female worm after localization with anti-SmDMT1 serum after depletion of KLH immunoreactivity. E, anti-SmDMT1-KLH serum gives a more intense reaction over tegument and parenchyma. Excretory ducts (arrowhead) display positive immunoreactivity with the non-depleted serum. F, preimmune serum.
Functional Expression in Yeast—To test whether SmDMT1A and -B could transport Fe2+, these molecules and ArIrt1 were cloned into pFL61 and transformed into DEY1453 iron transport mutant yeast. This strain grows poorly on media with low iron concentrations because high (fet3) and low (fet4) affinity Fe2+ transport has been disrupted (22). Yeast transformed with ArIrt1 and SmDMT1A and -B grew better than yeast that contained the empty cloning vector (Fig. 6A) on media either lacking or supplemented with 2 μM FeCl3. In liquid SD medium, supplemented with 20 μM FeCl3, cells transformed with ArIrt1 and SmDMT1A routinely entered the exponential growth phase earlier than empty vector or SmDMT1B (Fig. 6B). Despite the slower growth of yeast transformed with SmDMT1B, the cells still entered exponential growth earlier than control cells.
FIGURE 6.
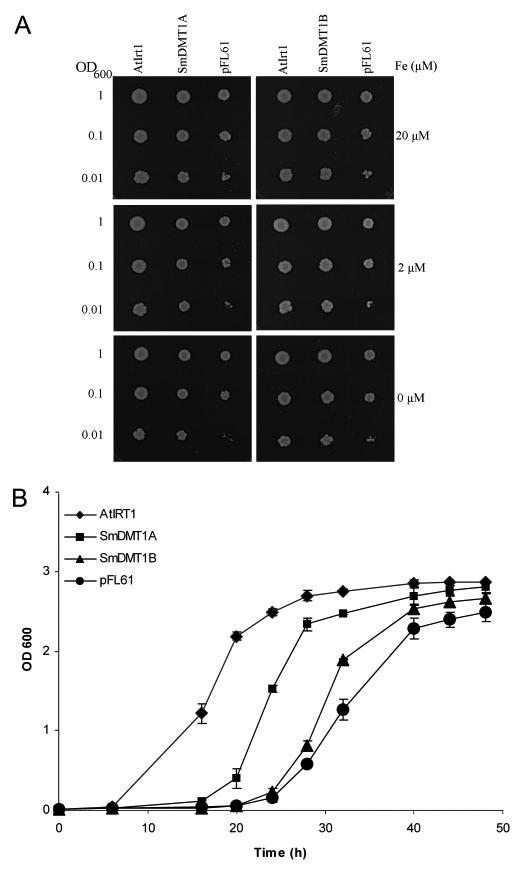
Functional analysis of SmDMT1A and SmDMT1B activity in yeast cells. fet3fet4-deficient yeast cells were transformed with either SmDMT1A, SmDMT1B, or positive control AtIrt1, which were all inserted in the expression vectorp FL61. Cells were also trans formed with the empty expression vector. A, growth of serially diluted cells after 6 days at 30 °C of SmDMT1A, SmDMT1B, AtIrt1, and control (pFL61) transformed fet3fet4 cells on synthetic-defined (SD) medium supplemented with 20, 2, or 0 μM FeCl3. B, growth in liquid SD medium supplemented with 20 μM FeCl3.
DISCUSSION
We have shown that homologues of the mammalian DMT1 proteins, which are involved in iron uptake in mammalian cells, also occur in schistosomes. Iron uptake assays demonstrated that SmDMT1s were able to rescue yeast growth in ferrous iron-transport deficient yeast (fet3fet4). This is the first description of a DMT1 homologue in a hematophagous helminth parasite. One EST with homology to the 3′-end of SmDMT1 has also been deposited in the Schistosoma japonicum data base (32). The Nramp family of proteins is highly conserved and is expressed in many species, and it is therefore not surprising that members also occur in platyhelminths. Schistosomes have a high demand for iron and are known to bind host transferrin at their surface (33). Female parasites express a ferritin in vitelline cells presumably as a iron store for embryogenesis (13, 34). Although much remains to be resolved about iron transport, storage, and utilization in schistosomes, the data of Clemens and Basch (33) and those presented here suggest two surface-linked pathways for iron absorption, a transferrin-dependent and transferrin-independent pathway.
Two S. mansoni DMT1 genes were identified in this study. The two sequences differed only in the N-terminal regions and had different expression profiles throughout the life cycle. SmDMT1A was expressed in all stages examined, whereas SmDMT1B expression appeared strongest in mammal-host-associated stages. The expression profiles thus suggest that the function of SmDMT1B is related specifically to parasitism in the mammalian host. Because the only sequence variation between the two isoforms is at the N terminus, a region predicted to be intracellular, it is possible that this variable region targets either transporter to distinct membrane domains with distinct sets of partner or regulatory proteins. Mammalian DMT1 is organized in the genome with 17 exons, containing alternative 5′- and 3′-exons (11, 35, 36). The functional significance of this variation, particularly that at the 5′-region is not understood but probably relates to the intracellular localization and regulation of DMT1 isoforms in different tissues (36).
Alternative 3′-exons in mammalian DMT1 differ with respect to the presence of an IRE in the untranslated region (35, 36). DMT1 isoforms of the duodenum, liver, and testes possess an IRE, whereas DMT1 isoforms of other tissues do not (36). This together with variation in 5′-regions, have led to the postulate that 5′- and 3′-ends of the mRNA may hold important clues for differences in iron response of different tissues (11). An IRE could not be demonstrated for either SmDMT1A or -B. Because IRE+ and IRE- forms are present in mammals, this observation is not surprising. In addition to this, both IRE+ and IRE- Nramps are variously expressed among different species (21), and there appears to be no one reason for this variability. Until more is known of the genomic organization of schistosome DMT1 genes, the possibility of an IRE+ 3′-exon in these organisms cannot be excluded.
In this study, immunolocalization of the schistosome DMT1 suggested that at least one of the DMT1 isoforms occurs in the tegument. In view of the strong hemoglobinolytic activity of the gastrodermis (2) and the abundance of iron-rich molecules in that organ, it might be anticipated that the gastrodermis is the primary site of iron absorption. However, despite one report to the contrary (37), schistosomes appear incapable of catabolizing heme and polymerize it instead into non-reactive hematin for egestion into the blood stream (38). It therefore seems unlikely then that iron is absorbed across the gut wall in schistosomes or, if it is, by a molecule other than DMT1. The tegument of schistosomes is in direct contact with the host blood and immune systems (39,40), and is involved in active transport of many small nutrients, but is not involved in degradation and ingestion of erythrocytes (41, 42). If DMT1 does play a role in iron absorption in schistosomes, then a surface-associated molecular pathway to liberate ferric iron from the host plasma transporter transferrin must exist. Other blood-dwelling parasites such as the trypanosomes (43) and Plasmodium (44) have strategies for receptor-mediated endocytosis of host transferrin. Transferrin receptors have been postulated for schistosomes (33), and it is possible that these act together with other molecules to provide ferric ions for uptake by SmDMT1.
The two primary sera used for Western blot analysis of native parasite proteins recognized bands corresponding to a size much larger than that predicted for the two protein isoforms. At the present time, we cannot explain this variation, although we noted that mouse DMT1 has been reported to range in size from 65 to 116 kDa (18, 27-29), even though the molecular mass is predicted to be 62 kDa (45). Size differences may likely reflect variation in sample source, preparation, glycosylation, and means of detection (11, 29).
The ability of schistosomes to survive for years in their mammalian host without exhibiting damage from iron-derived free radicals suggests that, along with the iron storage molecules already observed, these worms possess iron transport and regulation mechanisms critical for continued growth and maintenance. The mechanism of action of the artemisinin derivative artemether, an effective antischistosome compound, is thought to be dependant on its interaction with intraparasitic heme, although the precise mode of action remains controversial (46-49). With further identification of novel surface components involved in schistosome iron metabolism, work on new immunodiagnostic methods, potential vaccine candidates, and additional studies exploring the antischistosomal properties of artemether can be undertaken.
Acknowledgments
We thank Mary Duke for maintaining the schistosome lifecycle. We thank Professor David Day and Joanne Castelli for providing yeast strains and for excellent advice.
Footnotes
This work was supported in part by grants from the National Health and Medical Research Council (NHMRC), Wellcome Trust, and the Australian Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: DMT, divalent metal transporter; RT-PCR, reverse transcription PCR; KLH, keyhole limpet hemocyanin; RACE, rapid amplification of cDNA ends; IRE, iron-responsive element; UTR, untranslated region.
REFERENCES
- 1.Lawrence J. J. Parasitol. 1973;59:60–63. [PubMed] [Google Scholar]
- 2.Brindley PJ, Kalinna BH, Wong JY, Bogitsh BJ, King LT, Smyth DJ, Verity CK, Abbenante G, Brinkworth RI, Fairlie DP, Smythe ML, Milburn PJ, Bielefeldt-Ohmann H, Zheng Y, McManus DP. Mol. Biochem. Parasitol. 2001;112:103–112. doi: 10.1016/s0166-6851(00)00351-0. [DOI] [PubMed] [Google Scholar]
- 3.Kasschau MR, Dresden MH. Exp. Parasitol. 1986;61:201–209. doi: 10.1016/0014-4894(86)90153-0. [DOI] [PubMed] [Google Scholar]
- 4.Aisen P, Enns C, Wessling-Resnick M. Int. J. Biochem. Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 5.Crichton RR, Charloteaux-Wauters M. Eur. J. Biochem. 1987;164:485–506. doi: 10.1111/j.1432-1033.1987.tb11155.x. [DOI] [PubMed] [Google Scholar]
- 6.Ponka P, Beaumont C, Richardson DR. Semin. Hematol. 1998;35:35–54. [PubMed] [Google Scholar]
- 7.Frazer D, Anderson G. Hematology. 2001;6:193–203. doi: 10.1080/10245332.2001.11746572. [DOI] [PubMed] [Google Scholar]
- 8.McCord JM. Semin. Hematol. 1998;35:5–12. [PubMed] [Google Scholar]
- 9.Samokyszyn VM, Thomas CE, Reif DW, Saito M, Aust SD. Drug. Metab. Rev. 1988;19:283–303. doi: 10.3109/03602538808994137. [DOI] [PubMed] [Google Scholar]
- 10.Dietzel J, Hirzmann J, Presis D, Symmons P, Kunz W. Mol. Biochem. Parasitol. 1992;50:245–254. doi: 10.1016/0166-6851(92)90221-5. [DOI] [PubMed] [Google Scholar]
- 11.Mims MP, Prchal JT. Hematology. 2005;10:339–345. doi: 10.1080/10245330500093419. [DOI] [PubMed] [Google Scholar]
- 12.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 13.Schussler P, Potters E, Winnen R, Bottke W, Kunz W. Mol. Reprod. Dev. 1995;41:325–330. doi: 10.1002/mrd.1080410307. [DOI] [PubMed] [Google Scholar]
- 14.Dalton JP, Day SR, Drew AC, Brindley PJ. Parasitology. 1997;115:29–32. doi: 10.1017/s0031182097001091. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Tamura K, Nei M. Brief. Bioinf. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 17.Hooker C, Brindley P. Mol. Biochem. Parasitol. 1996;82:265–269. doi: 10.1016/0166-6851(96)02748-x. [DOI] [PubMed] [Google Scholar]
- 18.Trinder D, Oates PS, Thomas C, Sadleir J, Morgan EH. Gut. 2000;46:270–276. doi: 10.1136/gut.46.2.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith AM, Dalton JP, Clough KA, Kilbane CL, Harrop SA, Hole N, Brindley PJ. Mol. Biochem. Parasitol. 1994;67:11–19. doi: 10.1016/0166-6851(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 20.Eide D, Broderius M, Fett J, Guerinot ML. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. Plant J. 2003;35:295–304. doi: 10.1046/j.1365-313x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- 22.Eide D, Davis-Kaplan S, Jordan I, Sipe D, Kaplan J. J. Biol. Chem. 1992;267:20774–20781. [PubMed] [Google Scholar]
- 23.Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee PL, Gelbart T, West C, Halloran C, Beutler E. Blood Cells Mol. Dis. 1998;24:199–215. doi: 10.1006/bcmd.1998.0186. [DOI] [PubMed] [Google Scholar]
- 25.Hentze MW, Caughman SW, Casey JL, Koeller DM, Rouault TA, Harford JB, Klausner RD. Gene (Amst.) 1988;72:201–208. doi: 10.1016/0378-1119(88)90145-x. [DOI] [PubMed] [Google Scholar]
- 26.Robledo JAF, Courville P, Cellier MFM, Vasta GR. J. Parasitol. 2004;90:1004–1014. doi: 10.1645/GE-240R. [DOI] [PubMed] [Google Scholar]
- 27.Roth JA, Horbinski C, Feng L, Dolan KG, Higgins D, Garrick MD. J. Neurosci. 2000;20:7595–7601. doi: 10.1523/JNEUROSCI.20-20-07595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruenheid S, Canonne-Hergaux F, Gauthier S, Hackam DJ, Grinstein S, Gros P. J. Exp. Med. 1999;189:831–841. doi: 10.1084/jem.189.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canonne-Hergaux F, Gruenheid S, Ponka P, Gros P. Blood. 1999;93:4406–4417. [PubMed] [Google Scholar]
- 30.Thors C, Linder E. J. Histochem. Cytochem. 2003;51:1367–1373. doi: 10.1177/002215540305101013. [DOI] [PubMed] [Google Scholar]
- 31.Robijn ML, Wuhrer M, Kornelis D, Deelder AM, Geyer R, Hokke CH. Parasitology. 2005;130:67–77. doi: 10.1017/s0031182004006390. [DOI] [PubMed] [Google Scholar]
- 32.Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus DP, Xue CL, Feng Z, Chen Z, Han ZG. Nat. Genet. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- 33.Clemens LE, Basch PF. J. Parasitol. 1989;75:417–421. [PubMed] [Google Scholar]
- 34.WoldeMussie E, Bennett JL. J. Parasitol. 1982;68:48–52. [PubMed] [Google Scholar]
- 35.Tchernitchko D, Bourgeois M, Martin M, Beaumont C. Biochem. J. 2002;363:449–455. doi: 10.1042/0264-6021:3630449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubert N, Hentze MW. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu WQ, Li YL, Ruppel A. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2001;19:84–86. [PubMed] [Google Scholar]
- 38.Kloetzel K, Lewert R. Am. J. Trop. Med. Hyg. 1966;15:28–31. doi: 10.4269/ajtmh.1966.15.28. [DOI] [PubMed] [Google Scholar]
- 39.Gobert GN, Stenzel DJ, McManus DP, Jones MK. Int. J. Parasitol. 2003;33:1561–1575. doi: 10.1016/s0020-7519(03)00255-8. [DOI] [PubMed] [Google Scholar]
- 40.Jones MK, Gobert GN, Zhang L, Sunderland P, McManus DP. BioEssays. 2004;26:752–765. doi: 10.1002/bies.20058. [DOI] [PubMed] [Google Scholar]
- 41.Smyth JD, Halton DW. The Physiology of Trematodes. 2nd Edition Cambridge University Press; Sydney: 1983. [Google Scholar]
- 42.Shuhua X, Binggui S, Chollet J, Utzinger J, Tanner M. J. Parasitol. 2000;86:1125–1132. doi: 10.1645/0022-3395(2000)086[1125:TCIASM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 43.Fast B, Kremp K, Boshart M, Steverding D. Biochem. J. 1999;342:691–696. [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez MH, Jungery M. Nature. 1986;324:388–391. doi: 10.1038/324388a0. [DOI] [PubMed] [Google Scholar]
- 45.Kishi F, Tabuchi M. Mol. Immunol. 1997;34:839–842. doi: 10.1016/s0161-5890(97)00110-7. [DOI] [PubMed] [Google Scholar]
- 46.Meshnick SR. Med. Trop. (Mars) 1998;58:13–17. [PubMed] [Google Scholar]
- 47.Meshnick SR, Taylor TE, Kamchonwongpaisan S. Microbiol. Rev. 1996;60:301–315. doi: 10.1128/mr.60.2.301-315.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cumming JN, Ploypradith P, Posner GH. Adv. Pharmacol. 1997;37:253–297. doi: 10.1016/s1054-3589(08)60952-7. [DOI] [PubMed] [Google Scholar]
- 49.Li W, Mo W, Shen D, Sun L, Wang J, Lu S, Gitschier JM, Zhou B. PLoS Genet. 2005;1:e36. doi: 10.1371/journal.pgen.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bengert P, Dandekar T. Nucleic Acids Res. 2003;31:3441–3445. doi: 10.1093/nar/gkg568. [DOI] [PMC free article] [PubMed] [Google Scholar]