Abstract
Phospholamban (PLB) physically interacts with Ca2+-ATPase and regulates contractility of the heart. We have studied this interaction using electron microscopy of large two-dimensional co-crystals of Ca2+-ATPase and the I40A mutant of PLB. Crystallization conditions were derived from those previously used for thin, helical crystals, but the addition of a 10-fold higher concentration of magnesium had a dramatic effect on the crystal morphology and packing. Two types of crystals were observed, and were characterized both by standard crystallographic methods and by electron tomography. The two crystal types had the same underlying lattice, which comprised antiparallel dimer ribbons of Ca2+-ATPase molecules previously seen in thin, helical crystals, but packed into a novel lattice with p22121 symmetry. One crystal type was single-layered, whereas the other was a flattened tube and therefore double-layered. Additional features were observed between the dimer ribbons, which were substantially farther apart than in previous helical crystals. We attributed these additional densities to PLB, and built a three-dimensional model to show potential interactions with Ca2+-ATPase. These densities are most consistent with the pentameric form of PLB, despite the use of the presumed monomeric I40A mutant. Furthermore, our results indicate that this pentameric form of PLB is capable of a direct interaction with Ca2+-ATPase.
INTRODUCTION
Since the initial discovery of small two-dimensional (2D) crystals of the Na+ pump more than 20 years ago (1), the P-type ion pumps have proved to be remarkably amenable to the techniques of electron microscopy and x-ray crystallography. Structural insight into the sarcoplasmic reticulum (SR) Ca2+-ATPase has developed more rapidly, with several crystal forms studied by electron microscopy and the recent leap to atomic resolution by x-ray crystallography. Initial comparisons of calcium-bound Ca2+-ATPase from x-ray crystallography and calcium-free Ca2+-ATPase from electron microscopy suggested that dramatic domain rearrangements were responsible for coupling the energy of ATP hydrolysis with calcium transport (2–4). The details of these conformational changes have more recently been elaborated by a series of x-ray crystallographic structures of Ca2+-ATPase stabilized in five different reaction intermediates (3,5–9). As a result, we now have a reasonably sophisticated understanding of how the structural dynamics of the pump contribute to energy coupling.
Much less clear is the way in which phospholamban (PLB) regulates the activity of Ca2+-ATPase. PLB is a 52-residue, transmembrane protein present in cardiac and smooth muscle that confers β-adrenergic regulation on the calcium uptake by the SR. Specifically, PLB appears to physically bind to Ca2+-ATPase and thus reduce its apparent calcium affinity by severalfold. This reduction is sufficient to alter the proportion of calcium transported into the SR relative to that exported from the cell by the Na+/Ca2+ exchanger. Stimulation by β-adrenergic agonists leads to phosphorylation of PLB by protein kinase A, thereby reversing its inhibition of Ca2+-ATPase. The increased activity leads to an increased calcium load within the SR and to an enhanced cardiac contractility, which is an important physiological response to exercise and heart disease.
The structural basis for PLB regulation has been difficult to establish, due to its character as a small, membrane-bound peptide. A persistent ambiguity has been its oligomeric state, which has been shown in many conditions to be pentameric but which is postulated to be monomeric while interacting with and inhibiting Ca2+-ATPase. The pentamer appears to be a very stable structure that is routinely observed in SDS polyacrylamide gels. The pentamer has also been documented in lipid bilayers by spectroscopic measurements (10,11) and very recently observed in dodecylphosphocholine micelles by NMR (12). Scanning mutagenesis identified a number of mutants that are primarily monomeric by SDS-PAGE, and highlighted a series of Leu and Ile residues along one side of the transmembrane helix that engage in a coiled-coil interaction, thus stabilizing the pentamer (13,14). These monomeric mutants tend to have enhanced inhibitory properties with respect to Ca2+-ATPase, leading to the hypothesis that the monomeric form of PLB interacts with and inhibits Ca2+-ATPase, whereas the pentameric form represents storage for inactive or excess PLB.
The structure of PLB has been addressed by a variety of methods, producing a range of results and generally suggesting a flexible molecule that is able to adapt to its environment. A constant feature of all models is a very hydrophobic transmembrane helix (termed domain II) that spans from residue 31 to the C-terminus (12,15–17). There is a general consensus for an α-helix at the hydrophilic, N-terminal end (termed domain Ia) of the molecule that runs for 10–15 residues and ends near the phosphorylation site at Ser16 (12,16–18). Most structures have an unstructured loop after this helix, and some groups have postulated that phosphorylation influences the proportion of secondary structure. The region between the phosphorylation site and the transmembrane helix is known as domain Ib and there is little consensus on its structure. Solution NMR shows a helical extension of the transmembrane domain with only a few nonhelical residues surrounding Pro21, whereas models for the inhibitory interaction show a largely extended peptide chain wrapping around the cytoplasmic domain of Ca2+-ATPase (19,20). Solid-state NMR of PLB in lipid bilayers suggests that the entire cytoplasmic domain is dynamically disordered (15) and electron paramagnetic resonance provides evidence for a structural transition in this cytoplasmic domain upon binding to Ca2+-ATPase (21,22).
We have previously grown two-dimensional co-crystals of Ca2+-ATPase and PLB and used cryoelectron microscopy to characterize the interaction between these two molecules (23,24). Crystallization conditions were based on those for thin, helical crystals of Ca2+-ATPase within both the SR (25) and reconstituted membranes (26), which have been used for three-dimensional (3D) reconstruction at 6.5 Å resolution (4). Although the presence of several PLB mutants in reconstituted membranes was documented both by functional assays and by gel electrophoresis, they had only minimal contribution to the electron density map from co-crystals (23,24), making a definitive description of the physical interaction difficult . In the work presented here, we have found new conditions for co-crystallization of the I40A mutant of PLB with Ca2+-ATPase. Two different crystal morphologies have been produced, resembling folded ribbons and wide, tubular crystals. By calculating projection maps and 3D tomograms of negatively stained crystals, we show that these two crystal morphologies correspond to single and double layered 2D crystals with plane group symmetry p22121. The Ca2+-ATPase molecules are arranged in “dimer ribbons” previously observed in the thin, helical crystals, though in this case the molecules forming adjacent dimer ribbons protrude from opposite sides of the membrane. Furthermore, a new set of densities are observed in between the dimer ribbons, which have a size and shape consistent with the PLB pentamer. These features are confirmed with a projection map from frozen-hydrated crystals at 10 Å resolution. Based on these features, we have constructed a model depicting the molecular interactions between pentameric PLB and Ca2+-ATPase.
EXPERIMENTAL PROCEDURES
Materials
Octaethylene glycol monododecyl ether (C12E8) was obtained from Barnet Products (Englewood Cliffs, NJ). SM-2 Bio-Beads were obtained from Bio-Rad (Hercules, CA). Egg yolk phosphatidylcholine (EYPC), egg yolk phosphatidylethanolamine (EYPE) and egg yolk phosphatidic acid (EYPA) were obtained from Avanti Polar Lipids (Alabaster, AL). All the reagents used in the coupled enzyme assay (27) for measuring ATPase activity were of highest purity available (Sigma-Aldrich, Oakville, Ontario, Canada).
Reconstitution of Ca2+-ATPase and PLB
Skeletal muscle SR vesicles were prepared from rabbit hind leg (28), and the skeletal SR Ca2+-ATPase was purified by Reactive-Red affinity chromatography (29). Recombinant PLB was prepared as described (30) and, for these studies, the super-inhibitory PLB mutant Ile40-to-Ala (I40A) was used (13,31). Co-reconstitution of Ca2+-ATPase and PLB followed established protocols (26,32,33). Briefly, the appropriate amounts of PLB, EYPC, EYPE, and EYPA (8:1:1 lipid weight ratio) were solubilized in a solvent of eight parts trifluoroethanol and two parts water, dried to a thin film under nitrogen gas, and lyophilized. Buffer (20 mM imidazole, pH 7.0, 100 mM KCl, 0.02% NaN3) and detergent (C12E8) were added to solubilize the mixture, followed by the addition of 500 μg of detergent-solubilized, purified Ca2+-ATPase. The final concentrations were adjusted to obtain weight ratios of 1 protein:1 lipid:2 detergent (final molar ratio of 1 Ca2+-ATPase:3.5 PLB:150 lipid). The detergent was removed by incremental addition of SM-2 Bio-Beads over a 4-h time course. The co-reconstituted proteoliposomes containing Ca2+-ATPase and PLB were purified on a sucrose step-gradient (26), and the amounts of Ca2+-ATPase and PLB were determined by quantitative SDS-PAGE (23,31). Control samples were prepared simultaneously under identical conditions and consisted of Ca2+-ATPase reconstituted in the absence of PLB. ATPase activity of the co-reconstituted vesicles was measured by a coupled-enzyme assay that included free calcium concentrations from 0.1 μM to 10 μM (27,31). The KCa (calcium concentration at half-maximal activity) and Vmax (maximal activity) were determined by fitting the data to the Hill equation using Sigma Plot software (SPSS, Chicago, IL). Errors bars represent the standard error of the mean for a minimum of six independent reconstitutions.
Crystallization
After the sucrose gradient, proteoliposomes were collected by centrifugation, resuspended in sucrose-free buffer, and centrifuged again to wash away sucrose. To this pellet was added crystallization buffer (20 mM imidazole, pH 7.4, 100 mM KCl, 35 mM MgCl2, 0.5 mM EGTA, 0.25 mM Na3VO4, 30 μM thapsigargin) (34). The pellet was subjected to three freeze-thaw cycles (freezing in liquid N2, thawing in hand) and was gently resuspended with a micropipette, followed by two additional freeze-thaw cycles. Reconstituted samples were incubated at 4°C for several days to 1 week. Although crystallization occurred quickly, 3–5 days were optimal for the highest frequency and quality of 2D crystals.
Electron microscopy
Crystals were imaged in a 200kV electron microscope with a field emission gun (either a CM200FEG or an Tecnai F20 (FEI, Eindhoven, The Netherlands)) using either a standard room-temperature holder or a high-tilt tomography holder (E. A. Fischione Instruments, Export, PA) for negatively stained samples and a cryoholder (Oxford CT3500, Gatan, Pleasanton, CA) for frozen-hydrated samples. For the tomograms, images were recorded on a 4k × 4k charge-coupled device camera (F415 from Tietz Video and Imaging Processing GmbH, Gauting, Germany) using the program SerialEM (35) for automatic tilting, tracking, focusing, and image acquisition. For projection maps, low-dose images were recorded on film at 50,000× magnification and screened by optical diffraction. The best images were digitized at 14 μm intervals using a Zeiss SCAI densitometer (Intergraph, Madison, AL). All data were recorded with defocus levels between 0.5 and 1 μm.
Projection maps were determined using software from the MRC image processing suite (36). To start, image distortions were corrected before extracting amplitude and phase information from each image. Data from frozen-hydrated crystals were corrected for the contrast transfer function (CTF) after estimating defocus levels using the program PLTCTFX (37); correction was not required for negatively stained images because all data fell within the first zero of the CTF. Common phase origins were determined in the p22121 plane group, but merging was done with p1 symmetry (i.e., no assumed symmetry) using the program ORIGTILT considering reflections with IQ values <4. For averaging, data was weighted according to the signal/noise ratio (IQ) including data with IQ <7, and the corresponding phase residuals represent the inverse cosine of the figure of merit from this averaging. Maps were determined by Fourier synthesis from these averaged data using the CCP4 crystallographic program suite (38).
Tomograms were determined using software from Winkler and Taylor (39,40). Two tomograms were selected for presentation out of a total of six that were determined. The tilt series were recorded on a 4k × 4k charge-coupled device camera at 29k× magnification, producing a nominal pixel size of 6 Å after twofold binning. Images generally covered a tilt range of 135° and increments were calculated according to the cosine rule (i.e., smaller tilt intervals at high tilt). Alignment was accomplished by an iterative procedure of cross correlation between individual images and a reference produced by projection of intermediate 3D tomograms after applying low-pass and high-pass Fourier filters to the data. Improved alignment led to an improved tomogram that was used for the next round of refinement. Tomograms were calculated by R-weighted back projection. To determine an averaged map from these tomograms, a filtered image was first created using a Fourier mask that included the lattice lines generated by the 2D crystal lattice. A small motif comprising several unit cells was extracted from this filtered tomogram and used for cross correlation with the original tomogram. Correlation peaks were localized in the 3D cross-correlation map, and ∼2000 3D motifs centered on these peaks were extracted from the tomogram and averaged. This initial average was used for fine alignment of individual motifs (translational as well as ±5° of rotational alignment). A new average was calculated from 85% of motifs with the highest cross-correlation coefficient, followed by one additional round of alignment before calculating the final, averaged map. Resolution was judged by the Fourier shell correlation implemented in SPIDER (Wadsworth Center, Albany, NY) using the option for excluding the missing wedge of Fourier data. For this determination, the final data set was divided into two halves and used to calculate two independent maps.
RESULTS
Reconstitution
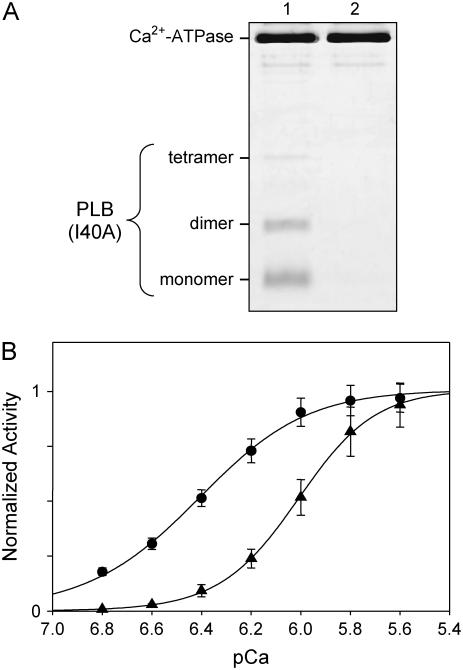
We have previously developed methods for reconstituting proteoliposomes with Ca2+-ATPase and PLB for both functional studies (31) and 2D crystallization (23,24,26). In the study presented here, we have applied these methods to Ca2+-ATPase reconstituted with the superinhibitory PLB mutant I40A (13,31). The resulting proteoliposomes have a lipid/protein molar ratio of ∼120:1, based on a starting ratio of 150:1, and a PLB/SERCA molar ratio of >3:1, based on quantitative SDS-PAGE (Fig. 1 A) (23,31). The I40A mutant of PLB has been described by others to be predominantly monomeric (13); indeed, the majority of our recombinant protein runs as a monomeric species, though a significant fraction is present as either dimers or tetramers (Fig. 1 A). Previous studies have shown that the conditions for SDS-PAGE affect this oligomeric equilibrium (41). Measurements of ATPase activity were used to determine apparent Ca2+ affinity (KCa) and maximal rate (Vmax) both in the presence and absence of PLB (Fig. 1 B). In particular, KCa was determined to be 0.98 μM (6.01 ± 0.01 pCa units, n = 6) in the presence of the I40A mutant of PLB and 0.38 μM (6.41 ± 0.01 pCa units, n = 43) in its absence. Statistical analysis indicated that this difference was significant with p < 10−3 (ANOVA between subjects, one-way analysis of variance followed by Tukey's highly significant difference post hoc test for pairwise comparisons). Values for Vmax were measured as 6.26 ± 0.09 μmoles/min/mg in the presence of I40A compared to 4.09 ± 0.15 μmoles/min/mg in its absence.
FIGURE 1.
Co-reconstitution of Ca2+-ATPase and PLB. (A) SDS-PAGE of reconstituted proteoliposomes used for crystallization. Proteoliposomes in lane 1 include both the I40A mutant of PLB and Ca2+-ATPase, whereas those in lane 2 include only Ca2+-ATPase. Gels were run at room temperature and samples were not boiled before loading. I40A runs predominantly as a monomer, with lesser amounts of dimeric and tetrameric species. (B) ATPase activity of Ca2+-ATPase reconstituted in the absence (•) and presence (▴) of I40A. KCa for each curve is reported in the text. Both curves have been normalized to the maximal activity of control samples (i.e., in the absence of PLB).
Crystallization
Like our previous work on co-crystallization of Ca2+-ATPase and PLB (23,26), crystals grew either from large vesicles or from lipidic aggregates produced by the freeze-thaw procedure and required the addition of decavanadate and EGTA. Thapsigargin was included to increase the propensity for crystallization. Also, crystallization was greatly enhanced by negatively charged lipids (23,33); specifically, we found that small amounts of EYPA were required for crystal formation (i.e., weight ratios of 9EYPC:1EYPA or 8EYPC:1:EYPE:1EYPA). Unlike previous work, we found that high Mg2+ concentrations (30–50 mM) produced large, flattened 2D crystals that were dependent on the presence of PLB for their formation. At lower Mg2+ concentrations (5–20 mM), we observed the thin, helical crystals that have previously been analyzed by helical reconstruction and which form even in the absence of PLB (4,23,42).
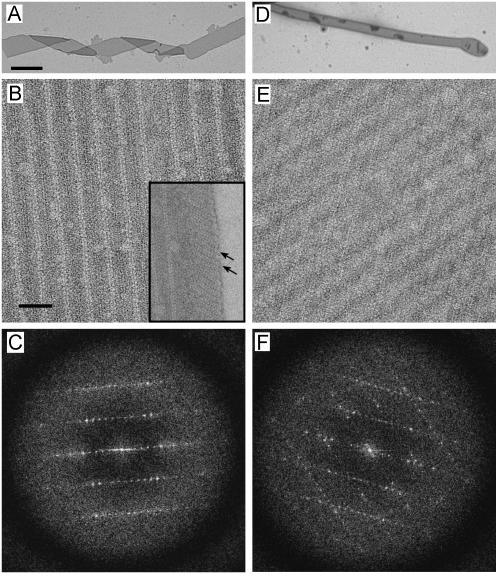
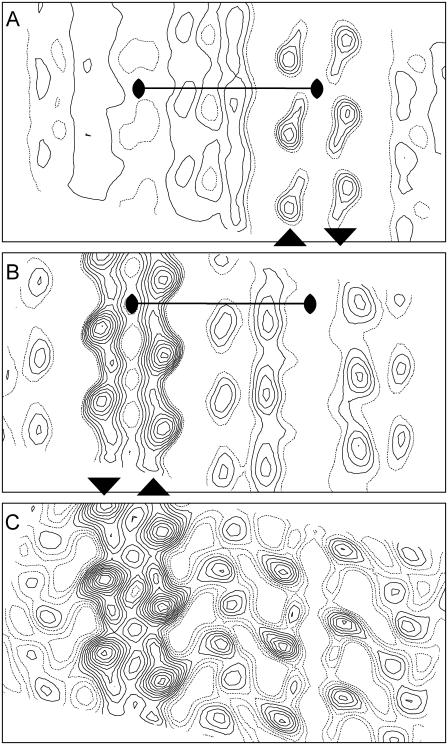
Closer examination of the flattened 2D crystals formed at high Mg2+ concentrations revealed two distinct morphologies. At low magnifications, these crystals resembled either a folded ribbon or a flattened tube, often emanating from a larger vesicle (Fig. 2, A and D). Both of these crystals are at least 10 times wider than the previous thin, helical crystals (>500 nm vs. 60 nm) with lengths ranging from 5–50 μm. Higher magnification images of negatively stained samples revealed that the ribbon-like crystals comprise a single lattice running parallel to the long axis of the ribbons, whereas the lattice of the tube-like crystals produces a criss-cross pattern (Fig. 2, B and E). This difference is also evident in the pattern of reflections in Fourier transforms, which are consistent with a single lattice from ribbon crystals (Fig. 2 C) and two overlapping lattices at an angle of ∼40° from tube-like crystals (Fig. 2 F). Furthermore, the longitudinal lattice from the ribbon crystals does not extend all the way to the edge, but is replaced by an oblique lattice with different dimensions that resembles the previous thin, helical crystals (Fig. 2 B, inset). These observations led us to hypothesize that ribbon-like crystals are single-layered and that tube-like crystals are double-layered (i.e., wide, tubular crystals that have been flattened). In the latter case, the two overlapping lattices would result from the “near” and “far” sides of the tube, which maintain a defined angle due to the underlying helical symmetry. Unit cell parameters for ribbon and tubular crystals were comparable and are shown in Table 1.
FIGURE 2.
Negatively stained co-crystals of Ca2+-ATPase and PLB reveal two different morphologies. (A–C) Some crystals resemble a folded ribbon with lines of molecules running parallel to the long axis. The edges of these crystals contain a different crystal lattice (arrows in B inset) that resembles previously analyzed dimer-ribbons of Ca2+-ATPase. The diffraction pattern (C) is consistent with a single lattice with reflections extending to ∼25 Å resolution. (D–F) Other crystals resemble a flattened tube with a disordered bulb on the end. A criss-cross lattice is evident at higher magnification (E) and two independent lattices are visible in the Fourier transform (F). Scale bars in A and B correspond to 1 μm and 50 nm, respectively. In C and F, the edges of the diffraction patterns correspond to 18 Å resolution, and the corners of the diffraction patterns correspond to 13.5 Å resolution.
TABLE 1.
Crystallographic parameters
| Negative stain
|
Frozen-hydrated
|
|||||
|---|---|---|---|---|---|---|
| Helical crystals* | Ribbon-like | Tubular near side | Tubular far side | Ribbon-like | Tubular† | |
| a (Å) | 56.9 | 350.0 | 334.4 | 337.7 | 359.0 | 359.2 |
| b (Å) | 117.1 | 69.9 | 70.7 | 70.6 | 72.0 | 71.9 |
| γ (°) | 63.9 | 89.5 | 89.4 | 88.8 | 89.3 | 90.3 |
| No. crystals | 70 | 2 | 2 | 2 | 1 | 5 |
| Phase residuals (°)‡ | – | 10.8/16.5 | 10.0/13.1 | 12.0/17.2 | – | 16.8/18.0 |
Data for thin, helical crystals taken from Xu et al. (4).
Only the best side was used from each crystal.
Two-phase residuals reported: first for comparing data from the various tubes during merging with p1 symmetry and second for comparing averaged data with symmetry-constrained centric phases.
Maps from negatively stained crystals
For each crystal form, projection maps of negatively stained samples were calculated after averaging Fourier data from two different crystals. Because wide, tubular crystals are double-layered, independent lattices were extracted from near and far sides of each crystal and were used to generate two independent projection maps. Fourier data generally extended to ∼20 Å resolution (Fig. 2, C and F). We used standard methods for correcting lattice distortions, which improved the sharpness of reflections and their signal/noise ratio. However, this procedure did not significantly improve the resolution, which is limited by specimen preservation in negative stain. All data were contained within the first zero of the CTF, so no corresponding correction was necessary. Examination of potential symmetries for each of these crystals using the program ALLSPACE suggested the plane group p22121 along with some spurious suggestions of three-, four-, or sixfold symmetry, which could be easily ruled out by inspection of unit cell parameters (Table 2). Nevertheless, we elected to be cautious about applying symmetry and therefore averaged data and calculated the projection map without assuming any symmetry at all (p1 plane group).
TABLE 2.
Crystal symmetry
| Ribbon crystals
|
Tubular crystals (near side)
|
Tubular crystals (far side)
|
||||
|---|---|---|---|---|---|---|
| Image No. | 373 | 374 | 377 | 378 | 377 | 378 |
| p2 | 18.4 | 25.0 | 16.2 | 15.4 | 12.0 | 24.0 |
| p12_b | 77.4 | 69.2 | 66.8 | 63.4 | 62.9 | 52.6 |
| p12_a | 79.8 | 75.4 | 71.0 | 72.8 | 79.3 | 74.2 |
| p121_b | 23.6 | 30.5 | 46.9 | 23.2 | 25.9 | 20.2 |
| p121_a | 18.8 | 19.4 | 34.8 | 16.2 | 17.5 | 11.5 |
| p222 | 61.1 | 59.8 | 48.9 | 62.3 | 49.2 | 68.4 |
| p2221_b | 58.0 | 59.1 | 55.0 | 56.6 | 57.7 | 46.1 |
| p2221_a | 54.0 | 58.7 | 41.5 | 55.4 | 42.8 | 59.3 |
| p22121 | 20.2 | 25.5 | 30.3 | 18.4 | 17.4 | 19.8 |
Phase residuals for symmetry related reflections as calculated by the program ALLSPACE.
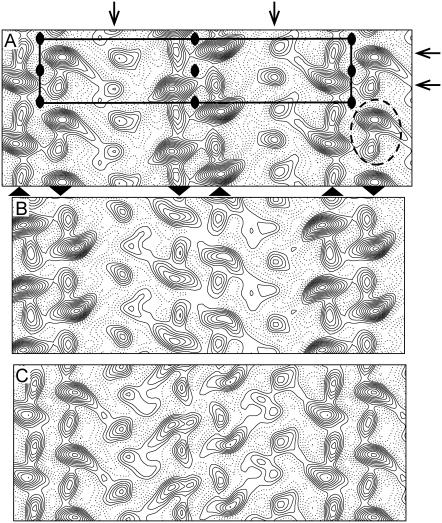
From these negatively stained crystals, we calculated three independent projection maps: one from the ribbon crystals and one each from the near and far side of wide, tubular crystals (Fig. 3). Each of these maps is dominated by a row of twofold related densities along the b axis (arrowheads at the bottom of Fig. 3 A), which resembles the “dimer ribbon” of Ca2+-ATPase molecules that has previously been described in thin, helical crystals of pure Ca2+-ATPase (43). These dimer ribbons are composed of two antiparallel lines of Ca2+-ATPase molecules, each of which produces a strong density flanked by a weaker density aligned with the axis of the dimer ribbons (dotted outline in Fig. 3 A). Although the maps were calculated without imposing symmetry, there is clear evidence of p22121 symmetry, especially in the ribbon-like crystals (Fig. 3 A). In the maps from tubular crystals (Fig. 3, B and C), densities from the central dimer ribbon are weaker, probably due to uneven staining. In particular, if these are indeed wide, tubular crystals with p22121 symmetry, then one set of molecules will be inside the tube and the other set will be outside the tube, thus producing different buildup of negative stain. The more symmetric appearance of ribbon-like crystals is consistent with more even staining from a single-layered structure.
FIGURE 3.
Projection maps from negatively stained crystals. (A) Map from ribbon-like crystals with a single unit cell outlined (a = 350 Å, b = 71 Å) and symmetry operators shown corresponding to the p22121 plane group. The density enclosed by the dotted ellipse has a profile consistent with a single Ca2+-ATPase molecule. The arrowheads at the bottom margin indicate the orientation of Ca2+-ATPase molecules within the three dimer ribbons shown in this map. (B–C) Two maps corresponding to the near and far sides of tubular crystals. Two sets of data were derived from each image by selecting independent lattices in Fourier transforms (see Fig. 2). Note that the two maps have opposite hands, consistent with lattices on opposite sides of a flattened tube. Also note that the central dimer ribbon of Ca2+-ATPase has lower contrast, consistent with a lower stain accumulation on the inside of the tube. All maps were calculated without imposing any particular symmetry (i.e., p1 plane group).
Accompanying the Ca2+-ATPase molecules, additional densities are observed in between the dimer ribbons. These additional densities were not previously observed in thin, helical crystals at lower Mg2+ concentrations (23). Indeed, the Ca2+-ATPase molecules are so far apart in the current co-crystals (175 Å between dimer ribbons compared to 105 Å in previous thin, helical crystals) that one would expect the presence of some additional factor to mediate the crystal contacts along the a axis (i.e., between adjacent dimer ribbons). These extra densities are relatively weak in the maps of negatively stained crystals, suggesting that they comprise considerably less mass than the Ca2+-ATPase molecule. A very likely candidate for these densities would be PLB.
Electron tomography of negatively stained crystals
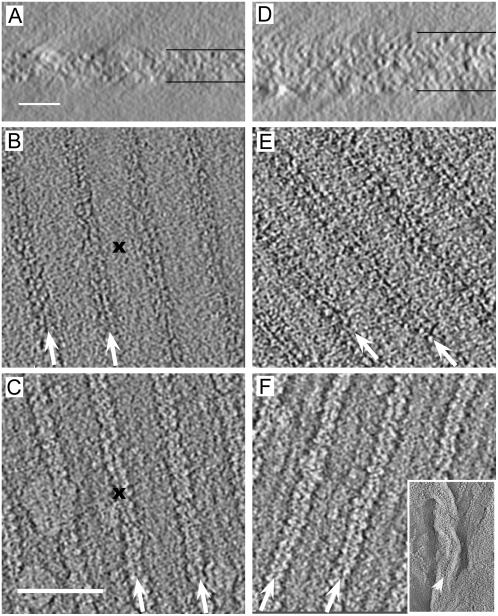
Although the ribbon-like crystals appeared to comprise a single layer, we wanted to eliminate the possibility that they were actually double-layered with lattices from the two layers lying in register. We therefore used electron tomography to produce 3D structures from both types of crystals, which was used not only to verify the number of layers, but also to confirm the crystal packing and plane group symmetry. To do this, we collected tilt series with >90 images from a given crystal tilted about a single axis and the resulting sections clearly resolve individual cytoplasmic domains of the Ca2+-ATPase molecules (Fig. 4). For ribbon-like crystals, the middle of the map appears featureless as expected for the stain-excluding region of the lipid bilayer. In contrast, the middle of maps from tubular crystals reveals a weak criss-cross pattern, consistent with poorly stained cytoplasmic domains in the center of the tube (not shown). Cytoplasmic domains of Ca2+-ATPase are clearly visible at the top and bottom of the tomograms and are organized in the dimer ribbons seen in the projection maps (Fig. 4, B–F). However, the distance between these dimer ribbons in tomograms is double that in projection maps (i.e., 350 vs. 175 Å). Furthermore, the molecules below the stain-excluding region in ribbon-like crystals are exactly out of register with those above the stain-excluding region (see X in Fig. 4, B and C), which is consistent with a twofold axis parallel to the membrane plane. In tubular crystals, dimer ribbons from the two surfaces run at ∼40°, consistent with their existence in two separate bilayers. Indeed, side views of tomograms show that the tubular crystals are roughly twice as thick as the ribbon-like crystals (Fig. 4, A and D). In particular, the overall thickness of the ribbon-like crystals is 170–180 Å, which compares favorably to the 190–200 Å thickness expected for two Ca2+-ATPase molecules protruding from opposite sides of a single bilayer. Furthermore, a fold in the tubular crystal shows what appears to be a double layer of stain-excluding material, presumably a pair of lipid bilayers (Fig. 4 F, inset). Thus, these tomograms unequivocally support the assignment of p22121 symmetry and confirm that ribbon-like and tubular crystals are single- and double-layered, respectively.
FIGURE 4.
Tomographic reconstruction of negatively stained crystals. A ribbon-like crystal is shown in A–C and a tubular crystal in D–F. Views parallel to the bilayer (A and D) show that tubular crystals are almost twice as thick as ribbon-like crystals. Top and bottom surfaces of crystals are shown in B and C for a ribbon-like crystal and in E and F for a tubular crystal. Rows of molecules correspond to the dimer ribbons seen in Fig. 3, but are separated by twice the distance (350 Å, arrows). The Xs in the center of B and C show that the dimer ribbons on the top and bottom are offset by half a unit cell, as one would expect for p22121 symmetry. On the other hand, lattices from the top and bottom of tubular crystals (E and F) are at 40° and independent of one another. The inset in F shows a fold in the tubular crystal (arrowhead) revealing two apposed membranes in the double-layered tubular crystal. The scale bars are 20 nm (A and D) and 50 nm (B, C, E, and F).
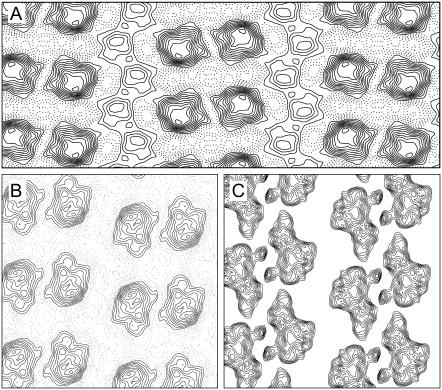
The periodic nature of the sample offered the possibility of enhancing the tomographic structure by employing real-space averaging. To do this, we first extracted ∼2000 unit cells from the tomogram of ribbon-like crystals and, after alignment, averaged the best ∼1500 unit cells to produce an averaged 3D map. The resolution of the map was judged from Fourier shell coefficients derived by dividing the data set in half, yielding either 20.8 Å or 19.3 Å for the 0.5 cutoff or 3 σ criteria, respectively. Although a similar 3D map could have been obtained by averaging Fourier data according to standard electron crystallographic methods, this tomographic approach is more generally applicable to poorly ordered or even nonperiodic samples and should be considered a serious alternative to the more cumbersome crystallographic approach. Sections from this averaged tomographic map through the cytoplasmic domains above and below the stain-excluding region are shown in Fig. 5, A and B. The pear-shaped molecular profile closely resembles the cytoplasmic domain previously determined from thin, helical crystals (see Fig. 7 C) and the arrangement further confirms p22121 symmetry. We calculated a projection of this 3D tomographic structure normal to the membrane plane and found that the resulting map (Fig. 5 C) closely resembles the projection maps determined by standard crystallographic methods (Fig. 3), showing both the dimer ribbons (arrowheads in Fig. 5, A and B) and the intervening densities that we attribute to PLB.
FIGURE 5.
Averaged map from a tomographic reconstruction of negatively stained ribbon-like crystals. Sections through the top (A) and bottom (B) of the map reveal that the pear-shaped cytoplasmic domain of Ca2+-ATPase in adjacent dimer ribbons protrude from opposite sides of the membrane. The arrowheads at the bottom margin indicate the orientation of Ca2+-ATPase molecules within the dimer ribbons. The contrast on the top surface is considerably lower due to uneven staining, which is also evident in comparing Fig. 4, B and C. The a unit cell axis (175 Å, half its length) together with its twofold symmetry operators, are indicated. A projection of the tomographic map (C) is consistent with that determined directly from projection images in Fig. 3.
FIGURE 7.
Map from frozen-hydrated crystals. (A) Projection map from co-crystals of Ca2+-ATPase and PLB analogous to Fig. 3. The region of the map shown is ∼460 Å by 173 Å. (B) Projection map from thin, helical crystals of Ca2+-ATPase alone showing dimer ribbons with a similar profile for individual Ca2+-ATPase molecules. The increased distance between dimer ribbons in co-crystals is filled with extra densities that we attribute to the PLB pentamer. (C) Section from map from thin, helical crystals through the cytoplasmic domain of Ca2+-ATPase reveals the pear-shaped profile seen in Figs. 3 and 5. This comparison illustrates that negative stain emphasizes the cytoplasmic domain relative to the transmembrane domain. In B and C, the regions of the maps shown are ∼225 Å × 225 Å.
Map from frozen-hydrated crystals
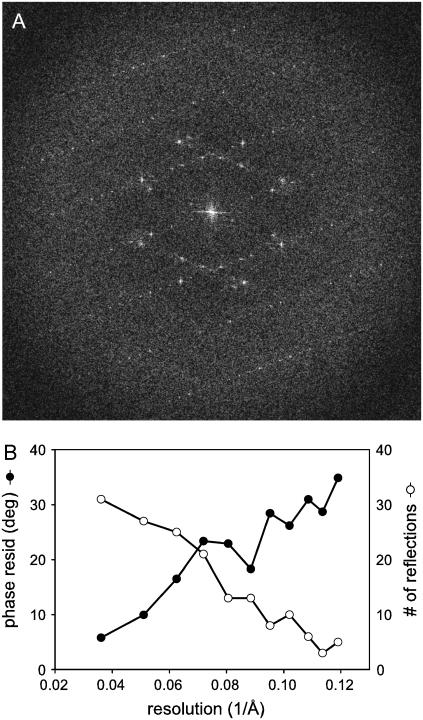
Although the negatively stained crystals are useful for characterizing symmetry and molecular packing, we have imaged crystals in the unstained, frozen-hydrated state to initiate structural characterization of PLB and its interaction with Ca2+-ATPase. Well-ordered crystals showed optical diffraction to ∼17 Å resolution and we chose the best five images from tubular crystals for calculating a projection map. Initially, we merged these data with no symmetry assumptions and noted that the projection map had the expected symmetry relationships (not shown). The data was then merged and averaged in p22121 and produced low phase residuals up to 8.5 Å resolution (Fig. 6). Although we did not include data from ribbon-like crystals in this map, we did compare phases from one such crystal with the merged set of phases from wide, tubular crystals. This phase comparison suggests that there are no differences in the lattice from these two crystal forms. In particular, the p1 phase residual between individual tubular crystals and the merged data set ranged between 13.2° and 19.4° with an average of 16.3°. When the data from a single ribbon crystal were compared with the merged data set, it produced a comparable phase residual of 21.2°, strongly supporting identical packing of molecules within the two lattices.
FIGURE 6.
Frozen-hydrated crystals. (A) Computed Fourier transform from a tubular crystal preserved in amorphous ice reveals the two lattices with Fourier reflections visible to ∼17 Å resolution. The edge of the diffraction pattern corresponds to 17 Å resolution and the corners of the diffraction pattern correspond to 12.8 Å resolution. (B) Statistics for merging five projection images indicate that phase residuals are <35° to a resolution of 8.5 Å, though the map was truncated to 10 Å due to low data completeness at the highest resolutions.
For calculation of the projection map (Fig. 7 A), we used data from five tubular crystals that were truncated at 10 Å resolution due to the relatively low data completeness at the higher resolutions (Fig. 6 B). This map shows the same Ca2+-ATPase dimer ribbons and PLB densities observed in negatively stained crystals, except that the shape of projected Ca2+-ATPase molecules is different. The projected shape will depend on the contrast mechanism, which is different for negatively stained and frozen-hydrated samples. For comparison, we generated a projection map of our earlier 3D reconstruction of frozen-hydrated Ca2+-ATPase from thin, helical crystals (Fig. 7 B) together with a section through the cytoplasmic domain (Fig. 7 C). The former is consistent with the projection of Ca2+-ATPase in our new map and the latter illustrates that the projection of negatively stained crystals is dominated by the cytoplasmic domains with relatively little contribution from the membrane domain.
DISCUSSION
We have discovered novel conditions for co-crystallization of PLB with Ca2+-ATPase and have generated projection maps of both negatively stained and frozen-hydrated crystals with densities that can be unambiguously assigned to Ca2+-ATPase and PLB. Crystallization conditions were similar to those previously used to make thin, helical crystals, but included an ∼8-fold increase in Mg2+ concentration (from 5 mM to 30–50 mM). This difference resulted in a new crystal form which, unlike previous crystals, has twofold screw axes parallel to the membrane plane, meaning that molecules protrude from both sides of the membrane. Despite the different orientations, the current crystals contain the same dimer ribbons found in previous helical crystals of Ca2+-ATPase from both native SR and reconstituted membranes. The dimer ribbons likely result from the use of decavanadate for crystallization, which has been shown to mediate specific intermolecular contacts between cytoplasmic domains of Ca2+-ATPase molecules (4). The thin, helical crystals required additional contacts between neighboring dimer ribbons, which were mediated by the luminal loops of adjacent Ca2+-ATPase molecules. In the new co-crystals, the distance between the dimer ribbons has increased significantly to ∼175 Å, compared to ∼105 Å in the previous helical crystals. This increased distance precludes direct interribbon interactions between Ca2+-ATPase molecules. Instead, contacts between dimer ribbons appear to be mediated by the new density that was not previously seen and that we have attributed to PLB.
The cross-sectional area of these extra densities suggests that they are too large for monomeric PLB, but most likely correspond to an oligomer. PLB contains 52 residues, which compose a single transmembrane helix and a cytoplasmic domain that has been described with a variable amount of α-helix. The extra densities in our projection map of frozen-hydrated crystals are ∼30 Å in diameter and separated by >40 Å. It is difficult to envision this dimension and packing arrangement occupied by a PLB monomer, which has only a single transmembrane helix. The strength of these PLB densities relative to the Ca2+-ATPase density (corresponding to 110 kD) would also support the presence of a PLB oligomer, although it is problematic to quantitate density levels in projection maps. In particular, a pentameric assembly of PLB is plausible because the dimension of the pentamer recently solved by NMR is quite consistent with the dimensions of our projection map and the pentamer has been repeatedly demonstrated to be a stable species by SDS-PAGE.
The observation of a pentameric PLB is surprising considering our use of the I40A mutant for these co-crystallization studies. In particular, the Ile40 residue is part of the “leucine-isoleucine zipper” along the transmembrane helix that has been postulated to stabilize the pentameric assembly. Indeed, previous publications (13,14,41) as well as Fig. 1 show that I40A PLB runs primarily as a monomer by SDS-PAGE, which has frequently been used as the diagnostic tool for assessing the oligomeric state. Our result illustrates that, although SDS-PAGE may be useful in assessing the relative strength of oligomeric associations, its denaturing conditions do not necessarily reflect the oligomeric state of PLB in the membrane. A similar conclusion could be reached from measurements of fluorescence energy transfer, which showed that mixing of labeled populations of PLB occurred at room temperature within membrane bilayers, but required boiling in detergent solutions (44). However, this same study concluded that the L37A mutant, which has similar properties to I40A, produces very little fluorescence energy transfer compared to wild-type PLB, consistent with L37A being monomeric within the membrane bilayer.
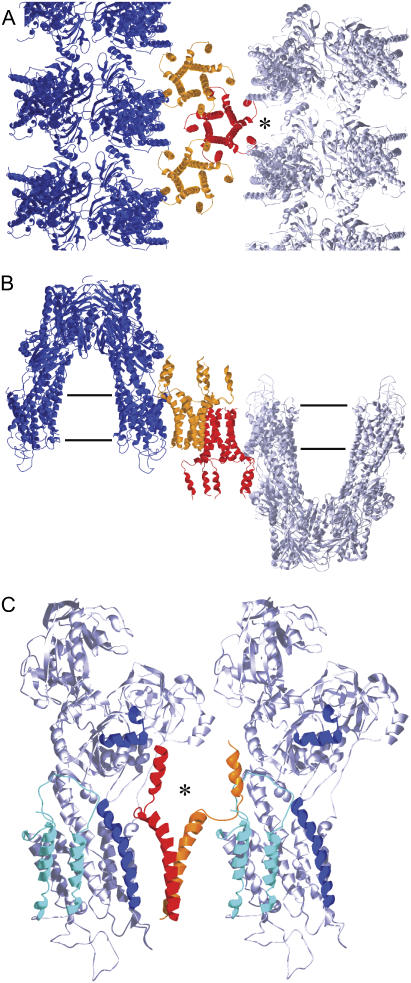
Although we do not yet have a 3D structure, we have built a 3D model based on the known structure of Ca2+-ATPase dimer ribbons (4,42) and on a recent NMR structure for the PLB pentamer (12) (Fig. 8). The dimer ribbons appear to represent stable building blocks of Ca2+-ATPase crystals, having been observed in thin, helical crystals of Ca2+-ATPase alone, in the current co-crystals and even as free-standing structures in partially crystalline proteoliposomes (26). Three types of intermolecular contacts stabilize the dimer ribbons: i), a strong bridge at the top of the cytoplasmic domain between twofold related molecules, ii), a decavanadate-mediated linkage between four molecules at the level of the A-domain ∼40 Å above the membrane surface, and iii), interactions between transmembrane domains. In thin, helical crystals, an additional intermolecular contact between luminal loops connects dimer ribbons into an extended 2D lattice. In the current crystal form, adjacent dimer ribbons protrude from opposite sides of the membrane and do not interact directly with one another. Rather, a row of PLB densities is intercalated between adjacent dimer ribbons. Given the twofold screw axes, PLB pentamers making up this row must adopt alternate orientations with respect to the membrane. As a result, interactions between adjacent PLB pentamers must involve transmembrane helices facing in opposite directions (Fig. 8 B), which cannot represent a situation found within a physiological context. Nevertheless, we hypothesize that each of these PLB molecules faces the same direction as the neighboring Ca2+-ATPase molecule and that the corresponding interactions are ones that could be found within the SR membrane.
FIGURE 8.
Packing model for co-crystals of Ca2+-ATPase and PLB. Ca2+-ATPase molecules (gray and blue) have been placed in dimer ribbons based on their arrangement in thin, helical crystals (Protein Data Bank code 1KJU). PLB pentamers (red and orange) are based on the NMR structure (Protein Data Bank code 1ZLL). Molecules have been placed at locations consistent both with the projection map in Fig. 7 and with the p22121 plane group. The Ca2+-ATPase dimensions are ∼150 Å × 100 Å × 80 Å. The PLB pentamer dimensions are ∼60 Å height × 55 Å width. (A) View normal to the membrane surface. (B) View parallel to the membrane surface (horizontal lines) showing molecules protruding from both sides. (C) Interaction between two Ca2+-ATPases and two monomers from a PLB pentamer. The location of the asterisk is the same in A and C. The monomer colored orange is close enough to interact with Ca2+-ATPase near its C-terminus; M6/M7 loop, M7, M8/M9 loop, and M10 are all colored cyan. The monomer colored red is close enough to interact with Ca2+-ATPase near M3; M3, and α4 from the P-domain are colored dark blue.
Our model suggests that pentameric PLB interacts with a pair of Ca2+-ATPase molecules within a single dimer ribbon. In terms of crystal formation, this pairwise interaction creates a more stable lattice. These two interactions involve the cytoplasmic domain of PLB and either the C-terminal region of Ca2+-ATPase or its P-domain immediately above the M3 helix. In particular, the former involves the C-terminus, the M6/M7 loop, and the M8/M9 loop, whereas the latter involves the cytoplasmic end of M3 and the P-domain loop including α4 (5). Within the membrane, the M3 helix from Ca2+-ATPase is closest to PLB, though the respective membrane domains seem to be too far away to interact. Within the cytoplasmic domains, we would imagine a conformational change in PLB induced by binding to Ca2+-ATPase both to account for the asymmetric distribution of density in our projection map and to optimize the binding surface. Indeed, there is ample evidence for flexibility in the PLB cytoplasmic domain (12,15), which would facilitate such a conformational change (e.g., a bending to provide a more specific interaction with Ca2+-ATPase). Although important for crystal stability, we cannot judge if either of these contacts are physiologically relevant. However, the interaction in the vicinity of M3 is consistent with our previous difference map derived from thin, helical crystals containing both Ca2+-ATPase and PLB (23,24).
These sites of contact are inconsistent with previous models based on site-directed mutagenesis and chemical cross-linking, in which monomeric PLB binds to a surface created by M2, M4, and M6 of Ca2+-ATPase (20,45,46). This surface is on the interior of the dimer ribbon, which we would expect to be accessible to monomeric PLB but which would be sterically prevented from binding to the larger pentameric species of PLB. It is possible that monomeric PLB is also bound to the interior site, a possibility that we will assess when we obtain a 3D structure. However, thapsigargin has been shown to block PLB cross-linking to the M4 site on Ca2+-ATPase (45–47) and its requirement for the current crystals may therefore preclude PLB binding to this site. Precise details of the stoichiometry, conformation, and interactions of PLB with Ca2+-ATPase will be best answered by a 3D structure from this crystal form, which represents our next goal.
Acknowledgments
We thank Jennifer L. Douglas for technical assistance with crystallization, as well as Jun Liu and Hanspeter Winkler for help with the tomographic reconstruction.
D.L.S. was funded by National Institutes of Health grant GM56960. H.S.Y. was funded by the Canadian Institutes of Health Research, the Alberta Heritage Foundation for Medical Research, the Canada Foundation for Innovation, and the Alberta Science and Research Investments Program. H.S.Y. is a scholar of the Alberta Heritage Foundation for Medical Research and a new investigator of the Canadian Institutes of Health Research.
References
- 1.Hebert, H., P. L. Jorgensen, E. Skriver, and A. B. Maunsbach. 1982. Crystallization patterns of membrane-bound Na+/K+-ATPase. Biochim. Biophys. Acta. 689:571–574. [DOI] [PubMed] [Google Scholar]
- 2.Stokes, D., M. Auer, P. Zhang, and W. Kuhlbrandt. 1999. Comparison of H+-ATPase and Ca2+-ATPase suggests that a large conformational change initiates P-type ion pump reaction cycles. Curr. Biol. 9:672–679. [DOI] [PubMed] [Google Scholar]
- 3.Toyoshima, C., and H. Nomura. 2002. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 418:605–611. [DOI] [PubMed] [Google Scholar]
- 4.Xu, C., W. Rice, W. He, and D. Stokes. 2002. A structural model for the catalytic cycle of Ca2+-ATPase. J. Mol. Biol. 316:201–211. [DOI] [PubMed] [Google Scholar]
- 5.Toyoshima, C., M. Nakasako, H. Nomura, and H. Ogawa. 2000. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 405:647–655. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima, C., and T. Mizutani. 2004. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 430:529–535. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima, C., H. Nomura, and T. Tsuda. 2004. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 432:361–368. [DOI] [PubMed] [Google Scholar]
- 8.Olesen, C., T. Sorensen, R. Nielsen, J. Moller, and P. Nissen. 2004. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 306:2251–2255. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen, T., J. Moller, and P. Nissen. 2004. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 304:1672–1675. [DOI] [PubMed] [Google Scholar]
- 10.Reddy, L., L. Jones, and D. Thomas. 1999. Depolymerization of phospholamban in the presence of calcium pump: a fluorescence energy transfer study. Biochemistry. 38:3954–3962. [DOI] [PubMed] [Google Scholar]
- 11.Cornea, R. L., L. R. Jones, J. M. Autry, and D. D. Thomas. 1997. Mutation and phosphorylation change the oligomeric structure of phospholamban in lipid bilayers. Biophys. J. 36:2960–2967. [DOI] [PubMed] [Google Scholar]
- 12.Oxenoid, K., and J. Chou. 2005. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc. Natl. Acad. Sci. USA. 102:10870–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura, Y., K. Kurzydlowski, M. Tada, and D. H. MacLennan. 1997. Phospholamban inhibitory function is enhanced by depolymerization. J. Biol. Chem. 272:15061–15064. [DOI] [PubMed] [Google Scholar]
- 14.Simmerman, H., Y. Kobayashi, J. Autry, and L. Jones. 1996. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 271:5941–5946. [DOI] [PubMed] [Google Scholar]
- 15.Andronesi, O., S. Becker, K. Seidel, H. Heise, H. Young, and M. Baldus. 2005. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Am. Chem. Soc. 127:12965–12974. [DOI] [PubMed] [Google Scholar]
- 16.Lamberth, S., H. Schmid, M. Muenchbach, T. Vorherr, J. Krebs, E. Carafoli, and C. Griesinger. 2000. NMR solution structure of phospholamban. Helv. Chim. Acta. 83:2141–2152. [Google Scholar]
- 17.Zamoon, J., A. Mascioni, D. Thomas, and G. Veglia. 2003. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys. J. 85:2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortishire-Smith, R. J., S. M. Pitzenberger, C. J. Burke, C. R. Middaugh, V. M. Garsky, and R. G. Johnson. 1995. Solution structure of the cytoplasmic domain of phospholamban: phosphorylation leads to a local perturbation of secondary structure. Biochemistry. 34:7603–7613. [DOI] [PubMed] [Google Scholar]
- 19.Hutter, M., J. Krebs, J. Meiler, C. Griesinger, E. Carafoli, and V. Helms. 2002. A structural model of the complex formed by phospholamban and the calcium pump of sarcoplasmic reticulum obtained by molecular mechanics. ChemBioChem. 3:1200–1208. [DOI] [PubMed] [Google Scholar]
- 20.Toyoshima, C., M. Asahi, Y. Sugita, R. Khanna, T. Tsuda, and D. MacLennan. 2003. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc. Natl. Acad. Sci. USA. 100:467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby, T. L., C. B. Karim, and D. D. Thomas. 2004. Electron paramagnetic resonance reveals a large-scale conformational change in the cytoplasmic domain of phospholamban upon binding to the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 43:5842–5852. [DOI] [PubMed] [Google Scholar]
- 22.Karim, C. B., T. L. Kirby, Z. Zhang, Y. Nesmelov, and D. D. Thomas. 2004. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc. Natl. Acad. Sci. USA. 101:14437–14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young, H. S., L. R. Jones, and D. L. Stokes. 2001. Locating phospholamban in co-crystals with Ca2+-ATPase by cryoelectron microscopy. Biophys. J. 81:884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young, H., and D. Stokes. 2004. The mechanics of calcium transport. J. Membr. Biol. 198:55–63. [DOI] [PubMed] [Google Scholar]
- 25.Dux, L., and A. N. Martonosi. 1983. Membrane crystals of Ca2+-ATPase in sarcoplasmic reticulum of normal and dystrophic muscle. Muscle Nerve. 6:566–573. [DOI] [PubMed] [Google Scholar]
- 26.Young, H. S., J. L. Rigaud, J. J. Lacapere, L. G. Reddy, and D. L. Stokes. 1997. How to make tubular crystals by reconstitution of detergent-solubilized Ca2+-ATPase. Biophys. J. 72:2545–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren, G. B., P. A. Toon, N. J. M. Birdsall, A. G. Lee, and J. C. Metcalfe. 1974. Reconstitution of a calcium pump using defined membrane components. Proc. Natl. Acad. Sci. USA. 71:622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eletr, S., and G. Inesi. 1972. Phospholipid orientation in sarcoplasmic reticulum membranes: spin-label ESR and proton NMR studies. Biochim. Biophys. Acta. 282:174–179. [DOI] [PubMed] [Google Scholar]
- 29.Stokes, D. L., and N. M. Green. 1990. Three-dimensional crystals of Ca-ATPase from sarcoplasmic reticulum: symmetry and molecular packing. Biophys. J. 57:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douglas, J., C. Trieber, M. Afara, and H. Young. 2005. Rapid, high-yield expression and purification of Ca2+-ATPase regulatory proteins for high-resolution structural studies. Protein Expr. Purif. 40:118–125. [DOI] [PubMed] [Google Scholar]
- 31.Trieber, C., J. Douglas, M. Afara, and H. Young. 2005. The effects of mutation on the regulatory properties of phospholamban in co-reconstituted membranes. Biochemistry. 44:3289–3297. [DOI] [PubMed] [Google Scholar]
- 32.Reddy, L. G., L. R. Jones, S. E. Cala, J. J. O'Brian, S. A. Tatulian, and D. L. Stokes. 1995. Functional reconstitution of recombinant phospholamban with rabbit skeletal Ca-ATPase. J. Biol. Chem. 270:9390–9397. [DOI] [PubMed] [Google Scholar]
- 33.Young, H., L. Reddy, L. Jones, and D. Stokes. 1998. Co-reconstitution and co-crystallization of phospholamban and Ca2+-ATPase. Ann. N. Y. Acad. Sci. 853:103–115. [DOI] [PubMed] [Google Scholar]
- 34.Dux, L., and A. Martonosi. 1983. Two-dimensional arrays of proteins in sarcoplasmic reticulum and purified Ca2+-ATPase vesicles treated with vanadate. J. Biol. Chem. 258:2599–2603. [PubMed] [Google Scholar]
- 35.Mastronarde, D. N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. [DOI] [PubMed] [Google Scholar]
- 36.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9–16. [DOI] [PubMed] [Google Scholar]
- 37.Toyoshima, C., K. Yonekura, and H. Sasabe. 1993. Contrast transfer for frozen-hydrated specimens: II. Amplitude contrast at very low frequencies. Ultramicroscopy. 48:165–176. [Google Scholar]
- 38.Potterton, E., S. McNicholas, E. Krissinel, K. Cowtan, and M. Noble. 2002. The CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 58:1955–1957. [DOI] [PubMed] [Google Scholar]
- 39.Winkler, H., and K. Taylor. 1996. Three-dimensional distortion correction applied to tomographic reconstructions of sectioned crystals. Ultramicroscopy. 63:125–132. [DOI] [PubMed] [Google Scholar]
- 40.Winkler, H., and K. Taylor. 1996. Software for 3-D reconstruction from images of oblique sections through 3-D crystals. J. Struct. Biol. 116:241–247. [DOI] [PubMed] [Google Scholar]
- 41.Cornea, R., J. Autry, Z. Chen, and L. Jones. 2000. Re-examination of the role of the leucine/isoleucine zipper residues of phospholamban in inhibition of the Ca2+-pump of cardiac sarcoplasmic reticulum. J. Biol. Chem. 275:41487–41494. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, P., C. Toyoshima, K. Yonekura, N. Green, and D. Stokes. 1998. Structure of the calcium pump from sarcoplasmic reticulum at 8-Å resolution. Nature. 392:835–839. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, K., L. Dux, and A. Martonosi. 1984. Structure of the vanadate-induced crystals of sarcoplasmic reticulum Ca2+-ATPase. J. Mol. Biol. 174:193–204. [DOI] [PubMed] [Google Scholar]
- 44.Li, M., L. G. Reddy, R. Bennett, N. D. Silva, Jr., L. R. Jones, and D. D. Thomas. 1999. A fluorescence energy transfer method for analyzing protein oligomeric structure: application to phospholamban. Biophys. J. 76:2587–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen, Z., D. Stokes, W. Rice, and L. Jones. 2003. Spatial and dynamic interactions between phospholamban and the canine cardiac Ca2+ pump revealed with use of heterobifunctional cross-linking agents. J. Biol. Chem. 278:48348–48356. [DOI] [PubMed] [Google Scholar]
- 46.Chen, Z., D. Stokes, and L. Jones. 2005. Role of leucine 31 of phospholamban in structural and functional interactions with the Ca2+ pump of cardiac sarcoplasmic reticulum. J. Biol. Chem. 280:10530–10539. [DOI] [PubMed] [Google Scholar]
- 47.Jones, L., R. Cornea, and Z. Chen. 2002. Close proximity between residue 30 of phospholamban and cysteine 318 of the cardiac Ca2+ pump revealed by intermolecular thiol cross-linking. J. Biol. Chem. 277:28319–28329. [DOI] [PubMed] [Google Scholar]