Abstract
The white rot fungus Schizophyllum commune is used for the analysis of mating and sexual development in homobasidiomycete fungi. In this study, we isolated the gene gap1 encoding a GTPase-activating protein for Ras. Disruption of gap1 should therefore lead to strains accumulating Ras in its activated, GTP-bound state and to constitutive Ras signaling. Haploid Δgap1 monokaryons of different mating types did not show alterations in mating behavior in the four different mating interactions possible in fungi expressing a tetrapolar mating type system. Instead, the growth rate in Δgap1 monokaryons was reduced by ca. 25% and ca. 50% in homozygous Δgap1/Δgap1 dikaryons. Monokaryons, as well as homozygous dikaryons, carrying the disrupted gap1 alleles exhibited a disorientated growth pattern. Dikaryons showed a strong phenotype during clamp formation since hook cells failed to fuse with the peg beside them. Instead, the dikaryotic character of the hyphae was rescued by fusion of the hooks with nearby developing branches. Δgap1/Δgap1 dikaryons formed increased numbers of fruitbody primordia, whereas the amount of fruitbodies was not raised. Mature fruitbodies formed no or abnormal gills. No production of spores could be observed. The results suggest Ras involvement in growth, clamp formation, and fruitbody development.
The homobasidiomycetous white-rot fungus Schizophyllum commune has been used as a model system for the investigation of mating and sexual development for decades since it can be grown from spore to spore through its entire life cycle within 14 days on defined media, and it shows easily distinguished phenotypes for a tetrapolar mating behavior (32, 53). The tetrapolar mating system consists of two sets of mating type genes. The A mating type genes encode homeodomain transcription factors that are assumed to directly regulate gene expression (39, 67). A multiallelic pheromone/receptor system is encoded by the B genes. Homology to the yeast pheromone system has led to the expectation that mating is in part controlled by a mitogen-activated protein kinase signal transduction cascade that is activated after stimulation of the G protein-coupled pheromone receptor (20, 76, 82).
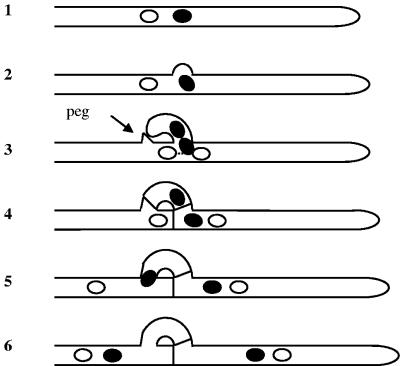
A fully compatible mating between two strains of S. commune occurs when both differ in their A and B gene specificities (A ≠ B ≠). The specificity of a locus is defined by a lack of activation of downstream developmental processes after crossing two strains with identical specificities in their mating type genes (20). Several steps of subsequent development can be distinguished. After cell fusion, septal breakdown and fast nuclear migration allow reciprocal nuclear exchange between the two mates. Migrant and resident nuclei pair, and dikaryotic hyphal tips are established. Subsequent conjugate nuclear division is accompanied by formation of clamp connections. Clamp connections are short, backwardly directed branches that fuse with the subapical cell and provide a bypass for one of the nuclei produced during synchronous division of the dikaryon, ensuring the equal distribution of the two different nuclei between mother and daughter cells (Fig. 1). Initiation of conjugate nuclear division is accompanied by formation of a lateral branch, the hook. After nuclear division and septum formation, one nucleus is temporarily entrapped in the hook until the hook cell fuses with the subapical cell, forming a clamp connection. The hook cell does not fuse with the subapical cell directly but with a peg formed by the subapical cell growing toward the hook (5, 11). Finally, the entrapped nucleus migrates from the clamp back into the subapical cell to restore the nuclear pairing (Fig. 1). Clamp formation is repeated at each subsequent cellular division (31, 52).
FIG. 1.
Scheme of clamp formation. At the site where mitosis will take place a hook is formed (diagrams 1 and 2). The hook grows backward toward the main hyphae, where a peg marks the intended fusion point. During mitosis one nucleus divides in direction of the hook, whereas the other nucleus divides along the main hyphal axis (diagram 3). Septae are formed between apical and subapical cell and at the basis of the hook between apical and hook cell. In this way, one nucleus stays temporarily entrapped into the hook cell (diagram 4). The hook cell fuses with the penultimate cell and releases the entrapped nucleus, restoring the dikaryotic character of the subapical cell (diagrams 5 and 6). Black and white nuclei represent nuclei with different A and B specificities. Alternate positions with respect to the hyphal tip are indicated according to the observations made in C. cinereus by Iwasa et al. (28).
If both mates differ only in their A gene specificities forming a semicompatible mating interaction, nuclei are paired in the apical cells, but nonapical cells are uninucleate, because hook cells fail to fuse with the subapical cells, keeping one nucleus entrapped. This heterokaryon is unstable and, if not forced, both strains remain separate forming a “barrage” reaction on agar plates (52). The semicompatible mating interaction in which only the B morphogenetic pathway is activated due to different B specificities in the mates is characterized by constant nuclear migration and expression of cell wall-degrading enzymes. In this case, cells contain various numbers of nuclei and show protoplasmic protrusions due to partial breakdown of cell walls. Hyphae are profusely and irregularly branched. Macroscopically, these heterokaryons show little aerial mycelium, leading to the term “flat” for this phenotype (49, 52, 83).
The fully compatible dikaryon with its heteroallelic A and B mating type factors is able to form fruitbodies. Morphogenesis starts with the formation of initials, loose microscopic hyphal tufts that develop to the spherical to cylindrical primordia. These differentiate an apical pit, the initiation of hymenium formation, and continue to increase in mass. The pit expands laterally and forms the pileus. Finally, the mushroom-like fruitbody becomes fully expanded, exposing its spore-bearing gills (36). Fruiting is strictly controlled by environmental factors. In S. commune fruiting is controlled by depletion of carbon and nitrogen sources and by the concentration of carbon dioxide, temperature, and light and requires thiamine (47). The regulation of fruitbody development involves cyclic AMP (cAMP) signaling since the addition of extracellular cAMP causes many fruitbodies to cease development at the primordial stage, whereas abnormal gills are formed by fruitbodies that continue growth (65). Similarly, addition of caffeine that inhibits cAMP degrading phosphodiesterase stimulates production of hyphal knots. Intracellular cAMP levels during fruiting are highest during hyphal knot formation and low at the time of pileus formation (30).
In an attempt to identify genes transcriptionally regulated by the mating type genes, we isolated a cDNA fragment coding for a Ras GTPase-activating protein (GAP) from S. commune (62). GAPs are regulators for small GTP-binding proteins that increase the intrinsic GTPase activity of the G protein, thereby turning it from the active, GTP-bound to the inactive, GDP-bound form. Several differentially structured RasGAPs can be defined in fungi (Fig. 2) which made analysis of the phenotype of gap1 deletions interesting. The counterparts of GAPs are guanine nucleotide exchange factors (GEFs) which promote the release of bound GDP, thus allowing exchange for GTP (7-9).
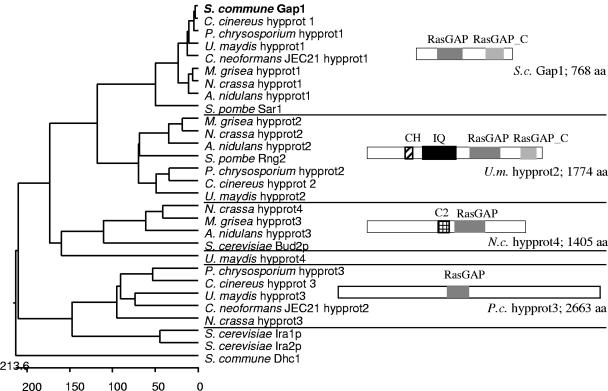
FIG. 2.
Phylogenetic tree of known and putative Ras specific GAPs that were found by BLAST analysis against eight fungal genomes using the S. commune Gap1 sequence. Protein IDs: Aspergillus nidulans hypprot1, EAA61076.1; A.n. hypprot2, EAA66795.1; A.n. hypprot3, EAA59943.1; Cryptococcus neoformans JEC21 hypprot1, EAL17992.1; C.n. hypprot2, CNF01820; Magnaporthe grisea hypprot1, EAA50087.1; M.g. hypprot2, EAA52105.1; M.g. hypprot3, EAA57136.1; Neurospora crassa hypprot1, EAA30748.1; N.c. hypprot2, EAA35739.1; N.c. hypprot3, EAA27028.1; N.c. hypprot4, EAA26628.1; Schizosaccharomyces pombe Sar1, NP_595370.1; S.p. Rng2, O14188; Saccharomyces cerevisiae Bud2p, NP_612831.1; S.c. Ira1p, NP_009698.1; S.c. Ira2p, NP_014560.1; Ustilago maydis hypprot1, EAK81710.1; U.m. hypprot2, EAK86071.1; U.m. hypprot3, EAK82145.1; and U.m. hypprot4, EAK85899.1. Coprinus cinereus and Phanerochaete chrysosporium ORFs were found in the following: C.c. hypprot1, cont1_192 scf9; C.c. hypprot2, cont1_198 scf10; C.c. hypprot3, cont1_258 scf15; P.c. hypprot1, scf31; P.c. hypprot2, scf20; and P.c. hypprot3, scf43. CH, calponin homology domain; IQ, IQ calmodulin-binding motif.
The small GTPase Ras is known to regulate various cellular processes in fungi. In the yeast Saccharomyces cerevisiae Ras regulates metabolism, proliferation, stress resistance, and pseudohyphal and haploid invasive growth (17, 18, 23, 29, 40, 42-44). The two Ras proteins in S. cerevisiae regulate the production of intracellular cAMP by activating adenylate cyclase. cAMP binding to the regulatory subunits of cAMP-dependent protein kinase A results in release of the catalytic subunits that phosphorylate target proteins. cAMP is hydrolyzed by phosphodiesterases (73, 74). Beside the Ras/cAMP pathway, Ras2p also activates a mitogen-activated protein kinase module and regulates cytoskeletal polarity perhaps with a third pathway not yet clearly defined (24, 44). In contrast, in the fission yeast Schizosaccharomyces pombe Ras1 does not activate adenylate cyclase (16).
For mushroom forming basidiomycetes little is known about the functions of Ras and its regulators. The isolation of Ras genes is published for Lentinus edodes, Coprinus cinereus, S. commune, and Laccaria bicolor and differential expression of the L. bicolor ras gene after interaction of the ectomycorrhizal fungus with host roots has been shown (25, 27, 55, 68). In addition, suppression of hyphal knot formation, the initial step of fruitbody development, and an altered hyphal growth pattern have been observed for C. cinereus strains expressing a dominant-active allele of ras (U. Kües, unpublished data). Recently, expression of the dominant active RasQ61L allele has been reported for S. commune to result in the reduction of monokaryotic growth rate and fruitbody initiation (87).
We describe here Ras-dependent development by investigation of the function of gap1 during growth and sexual development of the mushroom-forming basidiomycete S. commune. We show that deletion of gap1 impairs the maintenance of hyphal growth direction, the failure of clamp connection formation in dikaryons, and enhanced production of fruitbody primordia with hampered hymenium formation and lack of spore production.
MATERIALS AND METHODS
Strains and growth conditions.
The coisogenic S. commune strains 4-40 (matA α4 β6; matB α1, β1; CBS 340.81), 4-39 (matA α1 β1; matB α3, β2; CBS 341.81), and W21 resulting from a cross of 4-40 × 4-39 (matA α1 β1; matB α1, β1), as well as strain C6 (matA α3 β1; matB α2, β2; ura1−; trp1−) (strain collection at the University of Jena) were used. Strain DSII-1 (matA α1 β1; matB α3, β2; ura1−; trp1−), obtained from the cross 4-39 × C6, carries the gap1 allele of strain 4-39 as determined by restriction fragment length polymorphism analysis with ApaLI. Strain DSΔgap1 was generated by transformation of strain DSII-1 with plasmid pΔgap1. Strains DSΔgap1F1 through DSΔgap1F33 were progenies from a cross between strains DSΔgap1 and 4-40. They were tested by PCR for the presence of the Δgap1 allele, and their mating types were determined by test crosses to strains 4-40, W21, and DSII-1. Strains DSΔgap1komp1 through DSΔgap1komp27 and DSΔgap1trp1 through DSΔgap1trp20 were generated by transformation of strain DSΔgap1 with plasmid p5-3revtrp or pSKtrp, respectively. Strains of S. commune were grown on minimal medium (MM) or complex yeast medium (CYM) with or without supplementation of 4 mM tryptophan (MMT/CYMT) in dark at 30°C as described earlier (53). For the formation of fruitbodies, strains were grown at room temperature in the light.
Escherichia coli K-12 DH5α (Bethesda Research Laboratories) was used for plasmid construction.
Isolation of gap1.
For the identification of gap1, genomic DNA from strain W21 was digested with restriction enzymes and analyzed by Southern hybridization using the gap1 cDNA fragment (62) or the 697-bp PstI-XbaI fragment of clone p5-3 (see below) as a probe. From two partial genomic libraries containing 5-kb PstI or 4-kb KpnI fragments (see Results for details) cloned in pBluescript SKII, the overlapping clones p5-3 and p5-3rev harboring the complete gap1 open reading frame (ORF) were isolated and sequenced. To detect the putative introns in gap1, clone pcgap1II was isolated from a cDNA library of strain 4-40 cloned in λZAP II (Stratagene) using the gap1 cDNA fragment as a probe and sequenced.
Plasmid constructions.
Plasmid pBluescript SKII (Stratagene) was used for cloning, subcloning, and sequencing, and plasmid pSL1180 (Amersham Biosciences) was used for subcloning.
pΔgap1 is a pSL1180 derivative, in which three fragments were successively ligated into NdeI and BamHI sites (see Results for details). The first was the 1.4-kb XbaI-NdeI fragment obtained from p5-3rev (3′ end of gap1). The second was a 2.4-kb PCR fragment obtained from p5-3 with primers KL5-3_5′_BamHI (CGCGGATCCAGCTGGTAGTCC) and KL5-3_XbaI (TGCTCTAGATATGTTCAACTTCGG) and cut with BamHI and XbaI (region 5′ upstream of gap1). The third was a XbaI cut fusion product obtained by PCR of the promoter of the tef1 gene and the ura1 gene. For this purpose the promoter of tef1 was amplified from plasmid ptefuraEco, a precursor of plasmid pTUT1 (19), with the primers tefXbaI (TGCTCTAGATTCGGCGCACGACC) and tefura (GCTTGTGGGCGGCGGTCATTTTGAATGTTTTCTAGG), and the ura1 gene was amplified from plasmid pChi (K. B. Lengeler, unpublished) with the primers uraATG (ACCGCCGCCCACAAGCTCACATAC) and uraXbaI (GCTCTAGAATTCAAGTTACTCTCCGC) using the high-fidelity Vent polymerase (New England Biolabs). Both fragments were fused in a PCR with primers tefXbaI and uraXbaI. Orientation of the third fragment in pΔgap1 was determined by cutting with PstI.
Plasmid pSKtrp is a pBluescript SKII derivative that harbors the EcoRI-HindIII fragment of cosmid pTC20 containing the trp1 gene (46).
Plasmid pgaptrp was derived from plasmid p5-3rev by inserting the 4.6-kb EcoRI-HindIII fragment of pSKtrp. Plasmid p5-3rev contains the complete gap1 gene and an additional 613 bp upstream of the 5′ untranslated region of gap1.
DNA and RNA procedures.
For DNA manipulations, Southern blot analyses and isolation of total RNA standard procedures were followed (61). S. commune DNA was isolated by using the CTAB (cetyltrimethylammonium bromide) method described for plants (4) with the following change in the protocol. Pulverized mycelium was incubated in DNA extraction buffer (0.1 M Tris-HCl [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate) for 1 h at 65°C. After centrifugation and recovery of the supernatant, NaCl concentration was adjusted to 0.7 M. Three extractions with CTAB-NaCl solution followed as described above. For preparation of small amounts of DNA aerial mycelium was ground with sand in 1.5-ml tubes in extraction buffer according to the protocol of Cenis (12). Transformation of S. commune was performed from 2-day-old mycelium grown in liquid culture as described earlier (66), using 100 mg of Caylase (Cayla, France) per 20 ml of concentrated mycelium. The primers gap1_del (TCCCGCGAAGCACCCGAGAC), gap3′1_del (ACCGCATGAAGAACCAGCACAAG), and ura1_del (GGTGGGCGGCGTACATTGAGA) were used to screen for gap1 disruption mutants. Whereas the first two amplified a fragment in strains carrying the wild-type allele, the latter two led to amplification after homologous integration of the disruption construct. Disruption of gap1 was confirmed in Southern blot analyses with the 1.3-kb HindIII-KpnI fragment of clone p5-3 and the 1.4-kb XbaI-NdeI fragment of clone p5-3rev as 5′ and 3′ probes, respectively. For competitive PCR (for a review, see reference 14), total RNA was isolated from 3-day-old liquid cultures of a monokaryon (strain 4-39), a dikaryon (4-39 × 4-40), and a heterokaryon derived from a semicompatible B mating interaction (strains 4-39 × W21). Poly(A) RNA was isolated from total RNA by using the Oligotex mRNA Minikit (QIAGEN). cDNA for use in competitive PCR was synthesized by using oligo(dT) primers and the Superscript II reverse transcriptase (Roche Diagnostics). Competitive PCR was performed with the primers gap1_7 (TCAACCAGATCGAAGAAGAGA) and gap1_8 (CCGTGAGGATGCGTAGAT) using 10 ng of cDNA and various amounts of plasmid p5-3rev as a template. The competitor was used in a range from 1.35 × 10−6 fmol to 9.1 × 10−8 fmol (being equivalent to 813 and 55 molecules, respectively). The primer combination used amplified fragments of 679 and 871 bp for the cDNA and competitor, respectively, with the following PCR program: 3-min time delay at 94°C; 33 cycles of 30 s at 94°C, 30 s at 54°C, and 50 s at 72°C; and finally 10 min at 72°C. Schubert et al. (62) showed that gap1 (termed KL5) amplification is in the exponential phase using this large number of cycles. The primers gap1-6ATG (CTCATCATGTCCTACCCG) and gap1Ende (GCGAACACCTTCTTCAAG) were used to amplify the complete gap1 gene in strains transformed with the plasmid pgaptrp. To control for 3′ mRNA fragments in deletion strains, primers Gap1 (GAGACGCCGGTGCAGGACATC) and Gap2 (CCGGCCAGCCAGCAAGAGAA) were used at a 58°C annealing temperature to produce an 807-bp cDNA fragment. The DNA fragment of 911 bp did not occur when mRNA was used as a template. To control for cDNA, a positive control was performed with the primers TEF1 (GGTCACCGTGACTTCATCAAG) and TEF2 (CTTGATGATACCGGTCTCGAC) amplifying 534 bp of the gene encoding translation elongation factor EF1α at a 58°C annealing temperature (81).
Sequence analysis.
Sequence information was obtained by using a Licor DNA sequencer 4000L and the Sequenase 7-deaza-dGTP DNA sequencing kit (Amersham Biosciences) or by MWG Biotech AG. Sequence data were analyzed with the Lasergene software (DNASTAR, Inc.). The nucleotide sequences of the genomic and cDNA clones of gap1 have been submitted to GenBank under accession numbers AY653306 and AY653307. Similarity searches were performed by using the NCBI BLASTP 2.2.9 program (2). Protein domains were identified by using the ProfileScan program (http://hits.isbsib.ch/cgi-bin/PFSCAN) (13) and the Lupas algorithm for detection of coiled-coil domains (http://myhits.isb-sib.ch/cgi-bin/motif-scan) (38).
Microscopy and documentation.
Pictures of fruitbodies were taken with stereo microscope Stemi 2000-C (Carl Zeiss AG, Jena, Germany). For microscopic observation, a Zeiss Axioplan 2 microscope with differential interference contrast optic was used. For visualization of nuclei, mycelium grown in liquid medium was collected by centrifugation, fixed for 10 min in Tris-buffered saline containing 4% formaldehyde, and mounted in mounting medium (0.1 M Tris-HCl [pH 8.0], 50% glycerol, 1 mg of phenylenediamine/ml; 1 μg of DAPI [4′,6-diamidino-2-phenylindole]/ml). DAPI staining was visualized by illumination with UV light (filter set 02; Carl Zeiss AG, Jena, Germany). Pictures were taken with a Junior Spot camera (Diagnostic Instruments, Inc., Munich, Germany). Pictures of clamp connections were taken with the digital imaging system MicroMax1024 (Princeton Instruments). For time-lapse photographs, strains were grown on slides covered with CYM agar (1.6%). Image processing was done with the Spot software and Metamorph 4.6r6 (Universal Imaging Corp.).
Statistics.
The Student t test was performed at a level of 95% by using the SigmaStat 2.03 software (Systat Software, Inc.).
RESULTS
Characterization of gap1 encoding a Ras GTPase-activating protein.
A cDNA fragment encoding a Ras GTPase-activating protein, gap1, had been identified previously in a screen for differentially expressed genes in B-regulated development of S. commune (62). In order to identify the entire gene, two overlapping fragments were cloned from subgenomic libraries of strain W21 that hybridized to the gap1 cDNA fragment and which together contained the entire gap1 gene. The gap1 cDNA fragment was also used to isolate gap1 from a cDNA library prepared from strain 4-40. No indications for a homologous gene were obtained, suggesting gap1 to be the only member of this RasGAP family. Comparison of the genomic and cDNA sequences revealed an ORF of 768 codons, separated by seven introns (Fig. 3A). Analysis of the deduced amino acid sequence using the ProfileScan program revealed that Gap1 contains a central RasGAP domain (amino acids 171 to 380) and a RasGAP_C domain (amino acids 555 to 695). These domains are characteristic for proteins belonging to the family of Ras-specific GAPs (7). The sequence clearly placed Gap1 into a cluster of RasGAPs with Schizosaccharomyces pombe Sar1 (Fig. 2). Using the method described by Lupas et al. (38), the N-terminal region between amino acids 39 and 58 was detected to form a coiled-coil domain with a probability of over 90% (Fig. 3B). Gap1 showed the highest sequence identity over its entire length to Gap1 (Sar1) from Schizosaccharomyces pombe with 40% identity and to RasGAP1 from Dictyostelium discoideum (31% identity). Since both known proteins are GAPs specific to Ras, we propose that Gap1 also acts on Ras. High sequence identity was also observed to the hypothetical proteins from Ustilago maydis (80% identity), Gibberella zeae (64%), Magnaporthe grisea (64%), Aspergillus nidulans (64%), and Neurospora crassa (61%) (Fig. 3B).
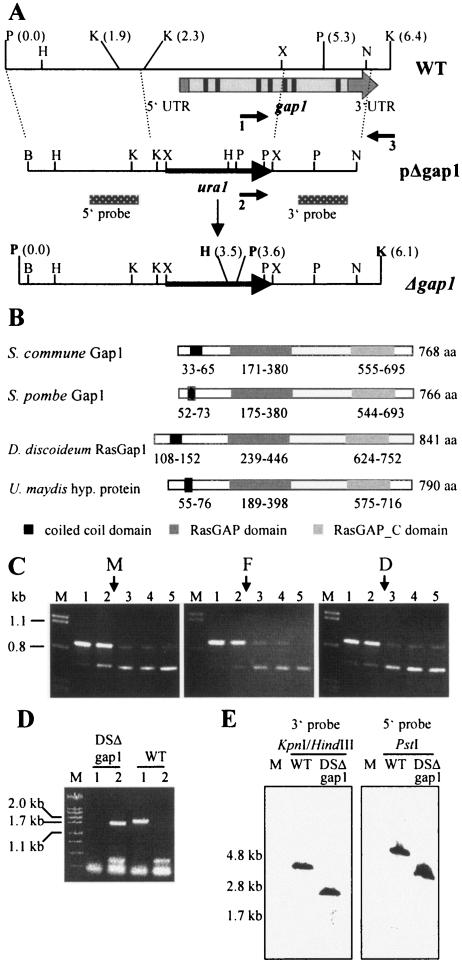
FIG. 3.
Sequence and expression analyses of gap1 and deletion strategy. (A) Restriction map of the genomic region of strain W21 harboring the gap1 gene (WT). The relative position of gap1 is indicated by the arrow. The cDNA of gap1 contains 5′ and 3′ untranslated regions (UTR) and seven introns (indicated in black) and eight exons (shown in light gray). Homologous recombination of the deletion construct (pΔgap1) leads to replacement of the 5′ part of gap1 by the ura1gene (Δgap1). The positions of primers used to identify the disruption mutant DSΔgap1 are indicated by arrows (1, gap1_del; 2, ura1_del; and 3, gap3′1_del). Location of probes 5′ and 3′ are shown by the dotted bars. The new restriction sites BamHI and XbaI in pΔgap1 were introduced during cloning and were used together with the existing XbaI and NdeI sites for construction of pΔgap1. B, BamHI; H, HindIII; K, KpnI; N, NdeI; P, PstI; X, XbaI. (B) Domain structures of Gap1 from S. commune and its homologues. Numbers represent the amino acid (aa) positions of each domain. Domain annotations: RasGAP, (smart) SM00323; RasGAP_C, (pfam)PF03836. Accession numbers: ScGap1, AAT74386.1; SpGap1, A40258; DdRasGap1, AAB39262.1; U. maydis hypothetical protein, UM00949.1 and EAK81710.1. (C) Expression of gap1 in monokaryon 4-39 (subpanel M), common A heterokaryon 4-39 × W21 (flat, subpanel F), and dikaryon 4-39 × 4-40 (subpanel D) of S. commune grown for 3 days in CYM liquid medium. A total of 10 ng of cDNA was used in each PCR. Competitive PCR led to coamplification of the gap1 cDNA fragment (680 bp) and the competitor fragment (870 bp). Arrows indicate the estimated equimolar concentrations of target and competitor fragments. Lanes: M, λ-DNA cut with PstI; 1, 1.35 × 10−6 fmol (813 molecules); 2, 6.9 × 10−7 fmol (416 molecules); 3, 3.5 × 10−7 fmol (211 molecules); 4, 1.8 × 10−7 fmol (108 molecules); 5, 9.1 × 10−8 fmol (55 molecules) competitor used in the reaction. (D) Agarose gel electrophoresis of PCR products that led to identification of DSΔgap1. Genomic DNA of strains indicated was used in the reactions. Lanes: M, λ-DNA cut with PstI; 1, primers 1 and 3; 2, primers 2 and 3 were used in the reaction. See panel A for primer names. WT, wild-type strain. (E) Southern blot analysis to confirm single, homologous integration of pΔgap1. Genomic DNA of wild-type strain (DSII-1) and strain DSΔgap1 was cut with the enzyme(s) indicated and hybridized to the 3′ and 5′ probe, respectively. Expected fragment sizes can be deduced from panel A.
The expression levels of gap1 were determined in monokaryons, dikaryons, and B-dependent, pheromone-induced semicompatible mating interactions of S. commune using competitive PCR, since the regulators of small G proteins, GAPs and GEFs, are not only activated via interaction with effector molecules, but they can also be regulated at the transcriptional level (26, 34). Only minor changes in gap1 cDNA levels were detected, meaning that gap1 expression does not depend on pheromone response during growth on complete medium (Fig. 3C).
Disruption of gap1 does not affect mating behavior.
In order to gain insight into the function of Ras in S. commune we constructed a Δgap1 disruption strain. The disruption was performed using the genomic sequence of gap1 and replacing the 5′ end (from positions −572 to +1512, with ATG at position 0), including the complete catalytic domain by the selective marker ura1 (Fig. 3A). To obtain high efficiencies in gene replacement, strain DSII-1 with the homologous gap1 and flanking sequences carrying the auxotrophy markers ura1− and trp1− was bred from a cross using strain 4-39. Transformation of S. commune yielded one replacement in 80 uracil prototrophic transformants carrying the homologous replacement which was identified by PCR (Fig. 3D) and confirmed by Southern blot analysis (Fig. 3E). Reverse transcription-PCR could not show any truncated 3′ mRNA which was expected because the promoter had been replaced.
The mutant strain DSΔgap1 was viable and showed normal appearance in all four tetrapolar mating interactions with wild-type strains. Heterozygous dikaryons resulting from fully compatible interactions developed fruitbodies that produced viable spores. Progeny of these crosses (DSΔgap1F1 to DSΔgap1F33) showed the expected segregation of the Δgap1 mutation with 50% as determined by PCR (data not shown). The Δgap1 mutation segregated independently of the mat loci, so that Δgap1 mutant strains with four different mating types could be isolated. Crosses between the mutant strains showed the expected barrage reactions in A≠ B= interactions, the flat phenotype for A= B≠ interactions and formation of dikaryons in compatible A≠ B≠ matings. Therefore, we conclude that the ability to mate is not affected in Δgap1 mutant strains.
Δgap1 mutants exhibit a reduced growth rate that can be reversed by reintroduction of the wild-type gap1 gene.
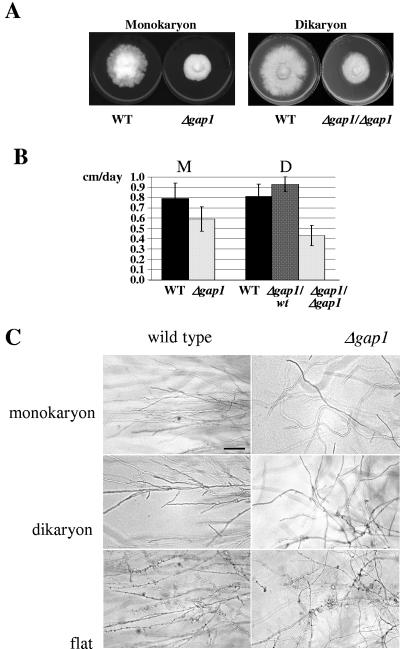
Growth rates of wild-type and Δgap1 mutant strains were determined by measuring the increase in colony diameter per day on complete medium. Whereas monokaryons of wild-type strains grew with 0.79 ± 0.15 cm/day, the Δgap1 mutant strains showed 0.59 ± 0.12 cm growth per day, which represents a reduction in growth rate by 25% (Fig. 4A and B). The reduction in growth rate was even more severe in dikaryons homozygous for Δgap1. The growth rate of wild-type dikaryons of 0.81 ± 0.12 cm/day was reduced by 47% in dikaryons homozygous for Δgap1 (0.43 ± 0.10 cm/day) (Fig. 4B). A slight elevation in growth rate was observed in dikaryons heterozygous for Δgap1 (growth rate = 0.93 ± 0.07 cm/day) (Fig. 4B). This heterosis effect might be depending on the limited amount of Ras sequestering Gap1 present in the heterokaryons. All differences are statistically significant at a level of 95% (P ≤ 0.001) as determined by the Student t test.
FIG. 4.
Growth rates and growth pattern of wild-type and Δgap1 strains. (A) Reduced growth rates were observed in monokaryotic and dikaryotic homoallelic Δgap1 strains. Strains were grown for 7 days (monokaryons) or 9 days (dikaryons) on CYMT agar at 30°C. (B) Growth rates of wild-type (n = 27 for monokaryons; n = 31 for dikaryons) and Δgap1 (n = 23 for monokaryons; n = 19 for heteroallelic dikaryons; n = 28 for homoallelic dikaryons) strains were determined by measuring the increase in colony diameter per day for 5 days. (C) Dikaryons and common A heterokaryons (flat) were homoallelic at the gap1 locus. Strains were grown on solid CYMT medium at 30°C. Bar, 100 μm.
The initial mutant strain DSΔgap1 was complemented by transformation with plasmid pgaptrp carrying the complete gap1 gene. Whereas 16 of 27 transformants with pgaptrp exhibited the wild-type phenotype (see below), none of the 20 transformants carrying only the vector control pSKtrp did. Comparison of growth rates of the 16 complemented strains with the 20 strains with vector only revealed the same reduction in growth rate of the mutant strains by 25% (0.72 ± 0.08 cm/day versus 0.54 ± 0.18 cm/day; P ≤ 0.001). Comparison of the complemented strains mated with compatible wild-type strains (growth rate = 0.72 ± 0.11 cm/day) to the noncomplemented control strains mated to compatible Δgap1 mutant strains (0.33 ± 0.07 cm/day) (P ≤ 0.001) led to a reduction of growth rate by 54%.
PCR of genomic DNA of eight strains transformed with pgaptrp each of the complemented and noncomplemented strains led to amplification of the complete gap1 ORF in the strains showing complementation but not in the noncomplemented strains (not shown), indicating that the failed complementation in these strains is due to only partial or total missing integration of the gap1 gene.
Δgap1 mutants are unable to maintain growth orientation and display altered clamp connections.
In addition to lower growth rate, Δgap1 mutant strains differed also in growth pattern of hyphae on solid media. While hyphae of wild-type strains grew straight keeping their growth axis, hyphae of Δgap1 mutants grew in a curved manner, changing their orientation during growth. This effect was observed during monokaryotic, homozygous Δgap1 dikaryotic, and homozygous Δgap1flat growth (Fig. 4C). All 16 complemented strains showed the wild type-like growth pattern, whereas the transformants containing vector pSKtrp only retained their disorientated growth.
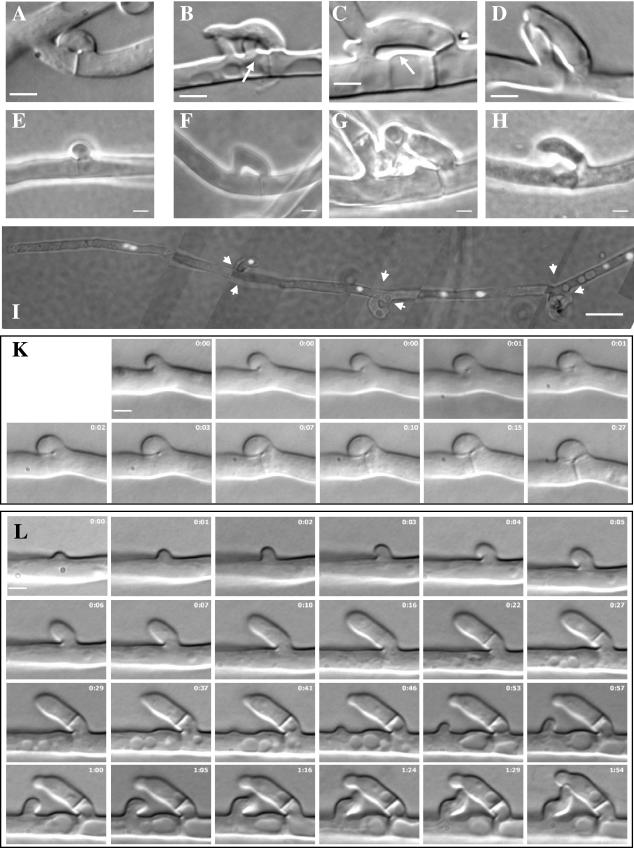
An even more severe effect was observed at clamp connections of dikaryons homozygous for Δgap1. In wild-type dikaryons, clamp connections are compact, the fusion point between hook and subapical cell being directly behind the newly synthesized septum with a distance of 0.7 ± 0.2 μm (n = 100) (Fig. 5A). In contrast, in Δgap1/Δgap1 dikaryons markedly changed, and variable structures were seen (Fig. 5B to D). The distance of the fusion points from the newly synthesized septum varied significantly with 3.0 ± 1.6 μm (n = 100). Most of the hook cells were delayed in fusion with the subapical cell. From the subapical cell grew a branch in a short distance from the peg (Fig. 5B, C, and E to H) that marked the normal point of hook cell fusion. This branch grew toward the elongating hook cell, and the two structures fused at variable points (Fig. 5B to D). After fusion, either the branch from the subapical cell or the tip of the elongated hook continued to grow in opposite direction to the nascent clamp-like structure in an outgrowing branch as opposed to true clamps, which rarely develop an outgrowing branch. As a consequence, more than 93% of the clamp-like structures in the mutant dikaryon showed a branch developing from them, whereas in wild-type dikaryons most of the clamp connections were unbranched (Table 1).
FIG.5.
Clamp connections, clamp-like structures, and pegs in wild-type and Δgap1/Δgap1 dikaryons and nuclear distribution in Δgap1/Δgap1 hyphae. Whereas in the wild type (A) the hook cell fused directly behind the newly synthesized septum, the distances between the septum and fusion site varied in mutant Δgap1/Δgap1 dikaryons (B to D). Arrows mark a peg or weakened cell wall. Pegs could be observed during clamp formation in wild-type (E) and Δgap1/Δgap1 (F to H) dikaryons. The dikaryotic character of Δgap1/Δgap1 hyphae was maintained despite the abnormal mode of clamp-like structure formation (I). The image is composed of seven single images, each representing an overlay of a differential interference contrast and a fluorescence image. Nuclei were stained with DAPI. Arrow heads indicate septae. (K) Time-lapse micrographs of dikaryons during clamp formation in the wild type show the growth of the hook cell continuously directed toward the mother cell (0 to 7 min). The ring visible at the fusion site between the hook and the mother cell (1 to 15 min) indicates that a peg is formed from the mother cell. The ring disappears when the hook and mother cell fuse (27 min). (L) In the Δgap1/Δgap1 dikaryon, the whole process is retarded. The hyphal extrusion representing the future hook initially also grows in the direction of the mother cell (0 to 4 min). Then, it changes its growth direction and grows away from the mother cell (5 to 16 min) and changes its growth direction again a second time (visible at 37 min). A swelling of the cell wall of the mother cell beside the hook indicates the localized weakening of the cell wall at the site of peg formation, where fusion originally was supposed to occur (27 min to 1 h 54 min). Close to this site, a branch develops from the mother cell (41 min) and grows toward the hook cell (41 min to 1 h 54 min) to rescue the failed clamp connection formation. The time is given in hours and minutes. Bars: 2 μm for images A to H, K, and L; 10 μm for image I.
TABLE 1.
Number of branches developing from clamp connectionsa
| Strain | No. (%) branchesb
|
|||||
|---|---|---|---|---|---|---|
| Tip/2nd cell
|
2nd/3rd cell
|
Older clamps
|
||||
| No branches | One branch | No branches | One branch | No branches | One branch | |
| Wild type | 96 (99) | 1 (1) | 96 (96) | 4 (4) | 200 (92) | 17 (8) |
| Δgap1/Δgap1 mutant | 7 (15) | 40 (85) | 6 (6) | 90 (94) | 10 (7) | 141 (93) |
Clamp connections were subdivided between the first clamp from the hyphal tip (tip/2nd cell), the next clamp (2nd/3rd cells), and other clamps on older parts of the hyphae (older clamps).
The percentages of branches formed near clamp connections were calculated from the total numbers counted for each septum type. The position of the clamp connection is indicated in the column subheadings.
The mode of clamp formation during mitosis in wild-type dikaryons leads to temporarily uninuclear subapical cells and hook cells prior to clamp fusion. Since the hook cell fusion was disturbed in Δgap1/Δgap1 dikaryons, DAPI staining of the nuclei was performed to reveal whether the delay in hook cell fusion disturbed the nuclear distribution of the hyphae. In spite of the special mode of clamp connection formation, hyphal cells were mainly dikaryotic (Fig. 5I and Table 2). However, the amount of tip cells containing three nuclei was slightly increased in the mutant compared to the wild type (15% versus 3%), as well as the number of temporarily uninucleate cells (Table 2). The latter resulted from the prolonged time needed in mutant dikaryons for hook cell fusion (Table 3).
TABLE 2.
Number of nuclei per cell
| Strain | No. of nuclei per cell (%)a
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tip
|
Penultimate
|
Other
|
||||||||||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 0 | 1 | 2 | 3 | 4 | 5 | 0 | 1 | 2 | 3 | 4 | 5 | |
| Wild type | 0 | 0 | 92 (92) | 3 (3) | 4 (4) | 1 (1) | 0 | 33 (33) | 65 (65) | 2 (2) | 0 | 0 | 1 (1) | 9 (5) | 157 (93) | 1 (1) | 1 (1) | 0 |
| Δgap1/Δgap1 mutant | 0 | 0 | 81 (77) | 16 (15) | 7 (7) | 1 (1) | 5 (5) | 52 (50) | 41 (39) | 4 (4) | 2 (2) | 0 | 1 (1) | 17 (18) | 71 (75) | 5 (5) | 1 (1) | 0 |
The percentages of the nuclei per cell were calculated from the total numbers of cells counted. Differences to 100% are due to rounding. The numbers of nuclei are indicated in the column subheadings.
TABLE 3.
Number of uncompleted clamp connections
| Strain | No. (%) of uncompleted clamp connectionsa
|
|||||
|---|---|---|---|---|---|---|
| Tip/2nd cell
|
2nd/3rd cell
|
Older clamps
|
||||
| NC | CC | NC | CC | NC | CC | |
| Wild type | 3 (3) | 97 (97) | 0 (0) | 100 (100) | 0 (0) | 217 (100) |
| Δgap1/Δgap1 mutant | 59 (56) | 47 (44) | 5 (5) | 96 (95) | 3 (2) | 151 (98) |
The percentages of completed versus uncompleted clamps were calculated from the total numbers of cells counted for each septum type. The position of the clamp connection is indicated in the column subheadings. NC, no clamp; CC, clamp completed.
Time-lapse microscopy shows branch development for rescue of clamp connection formation.
In order to look into more detail at the altered pattern of clamp connection formation, we took time-lapse micrographs of wild-type and Δgap1/Δgap1 dikaryons (Fig. 5K,L). In wild type, an extrusion appeared very rapidly at the site of clamp cell formation. This swelling grew rapidly backward, obtaining gradually the shape of a hook (Fig. 5K, 0 min). The polarized growth of the hook toward the subapical cell was observed throughout the formation of the clamp connection. At the site where the hook and the subapical cell met, a ring became visible (Fig. 5K, 1 to 15 min). This ring at the point where the tip of the hook met the hypha implicated that a peg was growing from the subapical toward the hook. A septum was formed in the hypha separating a subapical and tip cell and between the hook and apical cell (Fig. 5K, 7 min and 15 min). Finally, the fusion of the hook with the subapical cell was indicated by the disappearance of the ring (Fig. 5K, 27 min). The entire sequence was completed within 20 to 30 min.
In the Δgap1/Δgap1 dikaryon a hyphal swelling at the site of clamp connection formation became visible and was growing backward into the direction of the future subapical cell (Fig. 5L, 0 to 4 min). However, after 4 min, the hook cell changed its growth direction. It abandoned the growth toward the subapical cell and grew away from the main hypha. Septae were formed separating subapical and tip cell (less visible) and the hook from the main hypha (Fig. 5L, 16 to 29 min). The septum separating the hook from the hypha was not formed exactly at the base of the hook since it was during normal clamp connection formation but higher up. Under the hook, at the site where fusion of clamp and subapical cell would have occurred in the wild type, a peg or swelling of the main hypha could be observed (Fig. 5L, 27 to 37 min). Nevertheless, this peg did not come in contact with the hook and ceased growth. Instead, a little distance from this outgrowth a branch developed and grew toward the elongating “hook” cell (Fig. 5L, 41 min to 1 h 5 min). This site is atypical for the formation of new branches because in wild-type dikaryons side branches near septae occur opposite to clamp cells and not adjacent to them. The branch changed its orientation twice until it finally met the growing “hook.” Thus, it seemed that the alteration in clamp connection formation was not due to a failure in initiation of hook cell formation but rather to the inability of the hook cell to fuse with the subapical cell. This was caused by the inability of the “hook” to maintain its growth direction toward the subapical cell. The missing fusion of the hook with the main hypha resulted in continued growth of the hook cell like a normal side branch. The lack of hook cell fusion was then rescued by a branch growing out from the subapical cell toward the hook besides it and fusion of both uninucleate cells to restore the dikaryotic status.
Δgap1/Δgap1 dikaryons form increased numbers of fruitbody primordia but fail to produce spores.
Dikaryons homozygous for wild-type gap1 or the disrupted Δgap1, respectively, were investigated for fruitbody formation and spore production. After 6 weeks of cultivation under fruiting-inducing conditions, the numbers of hyphal knots, primordia, and fruitbodies were counted similar to the developmental stages described by Leonard and Dick (36). At stage I masses of aggregated cells were distinguished macroscopically, and at stage II spherical to cylindrical primordia without a visible pit were detectable. At stage III primordia had an apical pit, whereas stage IV represented fruitbodies with macroscopically visible gills but still with no spore production. Fruitbodies producing spores were counted separately as stage V.
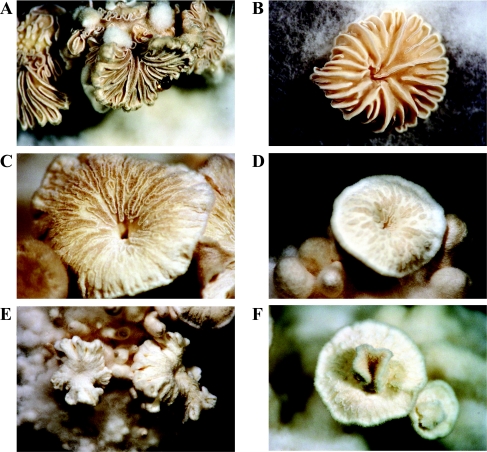
Comparison with wild-type dikaryons grown under the same conditions revealed that the mutant dikaryons formed about four times more hyphal knots and primordia (Table 4). Nevertheless, the amount of fruitbodies per colony (stages IV and V) remained the same, with 0.23 and 0.24 fruitbodies per colony for wild-type and Δgap1/Δgap1 dikaryons, respectively. In sum, in wild-type dikaryons 1 hyphal knot or primordium (stages I + II) out of 17 developed further to stage III, IV, or V, whereas in mutant dikaryons only 1 of 33 showed development to stage III or IV. Gills of fruitbodies from mutant dikaryons were less clearly developed and sometimes totally missing (Fig. 6). They appeared fluffy, indicating the presence of undifferentiated aerial hyphae. In rare cases, a second fruitbody occurred in the center of the first one (Fig. 6F). All fruitbodies from Δgap1/Δgap1 dikaryons failed to produce spores (Table 4).
TABLE 4.
Development of fruitbodies
| Stage of developmenta | Wild-type dikaryons (323 colonies)
|
Δgap1/Δgap1 dikaryons (376 colonies)
|
||
|---|---|---|---|---|
| Total no. | No./colony | Total no. | No./colony | |
| I+II | 5,045 | 15.62 | 22,158 | 58.9 |
| III | 234 | 0.72 | 578 | 1.54 |
| IV | 34 | 0.10 | 90 | 0.24 |
| V | 40 | 0.12 | 0 | 0.00 |
Stages of fruitbody development: I+II, hyphal knots and primordia without apical pit; III, primordia with porus; IV, fruitbodies without producing spores; V, fruitbodies producing spores.
FIG. 6.
Fruitbodies of wild-type and Δgap1/Δgap1 dikaryons. (A and B) Fruitbodies of wild-type strains exhibited clearly developed gills. (C to F) In the fruitbodies of Δgap1/Δgap1 strains the gills were only weakly (C) or partially (D) developed or were totally absent (E). In rare cases, a second fruitbody started to develop in the center of the first (F). Magnifications: ×0.65 (E), ×1 (A, B, and D), or ×1.6 (C and F).
DISCUSSION
In this study, we isolated the gene gap1 from filamentous homobasidiomycete S. commune encoding a GTPase-activating protein. The deduced amino acid sequence revealed a central RasGAP domain and similarities to other fungal Ras GAPs. Since there is no sequence relation between GAPs specific for other members of the Ras superfamily (85), we assume that Gap1 is a GAP acting specifically on Ras proteins. Ras proteins, as well as the functional domains of GAPs and GEFs, are highly conserved among eukaryotes, implicating a similar mechanism of interaction. For the GAPs Ira1p and Ira2p of Saccharomyces cerevisiae sequence and functional homology to their mammalian counterparts have been shown (50, 69, 71, 72, 86). The phenotypes observed after deletion of either IRA1 or IRA2 are typical of strains expressing the RAS2Val19 mutation, leading to reduced intrinsic GTPase activity rendering Ras constitutively active (10, 70, 71). In addition, the deletion of either of the IRA genes has been demonstrated to raise the proportion of RAS1 and RAS2 proteins bound to GTP (72). Therefore, we expect that deletion of gap1 in S. commune also leads to enhanced accumulation of Ras in its activated, GTP-bound form, and thereby to activation of Ras signaling.
The gene gap1 seems to be the single member of this type of RasGAPs. The presence of a member of one or more of the other families of RasGAPs is possible and phenotypes described for deletion of gap1 thus may overlap but not necessarily be identical to the phenotypes observed for constitutive Ras alleles expressed in S. commune.
gap1 deletion confers pleiotropic defects.
Since Ras is a component of different signaling pathways involved in the regulation of a variety of complex cellular processes, we expected pleiotropic effects after mutation of the gap1 gene. Indeed, phenotypes concerning different cellular processes such as growth rate, hyphal growth orientation, clamp-like structure formation, fruitbody development, and sporulation were observed.
However, during the initial events of mating in compatible and semicompatible interactions of Δgap1 strains of S. commune no differences to wild-type strains could be observed. Whereas Ras is not involved in the pheromone response pathway in S. cerevisiae, it is in fission yeast S. pombe, in the basidiomycetous yeast Cryptococcus neoformans and in the heterobasidiomycete Ustilago maydis. In C. neoformans, strains carrying a deletion of ras1 are unable to mate under nutrient starvation due to their failure to produce mating filaments in response to a compatible mating partner. They also fail to induce mating filament formation in the compatible wild-type partner (1, 79). In U. maydis, ras2 mutants fail to mate with a compatible mutant strain but show a reduced reaction when mated with a wild-type strain (35). The defects in mating behavior as described above were seen in strains exhibiting a reduction in Ras signaling, whereas we characterized strains exhibiting an activated Ras signaling pathway. In C. neoformans, strains carrying the dominant active RAS1Q67L allele are able to form filaments and undergo haploid fruiting in response to nutrient starvation without a mating partner (1). Formation of fruitbodies could also be observed in haploid Δgap1 strains of S. commune, but since the related wild-type strains also showed haploid fruiting, this phenotype could not be related to Ras signaling (data not shown).
The reduction in growth rate observed in Δgap1 strains is a phenotype that might be due to pleiotropic effects of the gap1 mutation. In S. cerevisiae Ras signaling causes an increase of intracellular cAMP levels, and cAMP is known to regulate cell proliferation and carbon metabolism (74, 84). However, since cAMP is not essential in U. maydis (21), the mode of action of Ras to regulate growth rate is rather speculative. Interestingly, the reduction in growth rate was nearly twice as strong in dikaryons homoallelic for Δgap1 compared to monokaryons, suggesting a more severe effect of gap1 deletion concerning growth rate in strains in which the mating type pathways are activated.
Similar to the reduction in growth rate, the failure of spore production might be associated with different effects of gap1 deletion. Hymenium formation is impaired in fruitbodies of mutant dikaryons, and gills are only weakly developed. Thus, prerequisites for basidium formation might be missing. Besides, altered Ras signaling might directly inhibit meiosis. Similar to the Δgap1/Δgap1 dikaryons, diploid S. cerevisiae strains homoallelic for ira1 or ira2 disruptions as well as diploid S. pombe strains homoallelic for the deletion of the GAP gene sar1 show severe sporulation defects (70, 72, 77).
Constant Ras signaling results in the failure of growth axis maintenance.
Altered growth pattern of Δgap1 strains was independent of activation of the A- and B-pathways or of only the B pathway. Mutant hyphae showed a disorientated growth with continuous changes of growth axes. A single change in growth axis is normally observed when a hypha grows toward another in order to form a tip-to-side fusion or when two hyphae grow toward each other to form tip-to-tip fusions. Several models that explain hyphal growth have been proposed, and a computer model based on the Spitzenkörper working as vesicle supply center was able to simulate hyphal meandering and changes of growth axis (6, 58). Experiments with drugs depolymerizing cytoskeletal structures in S. commune have indicated that an intact actin cytoskeleton at hyphal and hook cell tips and intact microtubules extending longitudinally through the hyphae toward the tips are necessary for the maintenance of polarized growth (56, 57, 59, 60). In C. neoformans and S. cerevisiae it has been shown that Ras is involved in the regulation of actin cytoskeleton polarization under mild temperature stress (24, 78). In the latter, the regulation of the actin cytoskeleton by Ras2p involved the stress response pathway that functions independently of the cAMP/protein kinase A pathway (24). In S. pombe, a multifunctional complex comprised of Ras1, the Rho GTPase Cdc42sp and Pak1/Shk1 (a homologue of the Ste20p protein kinase of S. cerevisiae) influences actin distribution and microtubule polymerization (15, 37, 41, 48, 51). In the homobasidiomycete Suillus bovinus Cdc42p is localized at the same sites as actin, at the hyphal tips, and at the sites of cross wall formation (22). For S. commune Cdc42, expression of constitutively active alleles under an inducible promoter could clearly show that Cdc42 is not involved in polar growth of the leading hypha, but rather polar tip growth in side branches was altered (80). The phenotype of enlargements resulting from isotropic growth in side hyphae described for constitutive Cdc42 signaling could not be observed for Δgap1 hyphae. Thus, the changed growth pattern of S. commune Δgap1 hyphae suggests that Ras signaling could be involved in the determination of hyphal growth axis in S. commune by regulating the polarization of the actin and/or microtubule cytoskeleton, while Cdc42 signaling especially influences side branch development and growth pattern.
The failure of clamp formation is rescued by lateral branch development.
A strong phenotype was observed in Δgap1 mutants of S. commune during the process of clamp formation. The failure of the hook to maintain the growth direction toward the subapical cell might result in growth of the hook away from the main hypha. The intended fusion point could often be seen by the peg formed at the subapical cell or by a protrusion beside the hook cell, indicating the localized activity of cell wall lysing enzymes. Therefore, the failure of hook cell fusion is not a result of missing peg formation as it is the case during pseudoclamp formation in heterokaryons with different A but similar B mating type genes (33). Perhaps a reduction in Ras signaling controlled by the B mating type genes, but not achieved in the absence of Gap1 in the mutant hyphae, is required for the continuous curved growth of the hook toward the subapical cell leading to the B regulated fusion of the hook with the subapical cell.
The failed fusion with the subapical cell was rescued by fusion with a side branch developing from the subapical cell near the site where the fusion should have originally occurred. The regular and very fast branch initiation beside the failed clamp connection, followed by the directed growth toward the hook cell, indicates some kind of communication between the hook cell and the subapical cell. Since both cells contain nuclei of different mating types, this communication is presumably mediated by the B factor-encoded pheromone/receptor system. Therefore, the formation of the clamp-like structures in Δgap1/Δgap1 dikaryons can be seen as intramycelial mating that suppresses the missing clamp formation and ensures the maintenance of the dikaryotic state. However, in S. commune hyphal attraction resulting in tip-to-tip or tip-to-side fusions occurs also independent of the mating type factors so that a generalization concerning the involvement of the pheromone/receptor system in hyphal attraction cannot be made thus far (for discussion, see reference 54). Remarkably, in many cases the two cells forming the clamp-like structures seemed to grow past each other, resulting not in tip-to-tip but rather in side-to-side fusions (peg-to-peg fusions, see reference 11). However, staining of nuclei revealed dikaryotic cells showing that cell fusion had occurred. Thus, dikaryotization is seen in gap1 deletion strains while dikaryotization was hampered in the transformants expressing constitutive cdc42 alleles (80) showing that different, if overlapping signaling cascades are triggered from Ras- and Rho-dependent cascades.
Deletion of gap1 affects fruiting.
Here, we could show that under fruiting-inducing conditions Δgap1/Δgap1 dikaryons produced elevated numbers of hyphal knots and primordia. Whereas they formed approximately four times more hyphal knots and primordia compared to dikaryons of the wild type, the number of primordia already showing an apical cavity that ceased development was only doubled and the number of fruitbodies (sum of stages IV and V) remained identical. The fruitbodies formed gills, which were not fully developed. They had only a partial gill structure or gills were totally absent. Spore production was never observed.
The phenotypes described here with increased numbers of primordia and partly developed septae resemble those described earlier by Schwalb (65) and Kinoshita et al. (30). They had added extracellular cAMP to dikaryons of S. commune, or caffeine, an inhibitor of cAMP degrading phosphodiesterase, both supplementations leading to raised intracellular cAMP levels. Since deletion of gap1 leads to activation of Ras signaling, it might be proposed that Ras regulates intracellular cAMP levels in S. commune. This would be similar to S. cerevisiae, where it has been shown that Ras activates adenylate cyclase (75). Interaction of Ras with cAMP signaling pathways has also been shown in C. neoformans and U. maydis (1, 35, 45). However, differences in cAMP levels between the Δgap1 mutant and the wild type were not observed in either monokaryons or dikaryons (data not shown). In accordance with this, Yamagishi et al. (87) did not detect changes in cAMP level in strains expressing a dominant-active RasQ61L allele. Yamagishi et al. could also show that transformation with a dominant-active Ras allele rendered the resulting transformants slow growing in monokaryons. They did not observe growth reduction in dikaryons and, in contrast to our results, observed fewer instead of higher amounts of fruitbody initials (87). A possible explanation for these obvious and unexpected discrepancies in phenotypes could be that expression of RasQ61L in the transformants was under control of a strong and developmentally regulated promoter of the aerial mycelium-specific hydrophobin Sc3 (3, 64). In addition, homology-dependent gene silencing in S. commune has been shown for strains expressing extra copies of the sc3 gene where gene silencing occurred in almost 90% of the transformants (63). The silencing was reversed when mycelia were stored at 4°C but occurred again after a few days of growth. Since the silencing was shown to act at the transcriptional level, it would also explain the striking differences in rasQ61L gene expression between monokaryotic and dikaryotic strains (87). Thus, the overlapping but not identical results from disruption of gap1 and introduction of a constitutive allele of ras are explainable.
A second possibility would be that the Ras/Gap1 interaction described here is not the same that has been studied by Yamagishi et al. (87). Either a different class of Gap proteins (see Fig. 2) performs the function described there, or a second, as-yet-unidentified Ras protein is under control of Gap1 in S. commune. Two ras genes have been described for C. neoformans and U. maydis. In C. neoformans one Ras protein, Ras1, seems to be the predominant one since no defect could be observed in strains lacking ras2 (78). In U. maydis, activation of the cAMP-protein kinase A pathway via Ras1 and of a mitogen-activated protein kinase cascade via Ras2 was demonstrated (35, 45).
The investigation of signaling in the sexual development of filamentous basidiomycetes reveals a striking fine tuning that relies on the interplay of many factors. For S. commune the first investigations of gap1, ras1 (87), and cdc42 (80) have already shown pleiotropic effects with somewhat overlapping, but nevertheless distinct phenotypes.
Acknowledgments
We thank P. Mitscherlich for technical assistance and J. Wendland for sharing techniqual equipment. We also thank M. Uuskallio (University of Helsinki) for isolating the full-length cDNA clone of S. commune.
This study was supported by the Deutsche Forschungsgemeinschaft and by a grant from the Academy of Finland to M.R. D.S. acknowledges support from TMWFK, Thuringia (graduate fellowship).
REFERENCES
- 1.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating, and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352-365. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ásgeirsdóttir, S. A., M. A. van Wetter, and J. G. H. Wessels. 1995. Differential expression of genes under control of the mating-type genes in the secondary mycelium of Schizophyllum commune. Microbiology 141:1281-1288. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Badalyan, S. M., E. Polak, R. Hermann, M. Aebi, and U. Kües. 2004. Role of peg formation in clamp cell fusion of homobasidiomycete fungi. J. Basic Microbiol. 44:167-177. [DOI] [PubMed] [Google Scholar]
- 6.Bartnicki-García, S. 2002. Hyphal tip growth: outstanding questions, p. 29-58. In H. D. Osiewacz (ed.), Molecular biology of fungal development. Marcel Dekker, New York, N.Y.
- 7.Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. [DOI] [PubMed] [Google Scholar]
- 8.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 348:125-132. [DOI] [PubMed] [Google Scholar]
- 9.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 10.Broek, D., N. Samily, O. Fasano, A. Fujiyama, F. Tamanoi, J. Northup, and M. Wigler. 1985. Differential activation of yeast adenylate cyclase by wild-type and mutant ras proteins. Cell 41:763-769. [DOI] [PubMed] [Google Scholar]
- 11.Buller, A. H. R. 1933. Researches on fungi. V. Hyphal fusions and protoplasmic streaming in the higher fungi, together with an account of the production and liberation of spores in Sporobolomyces, Tilletia, and Sphaerobolus. Hafner Publishing Co., New York, N.Y.
- 12.Cenis, J. L. 1992. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20:2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 15.Fukui, Y., and M. Yamamoto. 1988. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1−. Mol. Gen. Genet. 215:26-31. [DOI] [PubMed] [Google Scholar]
- 16.Fukui, Y., T. Kozasa, Y. Kaziro, T. Takeda, and M. Yamamoto. 1986. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell 44:329-336. [DOI] [PubMed] [Google Scholar]
- 17.Gancedo, J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:107-123. [DOI] [PubMed] [Google Scholar]
- 18.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast Saccharomyces cerevisiae lead to filamentous growth regulation by starvation and RAS. Cell 68:1077-1090. [DOI] [PubMed] [Google Scholar]
- 19.Gola, S., and E. Kothe. 2003. An expression system for the functional analysis of pheromone genes in the tetrapolar basidiomycete Schizophyllum commune. J. Basic Microbiol. 43:104-112. [DOI] [PubMed] [Google Scholar]
- 20.Gola, S., and E. Kothe. 2003. The little difference: in vivo analysis of pheromone discrimination in Schizophyllum commune. Curr. Genet. 42:276-283. [DOI] [PubMed] [Google Scholar]
- 21.Gold, S., G. Duncan, K. Barrett, and J. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8:2805-2816. [DOI] [PubMed] [Google Scholar]
- 22.Gorfer, M., M. T. Tarkka, M. Hanif, A. G. Pardo, E. Laitiainen, and M. Raudaskoski. 2001. Characterization of small GTPases Cdc42 and Rac and the relationship between Cdc42 and actin cytoskeleton in vegetative and ectomycorrhizal hyphae of Suillus bovinus. Mol. Plant-Microbe Interact. 14:135-144. [DOI] [PubMed] [Google Scholar]
- 23.Görner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, J., and A. Bretscher. 2001. Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell 12:1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori, K., S. Kajiwara, T. Saito, H. Miyazawa, Y. Katayose, and K. Shishido. 1991. Cloning, sequence analysis, and transcriptional expression of a ras gene of the edible basidiomycete Lentinus edodes. Gene 105:91-96. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, D. A., N. Yabana, and M. Yamamoto. 1994. Transcriptional regulation of a Ras nucleotide-exchange factor gene by extracellular signals in fission yeast. J. Cell Sci. 107:3635-3642. [DOI] [PubMed] [Google Scholar]
- 27.Ishibashi, O., and K. Shishido. 1993. Nucleotide sequence of a ras gene from the basidiomycete Coprinus cinereus. Gene 125:233-234. [DOI] [PubMed] [Google Scholar]
- 28.Iwasa, M., S. Tanabe, and T. Kamada. 1998. The two nuclei in the dikaryon of the homobasidiomycete Coprinus cinereus change position after each conjugate division. Fungal Genet. Biol. 23:110-116. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, Y., C. Davis, and J. R. Broach. 1998. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 17:6942-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinoshita, H., K. Sen, H. Iwama, P. P. Samadder, S. Kurosawa, and H. Shibai. 2002. Effects of indole and caffeine on cAMP in the ind1 and cfn1 mutant strains of Schizophyllum commune during sexual development. FEMS Microbiol. Lett. 206:247-251. [DOI] [PubMed] [Google Scholar]
- 31.Kniep, H. 1915. Beiträge zur Kenntnis der Hymenomyceten. III. Über die konjugierten Teilungen und die phylogenetische Bedeutung der Schnallenbildungen. Zeitschr. Botanik. 7:369-398. [Google Scholar]
- 32.Kothe, E. 1997. Solving a puzzle piece by piece: sexual development in the basidiomycetous fungus Schizophyllum commune. Bot. Acta 110:208-213. [Google Scholar]
- 33.Kües, U., P. J. Walser, M. J. Klaus, and M. Aebi. 2002. Influence of activated A and B mating-type pathways on developmental processes in the basidiomycete Coprinus cinereus. Mol. Genet. Genomics 268:262-271. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S., R. Escalante, and R. Firtel. 1997. A Ras GAP is essential for cytokinesis and spatial patterning in Dictyostelium. Dev. 124:983-996. [DOI] [PubMed] [Google Scholar]
- 35.Lee, N., and J. W. Kronstad. 2002. ras2 controls morphogenesis, pheromone response and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 1:954-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard, T. J., and S. Dick. 1968. Chemical induction of haploid fruiting bodies in Schizophyllum commune. Proc. Natl. Acad. Sci. USA 59:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, Y., C. Chen, and E. Chang. 2000. Fission yeast Ras1 effector Scd1 interacts with the spindle and affects its proper formation. Genetics 156:995-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupas, A., M. van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 39.Magae, Y., C. P. Novotny, and R. C. Ullrich. 1995. Interaction of the Aα Y and Z mating type homeodomain proteins of Schizophyllum commune detected by the two hybrid system. Biochem. Biophys. Res. Commun. 211:1071-1076. [DOI] [PubMed] [Google Scholar]
- 40.Marchler, G., C. Schüller, G. Adam, and H. Ruis. 1993. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcus, S., A. Polverino, E. Chang, D. Robbins, M. H. Cobb, and M. H. Wigler. 1995. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 92:6180-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morishita, T., H. Mitsuzawa, M. Nakafuku, S. Nakamura, S. Hattori, and Y. Anraku. 1995. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science 270:1213-1215. [DOI] [PubMed] [Google Scholar]
- 43.Mösch, H., E. Kübler, S. Krappmann, G. R. Fink, and G. H. Braus. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell 10:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mösch, H., R. L. Roberts, and G. R. Fink. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5352-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller, P., J. D. Katzenberger, G. Loubradou, and R. Kahmann. 2003. Guanyl nucleotide exchange factor Sql2 and Ras2 regulate filamentous growth in Ustilago maydis. Eukaryot. Cell 2:609-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Rivas, A., C. A. Specht, R. C. Ullrich, and C. P. Novotny. 1986. Isolation of the DNA sequence coding indole-3-glycerol phosphate synthetase and phosphoribosylanthranilate isomerase of Schizophyllum commune. Curr. Genet. 10:909-913. [DOI] [PubMed] [Google Scholar]
- 47.Niederpruem, D. J., and J. G. H. Wessels. 1969. Cytodifferentiation and morphogenesis in Schizophyllum commune. Bacteriol. Rev. 33:505-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ottilie, S., P. J. Miller, D. L. Johnson, C. L. Creasy, M. A. Sells, S. Bagrodia, S. L. Forsburg, and J. Chernoff. 1995. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 14:5908-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papazian, H. P. 1950. Physiology of the incompatibility factors in Schizophyllum commune. Bot. Gazet. 112:143-163. [Google Scholar]
- 50.Parrini, M. C., E. Jacquet, A. Bernardi, M. Jacquet, and A. Parmeggiani. 1995. Properties and regulation of the catalytic domain of Ira2p, a Saccharomyces cerevisiae GTPase-activating protein of Ras2p. Biochemistry 34:13776-13783. [DOI] [PubMed] [Google Scholar]
- 51.Qyang, Y., P. Yang, H. Du, H. Lai, H. Kim, and S. Marcus. 2002. The p21-activated kinase, Shk1, is required for proper regulation of microtubule dynamics in the fission yeast, Schizosaccharomyces pombe. Mol. Microbiol. 44:325-334. [DOI] [PubMed] [Google Scholar]
- 52.Raper, J. R., and C. A. Raper. 1968. Genetic regulation of sexual morphogenesis in Schizophyllum commune. J. Elisha Mitchell Sci. Soc. 84:267-273. [Google Scholar]
- 53.Raper, J. R., and R. M. Hoffman. 1974. Schizophyllum commune, p. 597-626. In R. C. King (ed.), Handbook of genetics, vol. I. Plenum Press, Inc., New York, N.Y. [Google Scholar]
- 54.Raudaskoski, M. 1998. The relationship between B-mating-type genes and nuclear migration in Schizophyllum commune. Fungal Genet. Biol. 24:207-227. [DOI] [PubMed] [Google Scholar]
- 55.Raudaskoski, M., A. G. Pardo, M. T. Tarkka, M. Gorfer, M. Hanif, and E. Laitiainen. 2001. Small GTPases, cytoskeleton, and signal transduction in filamentous homobasidiomycetes, p. 123-136. In A. Geitman, M. Cresti, and I. B. Heath (ed.), Cell biology of plant and fungal tip growth. IOS Press, Amsterdam, The Netherlands.
- 56.Raudaskoski, M., I. Rupes, and S. Timonen. 1991. Immunofluorescence microscopy of the cytoskeleton in filamentous fungi after quick-freezing and low-temperature fixation. Exp. Mycol. 15:167-173. [Google Scholar]
- 57.Raudaskoski, M., W. Mao, and T. Yli-Mattila. 1994. Microtubule cytoskeleton in hyphal growth: response to nocodazole in a sensitive and a tolerant strain of the homobasidiomycete Schizophyllum commune. Eur. J. Cell Biol. 64:131-141. [PubMed] [Google Scholar]
- 58.Riquelme, M., C. G. Reynaga-Peña, G. Gierz, and S. Bartnicki-García. 1998. What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24:101-109. [DOI] [PubMed] [Google Scholar]
- 59.Runeberg, P., and M. Raudaskoski. 1986. Cytoskeletal elements in the hyphae of the homobasidiomycete Schizophyllum commune visualized with indirect immunofluorescence and NBD-phallacidin. Eur. J. Cell Biol. 41:25-32. [Google Scholar]
- 60.Rupes̆, I., W. Mao, H. Aström, and M. Raudaskoski. 1995. Effects of nocodazole and brefeldin A on microtubule cytoskeleton and membrane organization in the homobasidiomycete Schizophyllum commune. Protoplasma 185:212-221. [Google Scholar]
- 61.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schubert, D., K. B. Lengeler, and E. Kothe. 2000. Identification of mating-type dependent genes by non-radioactive, arbitrarily primed PCR in Schizophyllum commune. J. Basic Microbiol. 40:65-70. [DOI] [PubMed] [Google Scholar]
- 63.Schuurs, T. A., E. A. M. Schaeffer, and J. G. H. Wessels. 1997. Homology-dependent silencing of the sc3 gene in Schizophyllum commune. Genetics 147:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuurs, T. A., H. J. P. Dalstra, J. M. J. Scheer, and J. G. H. Wessels. 1998. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fungal Genet. Biol. 23:150-161. [DOI] [PubMed] [Google Scholar]
- 65.Schwalb, M. N. 1974. Effect of adenosine 3′,5′-cyclic monophosphate on the morphogenesis of fruit bodies of Schizophyllum commune. Arch. Microbiol. 96:17-20. [DOI] [PubMed] [Google Scholar]
- 66.Specht, C. A., A. Muñoz-Rivas, C. P. Novotny, and R. C. Ullrich. 1988. Transformation of Schizophyllum commune: an analysis of parameters for improving transformation frequencies. Exp. Mycol. 12:357-366. [Google Scholar]
- 67.Stankis, M. M., C. A. Specht, H. Yang, L. Giasson, R. C. Ullrich, and C. P. Novotny. 1992. The Aα mating locus of Schizophyllum commune encodes two dissimilar multiallelic homeodomain proteins. Proc. Natl. Acad. Sci. USA 89:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sundaram, S., S. J. Kim, H. Suzuki, C. J. Mcquattie, S. T. Hiremath, and G. K. Podila. 2001. Isolation and characterization of a symbiosis-regulated ras from the ectomycorrhizal fungus Laccaria bicolor. Mol. Plant-Microbe Interact. 14:618-628. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka, K., B. K. Lin, D. R. Wood, and F. Tamanoi. 1991. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc. Natl. Acad. Sci. USA 88:468-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka, K., K. Matsumoto, and A. Toh-e. 1989. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka, K., M. Nakafuku, F. Tamanoi, Y. Kaziro, K. Matsumoto, and A. Toh-e. 1990. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol. Cell. Biol. 10:4303-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka, K., M. Nakafuku, T. Satoh, M. S. Marshall, J. B. Gibbs, K. Matsumoto, Y. Kaziro, and A. Toh-e. 1990. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell 60:803-807. [DOI] [PubMed] [Google Scholar]
- 73.Thevelein, J. M. 1994. Signal transduction in yeast. Yeast 10:1753-1790. [DOI] [PubMed] [Google Scholar]
- 74.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 75.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J., Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 76.Vaillancourt, L. J., M. Raudaskoski, C. A. Specht, and C. A. Raper. 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Y., M. Boguski, M. Riggs, L. Rodgers, and M. Wigler. 1991. Sar1, a gene from Schizosaccharomyces pombe encoding a protein that regulates ras1. Cell Regul. 2:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 79.Waugh, M. S., M. A. Vallim, J. Heitman, and A. J. Alspaugh. 2003. Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet. Biol. 38:110-121. [DOI] [PubMed] [Google Scholar]
- 80.Weber, M., V. Salo, M. Uuskallio, and M. Raudaskoski. 2005. Ectopic expression of a constitutively active Cdc42 small GTPase alters the morphology of haploid and dikaryotic hyphae in the filamentous homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 42:624-637. [DOI] [PubMed] [Google Scholar]
- 81.Wendland, J., and E. Kothe. 1997. Isolation of tef1 encoding translation elongation factor EF1α from the homobasidiomycete Schizophyllum commune. Mycol. Res. 101:798-802. [Google Scholar]
- 82.Wendland, J., L. J. Vaillancourt, J. Hegner, K. B. Lengeler, K. J. Laddison, C. A. Specht, C. A. Raper, and E. Kothe. 1995. The mating-type locus of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14:5271-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wessels, J. G. H. 1978. Incompatibility factors and the control of biochemical processes, p. 81-104. In M. N. Miles and P. G. Miles (ed.), Genetics and morphogenesis in the basidiomycetes. Academic Press, Inc., New York, N.Y.
- 84.Wittenberg, C., and S. I. Reed. 1996. Plugging it in: signaling circuits and the yeast cell cycle. Curr. Opin. Cell Biol. 8:223-230. [DOI] [PubMed] [Google Scholar]
- 85.Wittinghofer, A., K. Scheffzek, and M. R. Ahmadian. 1997. The interaction of Ras with GTPase-activating proteins. FEBS Lett. 410:63-67. [DOI] [PubMed] [Google Scholar]
- 86.Xu, G., B. Lin, K. Tanaka, D. Dunn, D. Wood, R. Gesteland, R. White, R. Weiss, and F. Tamanoi. 1990. The catalytic domain of the neurofibromatosis type1 gene product stimulates ras GTPase and complements ira mutants of Saccharomyces cerevisiae. Cell 63:835-841. [DOI] [PubMed] [Google Scholar]
- 87.Yamagishi, K., T. Kimura, M. Suzuki, H. Shinmoto, and K. Yamaki. 2004. Elevation of intracellular cAMP levels by dominant active heterotrimeric G protein alpha subunits ScGP-A and ScGP-C in the homobasidiomycete Schizophyllum commune. Biosci. Biotechnol. Biochem. 68:1017-1026. [DOI] [PubMed] [Google Scholar]