Abstract
MicroRNAs (miRNAs) are a class of 20–24 nt noncoding RNAs that regulate target mRNAs post-transcriptionally by binding with imperfect complementarity in the 3′-untranslated region (3′-UTR) and inhibiting translation or RNA stability. Current understanding of miRNA biology is limited, and antisense oligonucleotide (ASO) inhibition is a powerful technique for miRNA functionalization in vitro and in vivo, and for therapeutic targeting of miRNAs. Identification of optimal ASO chemistries for targeting miRNAs is therefore of great interest. We evaluated a number of 2′-sugar and backbone ASO modifications for their ability to inhibit miR-21 activity on a luciferase reporter mRNA. ASO modifications that improved target affinity improved miRNA ASO activity, yet the positioning of high-affinity modifications also had dramatically different effects on miRNA activity, suggesting that more than affinity determined the effectiveness of the miRNA ASOs. We present data in which the activity of a modified miRNA ASO was inversely correlated to its tolerability as an siRNA passenger strand, suggesting that a similar mechanism could be involved in the dissociation of miRNA ASOs and siRNA passenger strands. These studies begin to define the factors important for designing improved miRNA ASOs, enabling more effective miRNA functionalization and therapeutic targeting.
INTRODUCTION
MicroRNAs (miRNAs) are a class of endogenously expressed small regulatory noncoding RNAs thought to negatively regulate target mRNAs by binding with imperfect complementarity in their 3′-untranslated regions (3′-UTRs) (1–4). Based on early studies in invertebrates, miRNAs are expected to have roles in developmental regulation and cell differentiation in mammals, and roles for miRNAs in cardiogenesis (5) and lymphocyte development (6) have been demonstrated. Several studies suggest a strong connection between miRNA and human cancer (7–12). Recent reports implicate roles for mammalian miRNAs in metabolic pathways (13–16). MiRNAs have also been shown to suppress (17) and enhance (18) levels of viral RNA in cells. Despite this growing list of roles for mammalian miRNAs, most of the hundreds of miRNAs identified in mammals (19,20) have no reported function.
Conventional single strand antisense oligonucleotides (ASOs) (13,21–25) or double strand siRNAs (26–30) targeting coding genes have been successfully used to screen for gene function in high-throughput cellular assays. ASOs have also been widely used to evaluate gene function and validate gene targets in vivo (31–37) and several antisense therapeutics are in clinical trials (35,38–43). For many cellular applications and all in vivo studies, chemically modified oligonucleotides were used. Properties of modified oligonucleotides associated with improved activity include improved hybridization affinity for the target nucleic acid, resistance to endogenous nucleases, or improved activation of RNase H or other proteins involved in the terminating mechanism. In addition, alteration of protein binding properties with the aim of improving ASO delivery or altering toxicological profiles can be beneficial (44–56). Recently, anti-miRNA ASOs have been used to examine functions of miRNAs in flies (57–59), mammalian cells (13,14,60–63) and mice (15,16). Therapeutic potential for anti-miRNA ASOs has even been suggested (14–16,18,64). In contrast to the extensive optimization of oligonucleotide chemistry for ASOs targeting mRNA or pre-mRNA, little has been done to optimize the chemistry for ASOs targeting miRNAs. Most of the studies mentioned above employed 2′-O-methyl (2′-O-Me) substituted RNA oligonucleotides. There are also reports using DNA (57,59) or a mixed locked nucleic acid (LNA)-DNA ASO (62) and two studies have used 2′-O-methoxyethyl (2′-O-MOE) substituted RNA oligonucleotides (13,16). We set out to evaluate the effect of ASO structure and modification on anti-miRNA activity, using a ‘luciferase sensor’ assay for miR-21 activity in Hela cells. We report below effects of 2′-sugar and backbone modification on activity of anti-miRNA ASOs.
MATERIALS AND METHODS
Oligonucleotide synthesis
Oligonucleotides were prepared using conventional phosphoramidite chemistry and DNA synthesis equipment (Applied Biosystems). The 2′ modified phosphoramidites and succinate-linked solid support were obtained from commercial sources. The purity of all samples was >85% as monitored by electrospray ionization-mass spectrometry analysis. miR-21 ASO sequence: 5′-TCAACATCAGTCTGATAAGCTA-3′. For all modified ASOs except the 2′F, C was methylated at the 5 position. The 2′F ASO had unmethylated C and uracil replaced thymine.
siRNA preparation
Oligonucleotides for preparation of siRNA duplexes were ordered from Dharmacon or Integrated DNA Technologies. Oligonucleotides were purified using high-performance liquid chromatography (HPLC). Strands to be annealed were mixed at equal concentrations in 5× annealing buffer (Dharmacon) and incubated at 90°C for 2 min. The mixture was gradually cooled on a 37°C heat block to allow duplexes to form. The sequence of the siRNA targeting human PTEN: 5′-UUUGUCUCUGGUCCUUACUUCC-3′ (guide, unmodified RNA); 5′-GGAAGTAAGGACCAGAGACAAA-3′ (passenger, various chemistries as described in Results).
Luciferase sensor assay
The miR-21 luciferase sensor construct was engineered by inserting the full 22 bp sequence complementary to the mature miR-21 into the 3′-UTR of pGL3-Control (Promega). Hela cells were propagated in DMEM with High Glucose (Gibco) supplemented with 10% FBS (Gibco). 3.5 × 106 Hela cells in T-170 flasks (BD Falcon) were transfected in batch with 10 µg miR-21 luciferase sensor plasmid and 0.5 µg of phRL-CMV plasmid (Promega) for normalization using 20 µl/flask Lipofectamine 2000 (Invitrogen). After 4 h of transfection, cells were washed, trypsinized, and re-plated at 40 000 cells/well in 24-well plates (BD Falcon). The following day, cells were transfected in triplicate with ASOs for 4 h using Lipofectin (Invitrogen) at 2.5 µl Lipofectin/100 nM ASO/ml Opti-MEM I (Gibco). After ASO transfection, Hela cells were re-fed with complete media. Cells were lysed 24 h later, unless otherwise indicated, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Each experiment was performed at least three times and a representative example is shown. Control ASOs of matched chemistry but unrelated sequence were evaluated in each experiment and, in general, did not affect luciferase activity significantly compared to mock transfected control cells.
Cell proliferation assay
Hela cells were plated at 4000 cells/well in 96-well plates (BD Falcon). The following day, cells were transfected in triplicate with ASOs for 4 h using Lipofectin (Invitrogen) at 2.5 µl Lipofectin/100 nM ASO/ml Opti-MEM I (Gibco). After ASO transfection, Hela cells were re-fed with DMEM with high glucose and without phenol red (Gibco) supplemented with 10% FBS (Gibco). After 24 h, the cell viability was investigated using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega).
siRNA transfection
Hela cells were plated in 96-well plates (BD Falcon) at 4000 cells/well overnight. The following day, cells were treated with duplexes at various concentrations with 5 µl Lipofectamine 2000 (Invitrogen)/ml Opti-MEM I (Gibco) for 4 h. After the 4 h treatment, the Hela cells were re-fed with complete media and left overnight. The next day, cells were lysed and total RNA was isolated using RNeasy 96 columns (Qiagen) on a BioRobot 3000 (Qiagen). Levels of human PTEN mRNA were assayed using real-time Taqman RT–PCR on a Prism 7700 (Applied Biosystems). Forward PTEN Primer: 5′-AATGGCTAAGTGAAGATGACAATCAT-3′; Reverse PTEN Primer: 5′-TGCACATATCATTACACCAGTTCGT-3′; PTEN Probe: FAM-5′-TTGCAGCAATTCACTGTAAAGCTGGAAAGG-3′-TAMRA.
Northern blotting
RNA from treated cells was homogenized in Trizol (Invitrogen) and isolated according to the manufacturer's instructions. Total RNA was separated on a 14% acrylamide TBE 8 M urea mini-gel, then electroblotted onto Hybond N+ nylon filter (Amersham). An end-labeled (Promega) oligonucleotide probe for miR-21 was hybridized to the filter in Rapidhyb buffer (Amersham). The blot was reprobed for U6 to control for equal loading. Quantitation was done using a Storm 860 phosphoimager (Molecular Dynamics) and ImageQuant software.
Tm measurements
Absorbance versus temperature curves were measured at 260 nm using a Gilford Response II spectrophotometer. The buffer contained 100 mM Na+, 10 mM phosphate and 0.01 mM EDTA (pH 7.0) with sufficient Cl− to achieve ionic neutrality. Oligonucleotide concentration was 4 mM each strand determined from the absorbance at 85°C and extinction coefficients calculated according to Puglisi and Tinoco (65). Tm's of duplex formation were obtained from fits of data to a two state model with linear sloping baselines (66).
RESULTS
Selection of a sensitive assay for anti-miRNA activity
MiRNAs regulate their targets post-transcriptionally, either by inhibiting translation or causing mRNA degradation. Directly confirmed, translationally-regulated targets are few (67–69), and western blotting assays are cumbersome for rapid screening of anti-miRNA ASO activity. Changes in target mRNA levels after miRNA modulation have been reported (15,16,70), but the magnitude of the reported changes is small, often only 2- to 3-fold. We attempted to identify a miR-21 target gene upregulated in Hela cells at the mRNA level after ASO treatment. Taqman RT–PCR following ASO treatment was used to measure levels of 26 mRNAs predicted by the TargetScan algorithm (71) to be targets of miR-21. Two mRNAs were identified that appeared to be upregulated after miR-21 ASO treatment (data not shown), but each mRNA was increased no more than 2-fold, making it difficult to compare subtle differences in activity among modified ASOs.
For comparison of modified miRNA ASOs, we needed a convenient and sensitive system with a good dynamic range for evaluation of miRNA activity. Incorporation of perfectly complementary miRNA binding sites into the 3′-UTR of a reporter plasmid has been used previously to evaluate endogenous miRNA activity (72–75). We created a miR-21 luciferase ‘sensor’ plasmid for use in these studies, as miR-21 is abundant in Hela cells. When transfected into Hela cells, the endogenously-expressed miR-21 strongly represses the luciferase sensor expression (data not shown), presumably by binding with perfect complementarity and causing cleavage of the mRNA. Introduction of a miR-21 ASO can prevent this inhibition, resulting in increased luciferase expression. Using this system, the effect of various modified ASOs targeting the mature miR-21 sequence could be easily compared.
Effect of 2′-sugar substitution on anti-miRNA activity
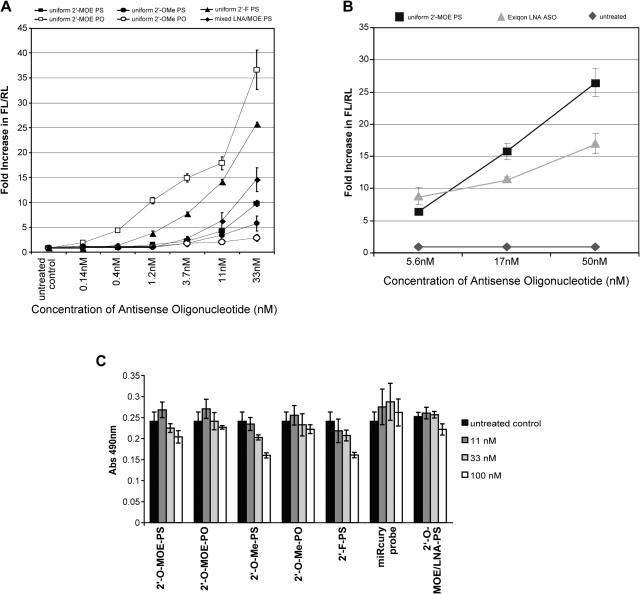
We first evaluated effects of several 2′-sugar modifications on miRNA ASO activity 24 h after ASO transfection. Phosphorothioate (PS) oligonucleotides with uniform 2′-O-MOE, 2′-O-Me or 2′-flouro (2′F) substitution were tested along with a 2′-O-MOE-PS compound containing seven high affinity LNA substitutions, one at every third position. The Tm's for these ASOs hybridized to a length matched complementary RNA were measured and are shown in Table 1. As predicted (50,56,76), all 2′ substitutions increased affinity significantly compared to DNA with 2′-O-Me ≅ 2′-O-MOE < 2′-F < 2′-O-MOE/LNA, and these high affinity ASOs were all more active than the 2′-deoxy-PS oligonucleotide which was inactive in this assay (data not shown).
Table 1.
Tm measurements for anti-miR-21 ASOs hybridized to complementary RNA
| ASO CHEMISTRY | Tm |
|---|---|
| DNA-PO | 49° |
| Alternating 2′-O-MOE-PS | 57° |
| MOE-DNA-MOE gapmer-PS | 58° |
| DNA-MOE hemimer-PS | 62° |
| MOE-DNA hemimer-PS | 62° |
| DNA-MOE-DNA gapmer-PS | 64° |
| 2′-O MOE-PS | 70° |
| 2′-O-Me-PS | 71° |
| 2′-F-PS | 75° |
| 2′-O-Me-PO | 76° |
| 2′-O-MOE-PO | 78° |
| MOE-LNA-PS | 86° |
Anti-miRNA ASOs modified with LNA are also commercially available from Exiqon. The miRCURY miRNA knockdown probe targeting miR-21 was evaluated in the luciferase sensor assay and compared to the 2′-O-MOE-PS ASO (Figure 1B). The 2′-O-MOE ASO showed better activity at the higher doses, although the compounds were similar at the lowest dose.
Figure 1.
Effect of 2′-sugar and phosphate backbone modifications on hybridization and anti-miR-21 activity of ASOs. Hela cells were treated in triplicate with anti-miR-21 ASOs containing a number of 2′-sugar and backbone modifications. At 24 h post transfection, cells were lysed and anti-miR-21 activity measured using the luciferase reporter assay. (A) 2′MOE, 2′-O-methoxyethyl; 2′-OMe, 2′-O-methyl; 2′-F, 2′-flouro deoxy; LNA, locked nucleic acid; PO, phosphate backbone; PS, phosphorothioate backbone. (B) miRCURY miRNA knockdown probe (LNA-Exiqon) compared to the 2′-O-MOE-PS ASO. (C) Proliferation of Hela cells 24 h after transfection with various modified ASOs measured with the colorimetric CellTiter 96® AQueous One Solution Cell Proliferation Assay, in which absorbance at 490 nm is an indication of cell viability.
We noticed that some modified ASOs, primarily 2′-O-Me-PS and 2′-F-PS, had negative effects on cell growth during the luciferase assay. In order to quantify the potential cytotoxic effects of the ASOs, a cell proliferation assay was performed 24 h after ASO treatment (Figure 1C). After treatment with 100 nM ASO, significant inhibition in cell proliferation was observed after treatment with the 2′-O-Me-PS and the 2′-F-PS ASOs. A minor inhibition was observed after treatment with the 2′-O-MOE-PS ASO. Similar effects were observed after treatment with control ASOs of unrelated sequence but with the same sugar and backbone modifications (data not shown), ruling out the possibility that the proliferation defect was related to miR-21 inhibition. After 33 nM ASO treatment, only minor effects on cell proliferation were observed for the 2′-O-Me and 2′-F modified ASOs. At 11 nM ASO treatment, no significant effects on cell proliferation were observed.
Effect of phosphodiester backbone on anti-miRNA activity
2′-O-MOE and 2′-O-Me ASOs with unmodified phosphate (PO) backbones were also evaluated. The PS modification is estimated to cause a 0.5–0.7°C decrease in Tm for each modification (50,76,77), so keeping the nucleosidic linkages as a phosphodiester was expected to result in improved target affinity. For these two ASOs, the Tm increase was only 4.4°C for the 2′-O-Me modified ASO, and 7.3°C for the 2′-O-MOE ASO. This increased Tm resulted in improved activity for the 2′-O-MOE-PO ASO, compared to the 2′-O-MOE-PS ASO. However, changing the backbone of the 2′-O-Me ASO from PS to PO gave no increase in activity. A 2′F phosphate backbone ASO was not tested, as it is expected to be susceptible to nuclease degradation within a short time (78) and a DNA-PO ASO had no activity (data not shown).
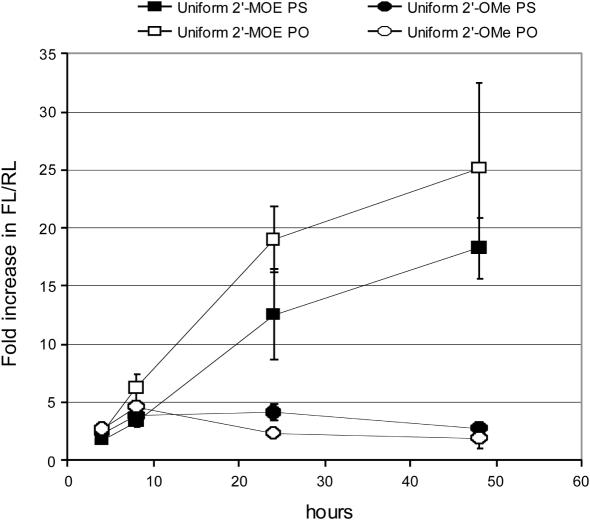
Onset and duration of action of 2′-O-MOE and 2′-O-Me modified ASOs
Stability of 2′-O-MOE ASOs to nucleases is significantly better than 2′-O-Me ASOs (48,49,76,79), which is one possible reason for the discrepancy between the 2′-O-MOE-PO and 2′-O-Me-PO ASO activities. To evaluate the onset and duration of action of these ASOs, luciferase sensor activity was measured at 4, 8, 24 and 48 h after transfection of 2′-O-MOE and 2′-O-Me modified ASOs with either PO or PS backbones (Figure 2). Minimal increase in luciferase activity was observed 4 h after transfection. Eight hours after transfection, some increase in luciferase sensor activity compared to untreated cells was observed for all ASOs tested. The fold change in luciferase ranged from 3- to 7-fold and differences between the four ASOs could not be reproducibly determined. For the 2′-O-MOE substituted ASOs, the fold change in luciferase compared to untreated cells increased from 12- to 18-fold at 24 h and increased even further at 48 h. For the 2′-O-Me substituted ASOs, activity did not increase beyond the 8 h time point.
Figure 2.
Duration of Action of Anti-miR-21 ASOs. Hela cells were treated in triplicate with 33 nM anti-miR-21 ASOs containing a number of 2′-sugar and backbone modifications and lysed 4, 8, 24 and 48 h post transfection initiation. Anti-miR-21 activity was then measured using the luciferase reporter assay. 2′MOE, 2′-O-methoxyethyl; 2′-OMe, 2′-O-methyl; 2′-F, 2′-flouro deoxy; LNA, locked nucleic acid; PO, phosphate backbone; PS, phosphorothioate backbone.
Anti-miRNA activity of ASOs with mixed DNA:2′-O-MOE backbones
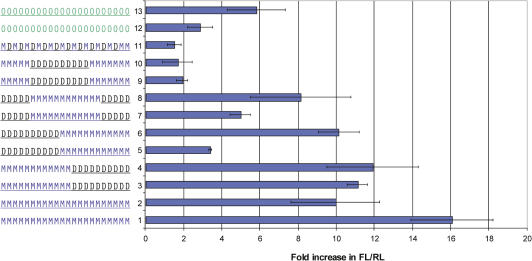
A 2′-O-MOE ‘gapmer’-PS ASO, with 2′-O-MOE modifications on the wings of the ASO and ten 2′-deoxy nucleotides in the center, is capable of triggering RNase H-dependent degradation of an RNA target. However, it was not effective at inhibiting miR-21 activity (Figure 3, Compound 9), suggesting that the miRNA-ASO complex may not be accessible to the RNase H enzyme. Apparently this gapmer ASO was also unable to work effectively using the non-RNase H mechanism(s) employed by the uniformly 2′ substituted ASOs. Substitution of a 2′-O-MOE residue with DNA in an ASO-RNA duplex reduces the Tm by 1–2°C per substitution (50,76,79). We therefore hypothesized that inactivity of the 2′-O-MOE gapmer-PS ASO was due to a decrease in target RNA binding affinity compared to the uniform 2′-O-MOE PS. To test this hypothesis, a series of ASOs was designed by varying positioning of the 2′-O-MOEs (Figure 3), and effects on Tm and miR-21 activity were evaluated. Like the original 5-10-7 gapmer, each ASO had twelve 2′-O-MOE and ten 2′-deoxy residues. Measured Tm's for the mixed DNA-MOE compounds were all between that of the full DNA and the uniform 2′-O-MOE compounds (Table 1). We were surprised to discover that compounds with the same number of 2′-O-MOE and deoxy residues could have dramatically different effects on anti-miRNA activity (Figure 3). For some compounds, the differences in activity correlated with the observed Tm. For example, inverting the 2′-O-MOE gapmer-PS design so that twelve 2′-O-MOE modifications were flanked by five 2′-deoxy residues on either side (Figure 3, Compound 7) resulted in a significant increase in activity compared to the original 2′-O-MOE gapmer-PS ASO (Figure 3, Compound 9), along with a Tm increase of 5.7°C. However, MOE-DNA-PS hemimer motifs with 2′-O-MOE residues on one end and 2′-deoxy on the other showed reproducibly different anti-miRNA activities, despite identical Tm's. Placement of 2′-O-MOE modifications on the 3′ end of the ASO (Figure 3, Compound 5) gave poor anti-miRNA activity, while the same number of 2′-O-MOE modifications on the 5′ end (Figure 3, Compound 3) resulted in activity almost as good as the uniformly-modified 2′-O-MOE-PS ASO (Figure 3, Compound 2). An ASO with an alternating MOE-DNA-PS design (Figure 3, Compound 11) had very poor activity, and a Tm very similar to the also inactive 2′-O-MOE gapmer.
Figure 3.
Placement of High Affinity Modifications: effect on activity of Anti-miR-21 ASOs. Hela cells were treated in triplicate with anti-miR-21 ASOs at 33 nM containing a number of 2′-sugar and backbone modifications. At 24 h post transfection, cells were lysed and anti-miR-21 activity measured using the luciferase reporter assay. The 2′-sugar and backbone modification of each anti-miR-21 ASO is indicated to the left of the graph, and each compound is referred to by the indicated number in the text. O, 2′-O-methyl; M, 2′-O-methoxyethyl; D, 2′deoxy; underline denotes phosphorothioate modified backbone.
For uniformly modified 2′-O-MOE ASOs, replacing the PS backbone with PO resulted in an increase in activity (Figure 1), so we next tested whether replacing the PS modifications with a PO backbone in the hemimer and gapmer motifs could improve their activity. Only the 2′-O-MOE modified regions of the ASOs were modified, as 2′-deoxy-PO ASOs are highly susceptible to nuclease degradation (47,80). Substituting PO into the backbone improved the anti-miRNA activity of the hemimer ASO with 2′-O-MOE in the 3′ part of the ASO (Figure 3, Compounds 5 and 6), and modestly improved activity for the gapmer with 2′-O-MOE modifications placed in the center (Figure 3, Compounds 7 and 8). However, it did little to improve the activity of the inactive 2′-O-MOE gapmer (Figure 3, Compounds 9 and 10) or the active hemimer with 2′-O-MOE in the 5′ part of the ASO (Figure 3, Compounds 3 and 4).
Comparison of anti-miRNA activity to siRNA activity
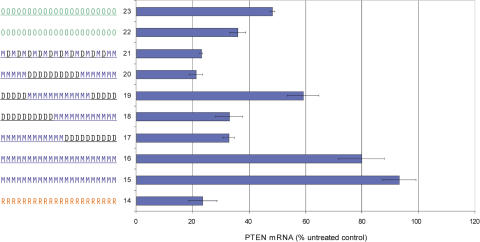
These data suggested that while affinity for target RNA is important for effective miRNA inhibition, other factors may also contribute. An anti-miRNA ASO binding to mature miRNA in RISC could be thought of as a passenger strand of the miRNA or an siRNA. There is now evidence that the double-stranded si/miRNA may associate with the Argonaute protein in RISC before passenger strand dissociation (81–85), and it is therefore possible that the activity responsible for the unwinding of the passenger strand is still associated with the complex when the anti-miRNA ASO binds. In this case, the same interactions influencing the tolerability of chemical modification of siRNA passenger strands may also hold true for modified anti-miRNA ASOs. Using a model siRNA targeting the PTEN mRNA, a set of modified sense, or passenger strands were paired with an unmodified RNA antisense, or guide strand, and tested for their ability to reduce PTEN mRNA levels in Hela cells (Figure 4). The best anti-miRNA ASO tested, the uniformly modified MOE-PO (Figure 4, Compound 15), was completely inactive as a PTEN siRNA passenger strand. The 2′-O-Me ASO, with a PO (Figure 4, Compound 22) or PS (Figure 4, Compound 23) backbone, was tolerated as a PTEN siRNA passenger strand, but with some loss of activity compared to unmodified RNA (Figure 4, Compound 14). Surprisingly, the MOE-DNA-MOE gapmer (Figure 4, Compound 20), and the alternating MOE-DNA motif (Figure 4, Compound 21) were both tolerated as siRNA passenger strands, with no loss of activity compared to unmodified RNA. These motifs were both ineffective as anti-miRNA ASOs. In general, anti-miR-21 activity for each modified passenger strand/miRNA ASO was inversely correlated with PTEN siRNA activity (Table 2).
Figure 4.
Placement of High-Affinity Modifications: effect on ability to function as a siRNA passenger strand. siRNA compounds were designed against human PTEN incorporating high-affinity modifications at various positions in the passenger strand. These modified passenger strands were each paired with an RNA guide strand, and the ability of each duplex (12 nM) to down regulate PTEN mRNA was measured using real-time RT–PCR. The 2′-sugar and backbone modification of each passenger strand is indicated to the left of the graph, and each compound is referred to by the indicated number in the text. R, RNA; O, 2′-O-methyl; M, 2′-O-methoxyethyl; D, 2′deoxy; underline denotes phosphorothioate modified backbone.
Table 2.
Modified oligonucleotides scored for their activity as anti-miR ASO and tolerability as an siRNA passenger strand
2′-MOE is represented in blue, DNA in black and 2′-OMe in red.
Mismatch specificity and length requirements for anti-miRNA ASOs
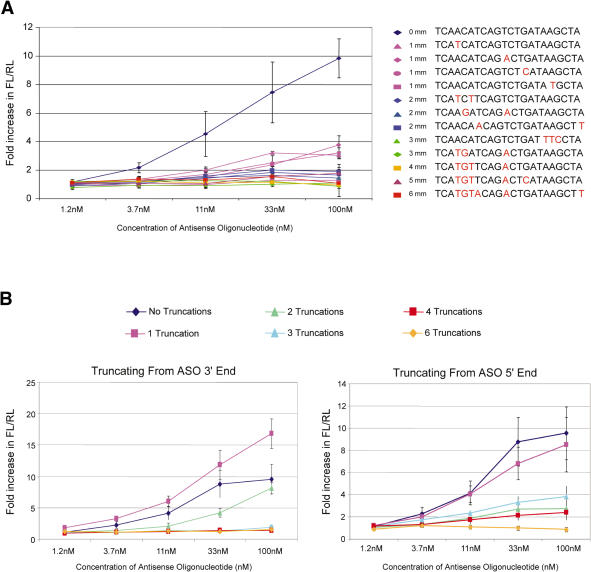
Finally, we examined the specificity and length requirements of the uniformly-modified 2′-O-MOE-PS ASOs. Many miRNAs come in families that share a 6–8 nt ‘seed’ sequence at the 5′ part of the miRNA, and some differ by only one or two nucleotides. As family members are predicted to regulate overlapping sets of target genes, it may be advantageous to be able to target multiple family members with a single ASO. Alternatively, there may be situations in which targeting a single family member is desired. Measuring levels and activities of multiple related miRNAs is technically challenging, but we can infer from the specificity of ASOs mismatched to miR-21 the likelihood of targeting multiple family members with a single ASO. A total of 1 to 6 nt mismatches were introduced into 2′-O-MOE-PS ASOs at a number of positions, and anti-miR-21 activity was measured 24 h after treatment (Figure 5A). Three of four ASOs evaluated with a single mismatch substantially reduced, but did not ablate, anti-miR-21 activity. The introduction of a single mismatch into the 3′ end of the ASO, which is complementary to the 5′ seed region of the miRNA, resulted in an additional loss of activity relative to the other ASOs containing a single mismatch. ASOs with two or more mismatches to miR-21 had poor activity, and three or more mismatches resulted in a complete loss of activity. These data suggest that anti-miRNA ASOs are likely to be specific for individual miRNA family members.
Figure 5.
Effect of Mismatches and Truncations on Activity of Anti-miR-21 ASOs. Hela cells were treated in triplicate with anti-miR-21 uniform 2′-MOE ASOs containing a number of mismatches. At 24 h post transfection, cells were lysed and anti-miR-21 activity measured using the luciferase reporter assay. (A) Effect of one to six mismatches on activity of anti-miR-21 ASOs. (B) Effect of truncations on activity of anti-miR-21 ASOs.
We also tested the effect of truncation on the ASOs' anti-miRNA activity. 2′-O-MOE-PS ASOs with one to five truncated nucleotides from either the 5′ or 3′ end were evaluated for their ability to inhibit miR-21 activity (Figure 5B). A 1 nt truncation from either end of the ASO was well tolerated with little loss of activity. Interestingly, truncating a single nucleotide from the 3′ end of the ASO modestly improved activity. In this case, the truncated nucleotide would be complementary to the first nucleotide of the miRNA, which is supposed to be tucked into a pocket of the Piwi domain of the Argonaute protein in RISC and not available to interact with target RNA (86,87). Truncating 2 nucleotides from either end of the ASO resulted in a loss of activity, although again a truncation from the 3′ end was better tolerated than a truncation from the 5′ end. Three or more truncations from either end resulted in a substantial loss of anti-miRNA activity.
Effect of inhibition of miRNA activity on miRNA levels
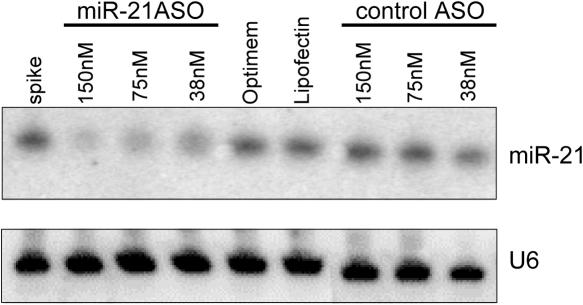
Degradation of the mature miRNA has been reported after in vivo inhibition of miR-122 with uniformly 2′-substituted ASOs (15,16). To test if the same effect was observed in vitro, we examined the effect of ASO inhibition on miR-21 levels in Hela cells after ASO treatment (Figure 6). There was a dose-dependent decrease in the levels of miR-21 detected on a northern blot 24 h after treatment with the 2′-O-MOE-PS ASO targeting miR-21, but not with a control ASO, indicating that degradation of the mature miRNA also occurred in vitro and on a similar time frame as the changes in luciferase sensor activity that we observed.
Figure 6.
Northern blotting for miR-21 in Hela cells after transfection of 2′-O-MOE-PS miR-21 ASO. miR-21 ASO or a control ASO were transfected into Hela cells and levels of miR-21 were measured 24 h later. A control experiment in which miR-21 ASO was spiked into Hela cell lysates showed that the ASO did not interfere with miR-21 detection (spike). Cells mock transfected with low-serum media only (Optimem) or with media containing the transfection reagent (Lipofectin) also showed no reduction in miR-21 levels.
DISCUSSION
In this study we set out to identify the optimal modified chemistries for inhibition of miRNA activity by ASOs. Few miRNAs have been assigned any function, and antisense oligos are a convenient method for inhibiting them. Antisense targeting may be the only possible approach for therapeutically inhibiting miRNAs, since the active molecule is a small RNA, which will be challenging to target specifically without using Watson–Crick base pairing. Therefore, identifying optimal chemistries for miRNA inhibition is crucial for effective functionalization and therapeutic targeting of miRNAs.
The ASOs evaluated in this study are complementary to mature miR-21, but could interfere with miRNA processing by binding to pri- or pre-miRNA. They could also act after Dicer processing, but before loading of the mature miRNA into Argonaute. At least one study suggested that ASOs targeting miRNA act primarily by binding to the mature miRNA and blocking its function in RISC (59). Krutzfeldt et al. (15) reported in vivo reduction of mature miR-122 with no effect on pre-miR-122 levels, also pointing to the mature miRNA as the primary site of action of the ASO. Two observations in this study are consistent with this model. First, inhibition of miRNA activity and reduction of mature miRNA levels occurred within 24 h. There is evidence to suggest that RISC-loaded mature miRNA is long-lived, as inhibition of Drosha processing, pre-miRNA nuclear export, or the PACT protein shown to be involved in RISC loading, had minimal effects on the steady-state levels of mature miRNAs (88–91). If the ASO were inhibiting miRNA processing or loading, effects on miRNA activity would not be observed until turnover of the RISC-loaded mature miRNA. Second, the correlation in Table 2 between active miRNA ASOs and poor siRNA passenger strands implies that the ASO is acting at the site of passenger strand unwinding, which has been reported to be in RISC (81–85). Thus, while we cannot rule out the possibility that the ASO inhibits miRNA processing or RISC loading, our data and others so far suggest the ASO acts primarily on mature miRNA in RISC.
The initial set of modified ASOs was designed to evaluate the role of affinity in anti-miRNA ASO activity. In the context of a PS modified backbone, the degree of miRNA inhibition correlated loosely with affinity. The two modifications with the highest Tm's, uniform 2′-F and the 2′-LNA/MOE mixed sugar, were most active. The 2′-O-Me and 2′-O-MOE substituted ASOs had similarly reduced Tm's and were less active at inhibition of miR-21 activity, although the 2′-O-Me-PS ASO was less active than expected based on affinity alone. Uniform DNA had the lowest Tm and was inactive. Replacement of a PS backbone modification with the diester, which should improve target affinity, resulted in an increase in activity for the 2′-O-MOE ASOs, but gave little or no improvement in activity for a 2′-O-Me ASO. This result was not surprising for the 2′-O-Me-PO ASO as this modification is rapidly degraded in cells and little to no cellular activity for ASOs containing 2′-O-Me residues with PO linkages has been reported (76,79). The timecourse data are consistent with this, as the 2′-O-MOE-PO and 2′-O-Me-PO ASOs showed similar activity at early timepoints, but the 2′-O-MOE-PO was much more active at later timepoints. Although 2′-O-Me-PS oligonucleotides are less resistant to degradation than 2′-O-MOE-PS oligonucleotides, ASOs with this modification are active in cellular assays (76,79). The unexpectedly reduced activity of the 2′-O-Me-PS ASO compared to the 2′-O-MOE-PS ASO in this miR-21 sensor assay was not likely only due to degradation.
That discrepancy, and the difference in activity between the 2′-O-MOE/2′-deoxy hemimer ASOs in spite of their almost identical Tm's, suggested that in some cases, more than affinity determined effective miRNA inhibition. This led us to the speculation that an anti-miRNA ASO, if it is acting primarily on mature miRNA in RISC, is analogous to a passenger strand of an siRNA or miRNA. Perhaps the same rules governing tolerability of chemical modification of siRNA passenger strands also hold true for modified anti-miRNA ASOs. Indeed, this is what we found. There was, generally speaking, an inverse correlation of PTEN siRNA activity with anti-miR-21 activity for each modified passenger strand/miRNA ASO. The best anti-miRNA ASO tested, the uniformly modified 2′-O-MOE-PO, was completely inactive as a PTEN siRNA passenger strand. The least active anti-miRNA ASOs, the MOE-DNA-MOE gapmer, and the alternating MOE-DNA motif, were the most active siRNA passenger strands. These data support the hypothesis that a similar mechanism could be involved in dissociation of an siRNA passenger strand and dissociation of an anti-miRNA ASO, which would result in ineffective inhibition of miRNA activity. It remains to be seen whether this hypothesis will be useful for predicting additional designs for active anti-miRNA ASOs.
Theoretically, the 2′-O-MOE gapmer ASO targeting the mature miRNA sequence could inhibit miRNA activity via an RNase H-based mechanism, acting on the mature, pre- or pri-miRNA. However, we observed little anti-miRNA activity in the luciferase sensor assay with the 2′-O-MOE gapmer. It is likely that the mature miRNA in RISC is not accessible to the RNase H enzyme. The possibility that the 2′-O-MOE gapmer ASO could target the pri- or pre-miRNA is still open, since this would inhibit production of new mature miRNA. Any measurable effects would depend on the rate of turnover of the already present mature miRNA, which may not be observable in the time-frame of these assays, as the half-life of mature miRNA is thought to be long (88–91). In addition, the mature miRNA is found in a structured hairpin in the pri- and pre-miRNA, and structured hairpin targets tend to reduce ASO activity (92). Targeting the pri-miRNA transcript outside the hairpin structure may be a better strategy for ASOs using an RNase H mechanism. Targeting the pri-miRNA transcript with siRNA is not likely to be an option, since pri-miRNA transcripts are found primarily in the nucleus, and siRNA targeting of pre-miRNAs has been reported to be inefficient, requiring much higher doses of siRNA than usually needed for targeting mRNAs (60). RNase H ASOs, on the other hand, have successfully been used to target noncoding RNAs in the nucleus (93).
These studies outline some of the factors important for effective targeting of miRNA with ASOs in cell culture. We evaluated effects of 2′-sugar and backbone ASO modifications, and their placement within the ASO. While affinity for the target miRNA was the most important determinant of anti-miRNA activity, our results suggested that to be an effective miRNA ASO, it may also be necessary to avoid dissociation by the factors responsible for si/miRNA passenger strand unwinding. Mismatches were not well tolerated, suggesting it will be difficult to target multiple miRNA family members with a single ASO. Truncation studies indicated that although significant activity was retained with truncation of just 1 nt, loss of two or more nucleotides resulted in a substantial loss of anti-miRNA activity.
These studies focused on a single miRNA in a single assay system. It will be essential to extend these studies to target other miRNAs, with other assays for miRNA activity. As several natural targets for miRNAs have now been identified, it will be possible to perform studies that measure ASO effects on miRNA target protein or mRNA levels directly. In addition, other factors will have to be considered in optimizing in vivo anti-miRNA activity for therapeutic targeting, as the pharmacokinetic properties of an ASO can dramatically influence its effectiveness in an animal. Data from in vivo targeting of miR-122 suggested that the outcome of miRNA ASO inhibition is degradation of the mature miRNA (15,16), and we also observed reduction in miR-21 levels after inhibition with a 2′-O-MOE-PS ASO in these studies. It's not clear how ASO binding to a miRNA in RISC results in disappearance of the mature miRNA, but understanding the mechanism of degradation may lead to more effective methods of miRNA targeting, and it will be an important question for future studies.
Acknowledgments
The authors thank Thazha P. Prakash for his generous assistance with Tm measurements and Tracy Reigle for help with preparing the figures. Funding to pay the Open Access publication charges for this article was provided by Isis Pharmaceuticals.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Du T., Zamore P.D. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y., Samal E., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 6.Chen C.-Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–87. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 7.Calin G.A., Liu C.G., Sevignani C., Ferracin M., Felli N., Dumitru C.D., Shimizu M., Cimmino A., Zupo S., Dono M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McManus M.T. MicroRNAs and cancer. Semin. Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 10.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Hammond S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev. 2005;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esau C., Kang X., Peralta E., Hanson E., Marcusson E.G., Ravichandran L.V., Sun Y., Koo S., Perera R.J., Jain R., et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 14.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., MacDonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 15.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 16.Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Lecellier C.H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saib A., Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 18.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahi P., Loukianiouk S., Bohne-Lang A., Kenzelmann M., Kuffer S., Maertens S., Eils R., Grone H.J., Gretz N., Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor M.F., Wiederholt K., Sverdrup F. Antisense oligonucleotides: a systematic high-throughput approach to target validation and gene function determination. Drug. Discov. Today. 1999;4:562–567. doi: 10.1016/s1359-6446(99)01392-6. [DOI] [PubMed] [Google Scholar]
- 22.Dean N.M. Functional genomics and target validation approaches using antisense oligonucleotide technology. Curr. Opin. Biotechnol. 2001;12:622–625. doi: 10.1016/s0958-1669(01)00270-1. [DOI] [PubMed] [Google Scholar]
- 23.Kramer R., Cohen D. Functional genomics to new drug targets. Nature Rev. Drug Discov. 2004;3:965–972. doi: 10.1038/nrd1552. [DOI] [PubMed] [Google Scholar]
- 24.Ho S.P., Hartig P.R. Antisense oligonucleotides for target validation in the CNS. Curr. Opin. Mol. Ther. 1999;1:336–343. [PubMed] [Google Scholar]
- 25.Koller E., Propp S., Zhang H., Zhao C., Xiao X., Chang M., Hirsch S.A., Shepard P.J., Koo S., Murphy C., et al. Use of a chemically modified antisense oligonucleotide library to identify and validate Eg5 (kinesin-like 1) as a target for antineoplastic drug development. Cancer Res. 2006;66:2059–2066. doi: 10.1158/0008-5472.CAN-05-1531. [DOI] [PubMed] [Google Scholar]
- 26.Aza-Blanc P., Cooper C.L., Wagner K., Batalov S., Deveraux Q.L., Cooke M.P. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
- 27.Berns K., Hijmans E.M., Mullenders J., Brummelkamp T.R., Velds A., Heimerikx M., Kerkhoven R.M., Madiredjo M., Nijkamp W., Weigelt B., et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 28.Zheng L., Liu J., Batalov S., Zhou D., Orth A., Ding S., Schultz P.G. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung R.K., Whittaker P.A. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol. Ther. 2005;107:222–239. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurreck J. Antisense and RNA interference approaches to target validation in pain research. Curr. Opin. Drug Discov. Devel. 2004;7:179–187. [PubMed] [Google Scholar]
- 31.Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 32.Zellweger T., Miyake H., Cooper S., Chi K., Conklin B.S., Monia B.P., Gleave M.E. Antitumor activity of antisense clusterin oligonucleotides is improved in vitro and in vivo by incorporation of 2′-O-(2-methoxy)ethyl chemistry. J. Pharmacol. Exp. Ther. 2001;298:934–940. [PubMed] [Google Scholar]
- 33.Watts L.M., Manchem V.P., Leedom T.A., Rivard A.L., McKay R.A., Bao D., Neroladakis T., Monia B.P., Bodenmiller D.M., Cao J.X., et al. Reduction of hepatic and adipose tissue glucocorticoid receptor expression with antisense oligonucleotides improves hyperglycemia and hyperlipidemia in diabetic rodents without causing systemic glucocorticoid antagonism. Diabetes. 2005;54:1846–1853. doi: 10.2337/diabetes.54.6.1846. [DOI] [PubMed] [Google Scholar]
- 34.Yu X.X., Murray S.F., Pandey S.K., Booten S.L., Bao D., Song X.Z., Kelly S., Chen S., McKay R., Monia B.P., et al. Antisense oligonucleotide reduction of DGAT2 expression improves hepatic steatosis and hyperlipidemia in obese mice. Hepatology. 2005;42:362–371. doi: 10.1002/hep.20783. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y., Vassilakos A., Feng N., Jin H., Wang M., Xiong K., Wright J., Young A. GTI-2501, an antisense agent targeting R1, the large subunit of human ribonucleotide reductase, shows potent anti-tumor activity against a variety of tumors. Int. J. Oncol. 2006;28:469–478. [PubMed] [Google Scholar]
- 36.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 37.Stepkowski S.M., Qu X., Wang M.E., Tian L., Chen W., Wancewicz E.V., Johnston J.F., Bennett C.F., Monia B.P. Inhibition of C-raf expression by antisense oligonucleotides extends heart allograft survival in rats. Transplantation. 2000;70:656–661. doi: 10.1097/00007890-200008270-00020. [DOI] [PubMed] [Google Scholar]
- 38.Chi K.N., Eisenhauer E., Fazli L., Jones E.C., Goldenberg S.L., Powers J., Tu D., Gleave M.E. A phase I pharmacokinetic and pharmacodynamic study of OGX-011, a 2′-methoxyethyl antisense oligonucleotide to clusterin, in patients with localized prostate cancer. J. Natl. Cancer Inst. 2005;97:1287–1296. doi: 10.1093/jnci/dji252. [DOI] [PubMed] [Google Scholar]
- 39.Dean N.M., Bennett C.F. Antisense oligonucleotide-based therapeutics for cancer. Oncogene. 2003;22:9087–9096. doi: 10.1038/sj.onc.1207231. [DOI] [PubMed] [Google Scholar]
- 40.Goodchild J. Oligonucleotide therapeutics: 25 years agrowing. Curr. Opin. Mol. Ther. 2004;6:120–128. [PubMed] [Google Scholar]
- 41.Crooke S.T. Antisense strategies. Curr. Mol. Med. 2004;4:465–487. doi: 10.2174/1566524043360375. [DOI] [PubMed] [Google Scholar]
- 42.Simoes-Wust A.P., Hopkins-Donaldson S., Sigrist B., Belyanskaya L., Stahel R.A., Zangemeister-Wittke U. A functionally improved locked nucleic acid antisense oligonucleotide inhibits Bcl-2 and Bcl-xL expression and facilitates tumor cell apoptosis. Oligonucleotides. 2004;14:199–209. doi: 10.1089/oli.2004.14.199. [DOI] [PubMed] [Google Scholar]
- 43.Da Ros T., Spalluto G., Prato M., Saison-Behmoaras T., Boutorine A., Cacciari B. Oligonucleotides and oligonucleotide conjugates: a new approach for cancer treatment. Curr. Med. Chem. 2005;12:71–88. doi: 10.2174/0929867053363603. [DOI] [PubMed] [Google Scholar]
- 44.Frieden M., Christensen S.M., Mikkelsen N.D., Rosenbohm C., Thrue C.A., Westergaard M., Hansen H.F., Orum H., Koch T. Expanding the design horizon of antisense oligonucleotides with alpha-L-LNA. Nucleic Acids Res. 2003;31:6365–6372. doi: 10.1093/nar/gkg820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frieden M., Hansen H.F., Koch T. Nuclease stability of LNA oligonucleotides and LNA-DNA chimeras. Nucleosides Nucleotides Nucleic Acids. 2003;22:1041–1043. doi: 10.1081/NCN-120022731. [DOI] [PubMed] [Google Scholar]
- 46.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 47.Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S.M., McGee D., Guinosso C.J., Cook P.D. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monia B.P., Johnston J.F., Sasmor H., Cummins L.L. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J. Biol. Chem. 1996;271:14533–14540. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- 49.Altmann K.H., Dean N.M., Fabbro D., Freier S.M., Geiger T., Haener R., Huesken D., Martin P., Monia B.P., Muller M., et al. Second generation of antisense oligonucleotides. From nuclease resistance to biological efficacy in animals. Chimia. 1996;50:168–176. [Google Scholar]
- 50.Freier S.M., Altmann K.H. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geary R.S., Watanabe T.A., Truong L., Freier S., Lesnik E.A., Sioufi N.B., Sasmor H., Manoharan M., Levin A.A. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 2001;296:890–897. [PubMed] [Google Scholar]
- 52.Lima W.F., Nichols J.G., Wu H., Prakash T.P., Migawa M.T., Wyrzykiewicz T.K., Bhat B., Crooke S.T. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 53.Henry S., Stecker K., Brooks D., Monteith D., Conklin B., Bennett C.F. Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J. Pharmacol. Exp. Ther. 2000;292:468–479. [PubMed] [Google Scholar]
- 54.Cramer H., Pfleiderer W. Nucleotides LXIV[1]: synthesis hydridization and enzymatic degradation studies of 2′-O-methyloligoribonucleotides and 2′-O-methyl/deoxy gapmers. Nucleosides Nucleotides Nucleic Acids. 2000;19:1765–1777. doi: 10.1080/15257770008045458. [DOI] [PubMed] [Google Scholar]
- 55.Sproat B.S., Lamond A.I., Beijer B., Neuner P., Ryder U. Highly efficient chemical synthesis of 2′-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989;17:3373–3386. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kierzek E., Ciesielska A., Pasternak K., Mathews D.H., Turner D.H., Kierzek R. The influence of locked nucleic acid residues on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 2005;33:5082–5093. doi: 10.1093/nar/gki789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boutla A., Delidakis C., Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leaman D., Chen P.Y., Fak J., Yalcin A., Pearce M., Unnerstall U., Marks D.S., Sander C., Tuschl T., Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Hutvagner G., Simard M.J., Mello C.C., Zamore P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y.S., Kim H.K., Chung S., Kim K.S., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 61.Cheng A.M., Byrom M.W., Shelton J., Ford L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan J.A., Krichevsky A.M., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 63.Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 64.Weiler J., Hunziker J., Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene. Ther. 2005;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 65.Puglisi J.D., Tinoco I., Jr Absorbance melting curves of RNA. Meth. Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 66.Petersheim M., Turner D.H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 67.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 68.Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 70.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 71.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes and microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 72.Brennecke J., Hipfner D.R., Stark A., Russell R.B., Cohen S.M. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 73.Mansfield J., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and othere developmentally regulated patterns of vertebrate microRNA expression. Nature Genet. 2004;36:1076–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 74.Meister G., Landthaler M., Dorsett Y., Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng Y., Yi R., Cullen B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baker B.F., Lot S.S., Condon T.P., Cheng-Flournoy S., Lesnik E.A., Sasmor H.M., Bennett C.F. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 77.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res. 1988;16:3209–3221. doi: 10.1093/nar/16.8.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawasaki A.M., Casper M.D., Freier S.M., Lesnik E.A., Zounes M.C., Cummins L.L., Gonzalez C., Cook P.D. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 79.McKay R.A., Miraglia L.J., Cummins L.L., Owens S.R., Sasmor H., Dean N.M. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 80.Hoke G.D., Draper K., Freier S.M., Gonzalez C., Driver V.B., Zounes M.C., Ecker D.J. Effects of phosphorothioate capping on antisense oligonucleotide stability, hybridization and antiviral efficacy versus herpes simplex virus infection. Nucleic Acids Res. 1991;19:5743–5748. doi: 10.1093/nar/19.20.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 82.Maniataki E., Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matranga C., Tomari Y., Shin C., Bartel D., Zamore P. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 84.Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parker J.S., Roe S.M., Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee Y., Hur I., Park S.Y., Kim Y.K., Suh M.R., Kim V.N. The role of PACT in the RNA silencing pathway. EMBO. J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 90.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 91.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vickers T.A., Wyatt J.R., Freier S.M. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]