Abstract
The ink cap Coprinopsis cinerea is a model organism for studying fruiting body (mushroom) formation in homobasidiomycetes. Mutant screens and expression studies have implicated a number of genes in this developmental process. Functional analysis of these genes, however, is hampered by the lack of reliable reverse genetics tools for C. cinerea. Here, we report the applicability of gene targeting by RNA silencing for this organism. Efficient silencing of both an introduced GFP expression cassette and the endogenous cgl1 and cgl2 isogenes was achieved by expression of homologous hairpin RNAs. In latter case, silencing was the result of a hairpin construct containing solely cgl2 sequences, demonstrating the possibility of simultaneous silencing of whole gene families by a single construct. Expression of the hairpin RNAs reduced the mRNA levels of the target genes by at least 90%, as determined by quantitative real-time PCR. The reduced mRNA levels were accompanied by cytosine methylation of transcribed and nontranscribed DNA at both silencing and target loci in the case of constitutive high-level expression of the hairpin RNA but not in the case of transient expression. These results suggest the presence of both posttranscriptional and transcriptional gene silencing mechanisms in C. cinerea and demonstrate the applicability of targeted gene silencing as a powerful reverse genetics approach in this organism.
The ink cap Coprinopsis cinerea is a model organism for studying fruiting body (mushroom) formation in homobasidiomycetes (reviewed in references 32 and 35). Mutant screens and expression studies have implicated a number of genes in this developmental process. An example for the latter case are the cgl1 and cgl2 isogenes, which code for two isogalectins that are highly induced during fruiting body formation (5). In addition, orthologues of genes involved in fruiting body formation in other fungi are revealed by the sequence of the C.cinerea genome (http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus) and its annotation, which is in progress (http://fungal.genome.duke.edu/cgi-bin/gbrowse/ccin/). Functional analysis of C. cinerea genes, however, is hampered by the lack of reliable tools for gene targeting. Although homologous recombination seems to occur in C. cinerea (3), targeted gene knockouts appear difficult to achieve, possibly due to very efficient nonhomologous DNA end joining, as has been shown for the filamentous ascomycete Neurospora crassa (47).
Recently, RNA-induced gene silencing (RNA silencing) has been emerging as a powerful tool for gene targeting in fungi, plants, and animals (reviewed in references 7, 11, and 13). This strategy exploits an endogenous gene regulatory mechanism of eukaryotic cells in which regulatory double-stranded RNAs (dsRNAs) interfere with homologous mRNA either by triggering its degradation or inhibiting its transcription or translation (see references 1 and 42 for recent reviews and specific references therein). For gene targeting, dsRNA homologous to the target gene is introduced into the organism either directly or indirectly as constructs leading to its endogenous expression. Both natural and introduced dsRNAs are cleaved into short pieces of 21 to 23 bp by a conserved bidentate RNase III-related RNase, Dicer, and can be amplified by RNA-dependent RNA polymerases (RdRP). These small dsRNAs are built in ribonucleoprotein complexes where the respective single strands target the activity (degradation, inhibition of transcription, or translation) of the complex to complementary mRNAs or transcriptionally active DNA regions. The activity of the complex is determined by the type of small dsRNA and the protein composition of the complex. All ribonucleoprotein complexes involved in RNA silencing known thus far appear to contain at least one member of the argonaute protein family, which shows structural homology to RNase H. Interestingly, in plants and worms, gene silencing both by exogenous and endogenous dsRNAs can be systemically transmitted within a multicellular organism, possibly by spreading of the small dsRNA as a silencing signal. In fungi, studies of transgene-induced gene silencing (quelling) in N. crassa were instrumental for the discovery of the mechanism and the genetic dissection of the underlying machinery (see reference 10 for a review). In the meantime, homology-based gene silencing induced by transgenes (cosuppression), antisense, or dsRNA has been demonstrated for many fungi including zygo-, asco-, and basidiomycetes (15, 19, 20, 22, 23, 29, 37, 43, 44, 46, 50, 54, 58, 64). For homobasidiomycetes, there is one report of transgene-induced silencing in Schizophyllum commune (53). Based on these results and the presence of candidate genes coding for orthologs of key components of RNA-mediated gene silencing in the C. cinerea genome (Dicer, argonaute, and RdRP), we tested the applicability of RNA silencing as a tool to target exogenous and endogenous genes in C. cinerea.
MATERIALS AND METHODS
Strains, growth, and transformation conditions.
Escherichia coli strain DH5α was used for cloning and amplification of plasmids. Transformation-competent cells were prepared as described previously (26). Plasmid-containing bacteria were selected at 37°C on Luria broth containing 100 mg of ampicillin/liter. Saccharomyces cerevisiae laboratory strain W303-1A (MATa ura3-1 trp1-1 his3-11,15 leu2-3,112 ade2-1 can1-100) was used for cloning by homologous recombination (see below for details). Transformants generated by the LiOAc method (27) were selected at 30°C on synthetic complete medium without uracil (30). C. cinerea strain AmutBmut (A43mut B43mut pab1.2) (41, 59) and one of its progenies, KK7 (A43mut B43mut trp1.1;1.6 pab1.2) (34), served as recipient of the various constructs. The pab1 allele in these strains was termed pab1.2 to distinguish it from another allele in the literature (12) and harbors a single point mutation (AAG to GAG) resulting in an amino acid substitution, K546E, at a highly conserved residue of the encoded bifunctional C. cinerea p-aminobenzoate (PABA) synthase (28) (C. Villalba and M. Künzler, unpublished results). The transformation of mononucleate asexual C. cinerea spores (oidia) was described previously (21). Selection for transformants was done on minimal medium (MM) supplied with Trp (100 mg/liter) or PABA (5 mg/liter) if necessary. Vegetative mycelium of strain KK7 was grown on cellophane disks placed on complete medium (YMG; supplemented with Trp for untransformed strain KK7) plates for 5 days (triple inocula on 90-mm petri dishes) at 37°C in darkness (in ventilated closed black boxes). Fruiting mycelium and primordia of strain AmutBmut were produced by precultivating vegetative mycelium on cellophane disks on YMG plates for 4 days (triple inocula) at 37°C in darkness and subsequent transferral to 25°C in a 12-h light/dark regime for another 3 days (containing secondary hyphal knots and primordia of up to 2 mm in diameter) and up to 10 days, respectively. The diameter of harvested primordia was 4 to 8 mm. Oidia for transformation and DNA isolation were produced by transferring vegetative mycelium grown on YMG (AmutBmut) or YMG+Trp (KK7) without cellophane disks at 37°C for 3 days in darkness (triple inocula) into constant white light (20 to 25 μE per m2 and s; emission spectrum of 275 to 780 nm) and incubating them at 37°C for another 4 and 5 days, respectively.
Construction of plasmids.
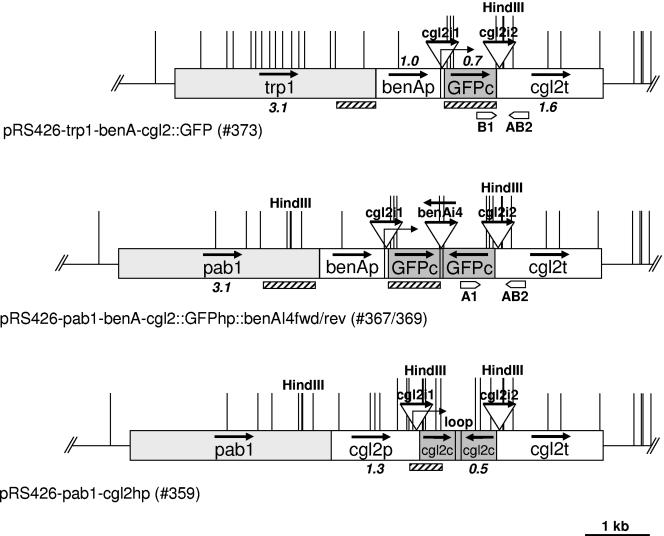
The plasmids used in the present study are listed in Table 1. The GFP and GFP-hairpin (GFPhp) RNA expressing plasmids were constructed in a stepwise procedure involving classical cloning in E. coli and homologous recombination in S. cerevisiae (see below). In a first step, a genomic 3.3-kb KpnI-SacI cgl2 fragment from C. cinerea strain AmutBmut (5) was cloned into the corresponding sites of S. cerevisiae-E. coli shuttle vector pRS426, resulting in plasmid 197 (pRS426-cgl2). In a second step, the cgl2 gene was put under the control of the C. cinerea benA-promoter by recombination of a PCR-generated (primers 426cgl2-BamHI-tub1000-fwd and 426cgl2-tub1000-rev on AmutBmut chromosomal DNA as a template) 1-kb benA promoter fragment into plasmid 197 opened with KpnI and AatII, resulting in plasmid 339 (pRS426-benA-cgl2) (Table 2). The design of the benA primers was based on the available benA sequence (GenBank no. AB007761) and the genome sequence of C. cinerea strain Okayama 7 (http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus). Analogously, the open reading frame (ORF) of cgl2 in plasmid 339 was replaced by the GFP ORF by homologous recombination of a PCR-generated (primers and on pBS-GFP as a template) GFP fragment into BsiWI-opened plasmid 339, resulting in plasmid 341 (pRS426-benA-cgl2::GFP). The GFP silencing constructs were made by using the same strategy by tandem recombination of two overlapping PCR fragments (primers CGL2p-GFP and benAInt4-GFPrev and primers benAInt4-GFPfwd and CGL2-GFPrev, respectively, on plasmid pBS-GFP as a template) into BsiWI-opened plasmid 339, resulting in plasmids 366 (pRS426-benA-cgl2::GFPhp::benAI4fwd) and 368 (pRS426-benA-cgl2::GFPhp::benAI4rev). The final construct codes for a hairpin RNA consisting of a stem formed by an inverted repeat of the complete GFP coding sequence in sense-antisense orientation and a loop formed by the fourth intron of the benA gene in a forward or reverse orientation (Fig. 1). Construction of the cgl2 silencing plasmid 222 (pRS426-cgl2hp) was done by recombination of three overlapping PCR fragments (primers CGL2-RNAi-II5 and CGL2-RNAi-OO3, CGL2-RNAi-O5 and CGL2-RNAi-I3, and CGL2-RNAi-I5 and CGL2-RNAi-O3, respectively, on plasmid 197 as a template) into KspAI-Pfl23II-opened 197. The final construct consists of an inverted repeat of the complete cgl2 coding sequence in sense-antisense orientation separated by a tandem repeat of the 9-bp spacer sequence (TTCAAGAGA) used in Ambion's silencing vectors pSilencer (Ambion, Austin, TX).
TABLE 1.
Plasmids used in this study
| Name | Description | Source or reference |
|---|---|---|
| pRS426 | 2μ-URA3 | 56 |
| pPAB1.2 | pTZ18R-pab1 | 21 |
| pCc1001 | pUC9-trp1 | 4 |
| pBS-GFP | pBSII-KS(+)-GFP | 18 |
| 197 | pRS426-cgl2 | This study |
| 336 | pRS426-pab1 | This study |
| 339 | pRS426-benA-cgl2 | This study |
| 341 | pRS426-benA-cgl2::GFP | This study |
| 354 | pRS426-pab1-benA-cgl2 | This study |
| 366 | pRS426-benA-cgl2::GFPhp::benAl4fwd | This study |
| 368 | pRS426-benA-cgl2::GFPhp::benAl4rev | This study |
| 367 | pRS426-pab1-benA-cgl2::GFPhp::benAl4fwd | This study |
| 369 | pRS426-pab1-benA-cgl2::GFPhp::benAl4rev | This study |
| 373 | pRS426-trp1-benA-cgl2::GFP | This study |
| 359 | pRS426-pab1-cgl2hp | This study |
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence | Purpose |
|---|---|---|
| CGL2p-GFP | GTATCACCAGTCTAACATCATGCTCAAGGGCGAGGAGCTGTTCACC | Cloning of cgl2::GFP fusion |
| CGL2 Stab-STOP-GFP | AGGGGGAAGTGGGGGGAGCAATCCATGGACCTACTTGTACAGCTCGTCCATGCCG | Cloning of cgl2::GFP fusion |
| 426cgl2-BamHI-tub1000-fwd | GCGTAATACGACTCACTATAGGGCGAATTGGGATCCAATAAGGATACACATGATCG | Cloning of benAp |
| 426cgl2-tub1000-rev | GGAAAGTCGAATGCCTACTTGGCTTTGAGCTGGGAACGCGAGGTCAGC | Cloning of benAp |
| Up-pRS426-Cc TRP1fwd | ACGACTCACTATAGGGCGAATCCATGATGATGACGGTAGAC | Cloning of trp1 |
| Down-benA-Cc TRP1rev | GATCATGTGTATCCTTATTAGATCTGAGTCGGTACTTCAAGTTCCC | Cloning of trp1 |
| Up-pRS426-pab1fwd | ACGACTCACTATAGGGCGAATTGCGAAGCAACTGAAGGAGC | Cloning of pab1 |
| Down-benA-pab1rev | GATCATGTGTATCCTTATTGGATCCTTCTTCTGGCATCTTTCCTC | Cloning of pab1 |
| Down-pRS426-pab1rev | GTCGACCTCGAGGGGGGGCCCTTCTTCTGGCATCTTTCCTC | Cloning of pab1 |
| benAlnt4-GFPrev | TAATTTTGGCCAGCACAACGCATGCTTGCGACAGGGATACATACCCTGCAGGCACTTGTACAGCTCGTCCATGC | Cloning of cgl2::GFPhp |
| benAlnt4-GFPfwd | AGCATGCGTTGTGCTGGCCAAAATTAATGATCATCACGTGATAGGCTTCCAGATCTTGTACAGCTCGTCCATGC | Cloning of cgl2::GFPhp |
| CGL2-GFPrev | AAGTGGGGGGAGCAATCCATGGACAAGGGCGAGGAGCTGTTC | Cloning of cgl2::GFPhp |
| 155 (GFP fwd real time) | ACATGGTCCTGCTGGAGTT | Genomic PCR and qRT-PCR of cgl2::GFP |
| 68 (CGL2-Term) | CGAACCGCTCTGAGGGAGG | Genomic PCR of cgl2::GFP and cgl2::GFPhp |
| 39 (GFP-(270)rev) | GCCTTCGGGCATGGCGG | Genomic PCR of cgl2::GFPhp |
| Down-CGL2-pab1rev | TCATTATCACGGTGGAACGGTTGCTGGATCCTTCTTCTGGCATCTTTCCTC | Cloning of pab1 |
| 185 (RT-PCR-rev-cgl2) | CCAGCGAGAATCCTAAGCA | qRT-PCR of cgl2::GFP |
| 186 (RT-PCR-fwd-benA) | GTCATGTCCGGTATCACCAC | qRT-PCR of benA |
| 187 (RT-PCR-rev-benA) | GGGAAAGGAACCATGTTGA | qRT-PCR of benA |
| CGL2-RNAi-II5 | GTTGGTCTCTTTTTGGATTCTTG | Cloning of cgl2hp |
| CGL2-RNAi-OO3 | GTATGAAGTCCGTTGGTGTCGC | Cloning of cgl2hp |
| CGL2-RNAi-O5 | CGGCGCCTGGGGCCCGGAGG | Cloning of cgl2hp |
| CGL2-RNAi-I3 | TCTCTTGAATTCTCTTGAAAGCAGGGGGAAGTGGGGGG | Cloning of cgl2hp |
| CGL2-RNAi-I5 | TTCAAGAGAATTCAAGAGAAGCAGGGGGAAGTGGGGGG | Cloning of cgl2hp |
| CGL2-RNAi-O3 | GCAAGAATCCAAAAAGAGACCAACAAACCTAATGCTCTACCACCTTTTCGTC | Cloning of cgl2hp |
| 61 (GFP) | ATGAGCAAGGGCGAGGAGC | GFP hybridization probe |
| 134 (GFPrev) | CTTGTACAGCTCGTCCATGC | GFP hybridization probe |
| 71 (Cgl2FusSeq2) | AGTGATATCCGGTGGTCAGC | cgl2 hybridization probe |
| 184 (CGL2ORFRev2) | TCAGCGTACGGGATGCGTTC | cgl2 hybridization probe |
| 119 (07pab2006fwd) | TTGTGGCGTTGAAGAGTACG | pab1 hybridization probe |
| 120 (07pab2637rev) | CATGGCTGATGCTTAATTGC | pab1 hybridization probe |
| 54 (TRP1F-Forward) | GCCGGTCTCGACTATCCAGGTGTAGG | trp1 hybridization probe |
| 38 (TRP1SeqTerm) | GACCCCCTCAAACACTATTGG | trp1 hybridization probe |
| 180 (Cgl3PromFwd) | TCTGCCTCTGCTAGCTTGTC | cgl3 hybridization probe |
| 126 (CGL3rev) | ACGGTTGATTCGAGTCTG | cgl3 hybridization probe |
FIG. 1.
Schematic representation of DNA constructs used in the present study. Promoter (p), terminator (t), and coding (c) regions of indicated genes are represented as boxes, and introns (i) as triangles. Bold arrows indicate the direction of transcription of the respective DNA element at the original locus, and the fine arrows indicate the transcriptional start sites of the constructs. The length of the longer DNA fragments is given in kilobases. Hatched boxes and block arrows below the boxes indicate hybridization probes used for Southern blot analyses and primers used for genomic PCR analyses, respectively. Restriction sites used for Southern blot analyses are indicated by thick (HindIII) or thin lines (HpaII/MspI).
Recombination was also used to supply plasmids pRS426, 222, 339, and 341 with C. cinerea marker genes. Plasmids were opened either with KpnI (pRS426 and plasmid 222) or with BamHI (plasmids 339 and 341) and recombined with PCR-generated pab1 (primers Up-pRS426-pab1fwd and Down-pRS426-pab1rev [pRS426], Up-pRS426-pab1fwd and Down-cgl2-pab1rev [plasmid 222], or Up-pRS426-pab1fwd and Down-benA-pab1rev [plasmid 339] on plasmid pPAB1.2 as a template) and trp1 (primers Up-pRS426-CcTRP1fwd and Down-benA-CcTRP1rev on plasmid pCc1001 as a template; for plasmid 341) fragments resulting in plasmids 336 (pRS426-pab1), 359 (pRS426-pab1-cgl2hp), 354 (pRS426-pab1-benA-cgl2), and 373 (pRS426-trp1-benA-cgl2::GFP). The final GFP silencing constructs were yielded by recombination of EcoRI-BamHI benA-cgl2::GFPhp::benAI4fwd and benA-cgl2::GFPhp::benAI4rev from plasmids 366 and 368, respectively, into BstXI-opened plasmid 354. All PCR-generated constructs were verified by DNA sequencing (Microsynth, Balgach, Switzerland). Plasmid DNA from E. coli was isolated by using a one-tube protocol as described previously (14).
Cloning by homologous recombination in S. cerevisiae.
Routinely, fragments with ends of 25 to 30 nucleotides (nt) homologous to regions on the recipient vector were generated by PCR and cotransformed into strain W303-1A, together with minor amounts of the recipient vector linearized in between the two recombining regions. Plasmids from single yeast transformants were rescued and amplified in E. coli as described previously (51). The plasmids were analyzed by restriction enzyme digestion, diagnostic PCR, and DNA sequencing.
PCR.
PCRs using Taq polymerase for diagnostic purposes or Pfu polymerase for preparative purposes were performed on a Robocycler (Stratagene, La Jolla, CA) according to standard protocols (52).
Southern blot analysis.
C. cinerea genomic DNA from oidia was isolated as described previously (63). Primordia were frozen in liquid nitrogen together with an equal volume of glass beads (400 to 600 μm in diameter) and ground by applying one pulse of 40 s at level 6 in a Fast-Prep machine (Bio 101 Savant; Savant Instruments, Inc., Holbrook, NY) before DNA isolation. GFP target and silencing loci were isolated by digesting genomic DNA of the corresponding transformants with HindIII, separating the fragments by agarose gel electrophoresis and isolating fragments of the respective sizes (∼12 and 3 kb, respectively; see Fig. 2B) from the gel. Southern blot analysis of isolated C. cinerea genomic DNA was performed according to standard protocols (52). Detection of specific DNA fragments was done by using the digoxigenin (DIG) system according to the manufacturer's recommendations (Roche Applied Science, Mannheim, Germany). PCR-generated hybridization probes were generated by standard PCR (GFP, primers 61 and 134 on pBS-GFP as a template; cgl2, primers 71 and 184 on plasmid 197; pab1, primers 119 and 120 on plasmid pPAB1.2; trp1, primers 54 and 38 on plasmid pCc1001; and cgl3, primers 180 and 126 on chromosomal AmutBmut DNA) using Taq polymerase and a DIG-dUTP-containing nucleotide mix (Roche). DIG-labeled DNA-Molecular-Weight-Marker VII (Roche) was used as a size standard.
FIG. 2.
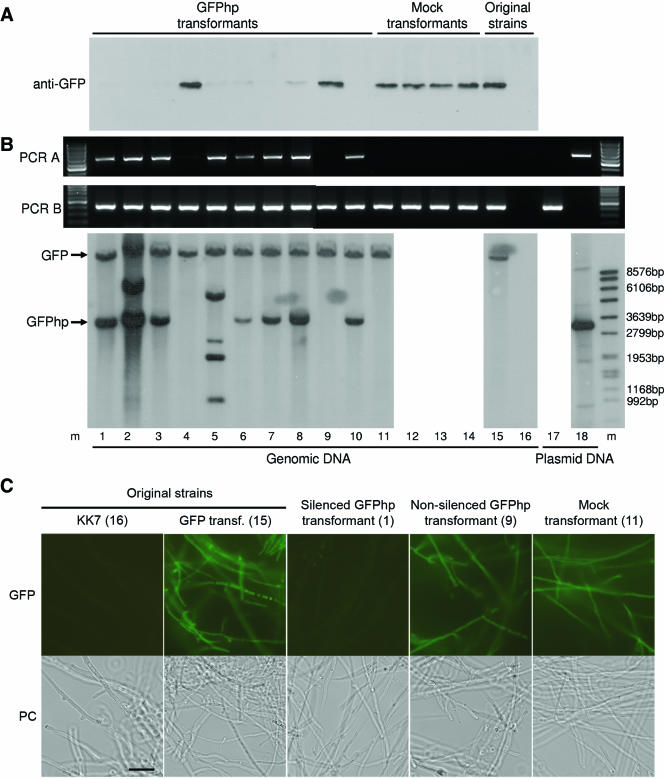
Molecular analysis of GFP silencing in C. cinerea strain KK7. Analysis is shown for the strain without (lane 16) and with the GFP-expression construct 373 (lane 15), as well as for individual transformants of the latter transformant with either a mock plasmid carrying only the marker gene (pPAB1.2; lanes 11 to 14) or the GFP-silencing construct 367 (lanes 1 to 10). (A) Analysis of GFP protein levels in WCEs. A total of 3 μg of WCE prepared from the individual strains or transformants was analyzed by immunoblotting using a specific anti-GFP antiserum (see the text for details). (B) Analysis of DNA integration by genomic PCR and Southern blotting. Genomic DNA was prepared from the individual strains or transformants and used as a template for two PCRs indicative for the integration of either the GFP silencing construct (PCR A, primers A1 and AB2) or the GFP expression construct (PCR B, primers B1 and AB2) in the genome. A 100-bp DNA ladder (Fermentas International, Inc., Burlington, Ontario, Canada) was used as a size standard (m). The bottom panel shows a Southern blot analysis of the same genomic DNAs with HindIII as restriction enzyme and a GFP hybridization probe (see Materials and Methods for details). DNA-Molecular-Weight-Marker VII (Roche) was used as a size standard (m). The positions of the expected GFP and GFPhp DNA fragments are indicated. Plasmids 373 (lane 17) and 367 (lane 18) were included as controls. (C) GFP fluorescence microscopy of silenced and nonsilenced KK7 transformants. A silenced and a nonsilenced transformant in comparison to the appropriate control strains or transformants (the numbering in the brackets refers to panels A and B) were examined by GFP fluorescence and phase-contrast microscopy (PC) as described in Materials and Methods. Bar, 20 μm.
RNA extraction and cDNA synthesis.
Mycelia were harvested, immediately frozen in liquid nitrogen, and stored at −80°C. Samples were lyophilized and total RNA was extracted by using an RNeasy Lipid Tissue Minikit (QIAGEN, Valencia, CA). Lyophilized mycelia (20-25 mg) were homogenized in a Fast-Prep machine (Bio 101 Savant) with three consecutive pulses of 45 s each at levels 4.5, 5.5, and 6.5 using a volume of about 200 μl of acid-washed glass beads (400 to 600 μm in diameter). Then, 1 ml of QIAzol lysis reagent was added to the homogenates, and all further steps were carried out according to the manufacturer's instructions, including an additional on-column DNase digestion using the RNase-Free DNase Set (QIAGEN). RNA purity and integrity were checked by determining the 260/280- and 260/230-nm absorption ratios and by visual inspection after separation of 10 μg of total RNA on a 6.7% formaldehyde-1% agarose gel.
cDNA was synthesized by using M-MLV Reverse Transcriptase RNase H Minus (Promega, Madison, WI) and oligo(dT) (20) primers according to the manufacturer's instructions. A total of 1 μg of RNA per reaction (25 μl) was used, and the obtained cDNA was stored at −20°C until use.
Quantitative real-time PCR analysis.
Real-time PCR was carried out by using a QuantiTect SYBR Green PCR kit (QIAGEN) with 0.9 μM forward and reverse primer concentrations each and a variable amount of cDNA (20 to 0.02 ng per reaction) in a final reaction volume of 20 μl. Thermocycling was performed by using a Rotor-Gene 3000 Real-Time Thermal Cycler (Corbett Research, Sydney, Australia) initiated by a 15-min incubation at 95°C, followed by 50 cycles of 15 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Fluorescence data were acquired during the elongation step in every cycle. Each run was completed with a melting curve analysis to confirm specificity of amplification and absence of primer dimers. The amplification of genomic DNA was prevented by designing primers on exon-exon junctions and by DNase digestion during RNA extraction. These measures proved to be effective since no amplification could be observed in a cDNA control where reverse transcriptase was omitted. benA detected with the primers 186 plus 187 served as a reference gene for the relative quantification of GFP, which was detected with the primers 155 and 185. PCR efficiencies were determined with the serial dilution method of cDNAs. Transcript quantification of individual samples is based on measurements in triplicate and analysis using the mathematical model of Pfaffl (49).
Preparation of C. cinerea whole-cell extracts (WCEs).
Vegetative or fruiting mycelia grown on cellophane disks or primordia were shock frozen in liquid nitrogen and stored at −80°C. For extraction, the material was lyophilized, and 20 μg was ground by adding 200 μl of glass beads (400 to 600 μm in diameter) and applying one pulse of 20 s at level 6 in a Fast-Prep machine. The resulting powder was extracted by adding 200 μl of ice-cold 0.5× phosphate-buffered saline (PBS) containing 1 mM phenylmethylsulfonyl fluoride and applying one pulse of 30 s at level 6 in the Fast-Prep machine, cooling on ice, and repeating the buffer addition and the pulsing. The resulting lysate was centrifuged twice at 15,000 rpm to remove particulate material. The protein concentration of the cleared lysate was determined by the Bradford method (Bio-Rad, Hercules, CA), mixed with an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (250 mM Tris-HCl [pH 6.8], 9.2% SDS, 40% [vol/vol] glycerol, 0.2% [wt/vol] bromophenol blue, 100 mM dithiothreitol), and boiled for 3 min.
Immunoblotting.
Equal amounts (3 to 5 μg) of C. cinerea WCE were separated on a SDS-15% PAGE gel (36) and transferred to a nitrocellulose membrane (Protran; Schleicher & Schuell, Keene, NH) by using a Trans-Blot Semi-Dry Transfer unit (Bio-Rad). Blots were blocked with 5% (wt/vol) dry milk powder in PBS containing 0.1% (vol/vol) Tween 20 (PBST) and hybridized with primary rabbit antiserum and secondary horseradish peroxidase (HRP)-coupled goat anti-rabbit immunoglobulin G (Santa Cruz, Santa Cruz, CA). Polyclonal rabbit antiserum raised against recombinant green fluorescent protein (GFP) was a gift from Walter Nickel (Biochemie-Zentrum, Heidelberg, Germany). Polyclonal rabbit antiserum raised against isogalectins CGL1 and CGL2 purified from C. cinerea has been described elsewhere (5). Polyclonal rabbit antiserum against recombinant CGL3, a C. cinerea lectin with high sequence similarity to CGL1 and CGL2, will be described elsewhere. HRP was detected either by enhanced chemiluminescence (Amersham Biosciences, Uppsala, Sweden) or developed by using chloronaphthol as a chromogen (24).
Microscopy.
C. cinerea strains or transformants were cultivated by inoculating liquid YMG (supplemented with Trp for KK7) with respective oidia stored in 10% (vol/vol) glycerol at −80°C, followed by incubation with vigourous agitation at 37°C until small fungal pellets were visible. The pellets were washed with deionized water, and hyphae were investigated for GFP fluorescence by using a Zeiss Axiophot equipped with a mercury lamp and a Zeiss filter set 09 (excitation BP, 450 to 490 nm; emission LP, 515 nm) (Carl Zeiss AG, Göttingen, Germany). Pictures were captured by using a Zeiss Axiocam MRc and Zeiss Axiovision software (version 4.4).
Nucleotide sequence accession number.
The nucleotide sequence of the lectin-encoding cgl3 gene from C. cinerea strain AmutBmut has been deposited in GenBank under accession number DQ408306.
RESULTS
Construction of a C. cinerea GFP expression cassette based on endogenous control elements.
As a first step toward evaluation of RNA silencing in C. cinerea, we constructed an expression cassette for GFP in C. cinerea based on endogenous control elements. Previous reports suggested that GFP expression in mushrooms was dependent on the presence of introns (6, 38, 39). In the only GFP expression cassette available for C. cinerea to date, the intron is located within the coding region resulting in a GFP fusion protein (6). The coding regions of the C. cinerea cgl1 and cgl2 genes are flanked by two introns, one immediately upstream of the translational start codon and one immediately downstream of the translational stop codon (5). We reasoned that such a gene structure would be the ideal basis for a C. cinerea expression cassette for heterologous coding sequences such as GFP. We cloned the cgl2 gene from C. cinerea strain AmutBmut including 1 kb upstream and 1.6 kb downstream of the coding sequence on a S. cerevisiae-E. coli shuttle vector in order to be able to use yeast recombination in subsequent modifications. Since the cgl1/2 genes are hardly transcribed during vegetative growth (2, 5), we exchanged the cgl2 promoter region with 1 kb of the promoter region of the AmutBmut benA gene coding for β1-tubulin by using yeast recombination. Using the same technique, most of the cgl2 coding region (except for a 36-bp sequence immediately upstream of the translational stop codon) was exchanged with the entire GFP coding region. Finally, the C. cinerea trp1 gene was inserted immediately upstream of the benA promoter region as a selection marker in C. cinerea (Fig. 1; see Materials and Methods for details). To check the construct for GFP expression, we transformed the plasmid into C. cinerea strain KK7. Trp-prototrophic transformants were analyzed for GFP expression by immunoblotting of WCE from vegetative mycelium (data not shown and Fig. 2A, lane 15), as well as for the presence and the copy number of the construct in the genome by Southern blot analysis (data not shown; Fig. 2B, lane 15). Most of the transformants with one or more copies of the construct in the genome showed a robust steady-state level of GFP (data not shown and Fig. 2A and B, lane 15, and as a control, lane 16). For unknown reasons, the GFP protein expressed in C. cinerea migrated consistently by 1 to 2 kDa faster in SDS-PAGE than did the same coding region expressed in S. cerevisiae (data not shown). Regardless of its smaller size, GFP protein expressed in C. cinerea was functional as shown by a strong green fluorescence in the cytosol of GFP-expressing transformants (Fig. 2C, panels 15 and panel 16 as a control).
Silencing of GFP expression by introduction of a homologous hairpin RNA.
In a second step we set out to silence the GFP expression in two of these transformants by introducing a construct expressing a homologous hairpin RNA. Such hairpin constructs were shown to be effective in gene silencing in animals, plants, and other fungi (see the introduction for references), including the dimorphic basidiomycetous fungus, Cryptococcus neoformans (37). For construction we introduced an inverted repeat of the entire GFP coding region in a sense-antisense orientation into the benA-cgl2 construct described above by recombination in yeast (Fig. 1; see Materials and Methods for details). As a loop sequence we introduced the fourth intron of the C. cinerea benA gene (GenBank no. AB007761). It was shown that a spliceable intron positioned in the loop region of the hairpin enhanced the silencing effect of hairpin RNAs in plants and flies (31, 57). To test whether the presence of a spliceable intron in the loop region of the hairpin acted as an enhancer of silencing in C. cinerea, we manufactured two constructs: one containing the intron in a normal orientation and the other one containing the intron in a reverse orientation (Fig. 1; see Materials and Methods for details). After supplying these constructs with the C. cinerea pab1 gene as a selection marker (see Materials and Methods for details), two different KK7 transformants carrying the benA-cgl2::GFP construct in a single copy were each transformed with the two different silencing constructs and plasmid pPAB1.2 harboring only the pab1 marker gene as a mock control. Ten different transformants each of the silencing constructs and four transformants each of the mock controls were analyzed by various means. First, all of the 48 transformants and the original strains were checked for GFP expression on the protein level by immunoblotting the WCEs of vegetative mycelium by using anti-GFP antiserum (see Materials and Methods for details). Figure 2A shows the results for a representative series of transformants, together with the control strains or transformants. In 25 of the 40 transformants to be silenced the GFP protein levels in the WCE were significantly reduced (Fig. 2A and data not shown). It is generally not predictable what part of a plasmid is integrated in the C. cinerea genome upon transformation and integrated DNA can also be lost (4, 21). The apparent lack of silencing could therefore arise by integration of the marker gene without the silencing cassette, whereas the apparent silencing could also be due to loss of the GFP expression cassette. We applied both genomic PCR and Southern blot analysis of oidial DNA to check all 48 transformants for the presence of the GFP expression and silencing constructs. Figure 2B shows the results of this analysis for the series of transformants shown in Fig. 2A. As a first result, genome analysis revealed that all of the analyzed transformants still contained the GFP expression construct. In addition, the majority (9 of 15) of the nonsilenced transformants lacked the diagnostic 3.2-kb HindIII fragment indicative of the complete silencing cassette and contained either no additional (to the one of the GFP expression construct) GFP-containing fragment or additional GFP-containing fragments of different sizes. There were only 6 of 15 nonsilenced transformants that contained an apparently complete GFP silencing cassette. These results suggest that integration of the complete silencing cassette leads to silencing with a probability of at least 25 of 31 (80%). None of the mock transformants which received only the pab1 marker gene showed any signs of silencing. On the other hand, the presence of a complete silencing cassette was a prerequisite for silencing in that only 2 of 25 silenced transformants contained, instead of the diagnostic 3.2-kb HindIII fragment, a GFP-containing fragment of a different size (Fig. 2B, lane 5, and data not shown). With regard to the orientation of the intron in the silencing construct, the percentage of nonsilenced transformants was almost the same (8 of 20 and 7 of 20 for the normal and reverse orientations, respectively) in either case, but the number of cases with a complete silencing cassette among the nonsilenced transformants was slightly higher (4 of 7 versus 2 of 8 in case of the normal orientation) in the case of the reverse orientation of the intron (data not shown). The two original GFP transformants slightly differed in their susceptibility to silencing in that one of the transformants yielded more nonsilenced GFPhp transformants (11 of 20 versus 4 of 20), of which a similar portion contained a complete silencing cassette (4 of 11 versus 2 of 4) (data not shown).
As independent means of examining the GFP protein levels in the various transformants, mycelial pellets of selected transformants and control strains were grown in liquid culture and visualized by fluorescence microscopy. In accordance with the immunoblotting results, fluorescence of silenced transformants was reduced to the background fluorescence of strain KK7 devoid of any GFP expression construct (Fig. 2C and data not shown).
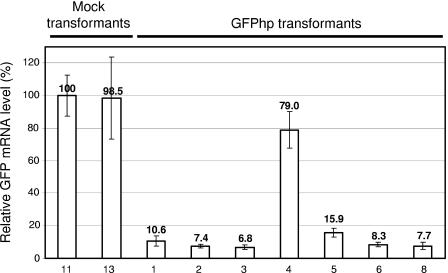
In principle, the observed silencing could occur at the levels of transcription or translation. In order to distinguish between these two possibilities and to determine the efficiency of silencing, the GFP mRNA levels in selected (at least six transformants of each series to be silenced and two of each mock transformant series) were determined by using quantitative real-time PCR. Figure 3 shows the results of the same series of transformants as shown in Fig. 2A and B together with the appropriate control strains. The results demonstrate that the GFP mRNA levels in the silenced transformants are reduced to 5 to 10% of those in the nonsilenced transformants, the mock transformants, and the original GFP transformant. These results show that the observed silencing is due to a decrease in the mRNA level. With regard to the orientation of the intron, no significant differences in the silencing efficiency were observed (data not shown). In addition, a single copy of the unchanged silencing construct was sufficient to confer this maximal degree of silencing since most of the silenced transformants revealed only one additional (to the one of the GFP expression construct) GFP-containing fragment in a BamHI or PmlI digest of genomic DNA (data not shown). In contrast to HindIII, these restriction endonucleases cleave the transformed plasmids only once: in the pab1 gene or in the GFP silencing cassette, respectively (Fig. 1 and data not shown).
FIG. 3.
Determination of GFP silencing efficiency using quantitative real-time PCR. Relative GFP mRNA levels were determined for the same representative series of transformants as in Fig. 2 (numbering refers to Fig. 2A and B). Values (above the histogram bars) are given as the percentage of a mock transformant. The analysis was performed as described in Materials and Methods. The error bars represent the standard deviations of three different measurements of the same cDNA.
In summary, our results suggest that expression of a homologous hairpin RNA is an efficient means to downregulate a specific mRNA in C. cinerea.
Simultaneous silencing of two endogenous isogenes by a single homologous hairpin RNA.
In order to test whether the RNA silencing approach would also be applicable for endogenous and developmentally regulated C. cinerea genes, we set out to silence the cgl2 gene coding for one of two isogalectins, CGL1 and CGL2, which are highly induced during fruiting body formation (2, 5). Due to the high sequence similarity of the cgl1 and cgl2 genes at DNA level (87% identity of the coding regions), we expected to see simultaneous silencing of both genes if the cgl2 silencing construct was functional. In the GFP silencing experiment, transcription of the hairpin RNA was under control of the same constitutive promoter as the target gene, and the presence of a spliceable intron in the loop region of the hairpin RNA did not significantly influence the silencing efficiency. Therefore, we designed a cgl2 silencing construct in which we introduced a loop region consisting of a tandem repeat of a 9-bp spacer region used by Ambion in commercial silencing vectors (pSilencer) upstream of an inverted copy of the complete cgl2 coding region between the last cgl2 codon and the translational stop. The resulting cgl2 hairpin construct was under the control of the endogenous cgl2 promoter region that comprised the complete intergenic region between cgl1 and cgl2 (Fig. 1), which was shown to be sufficient for developmental regulation (2). As in case of the GFP constructs, we placed the C. cinerea marker gene pab1 immediately upstream of the cgl2 silencing construct. Since we wanted to test the effect of eventual cgl1 and cgl2 silencing on fruiting, we transformed the cgl2 silencing construct and an appropriate vector control containing only the pab1 gene (plasmid 336) into the homokaryotic fruiting strain AmutBmut.
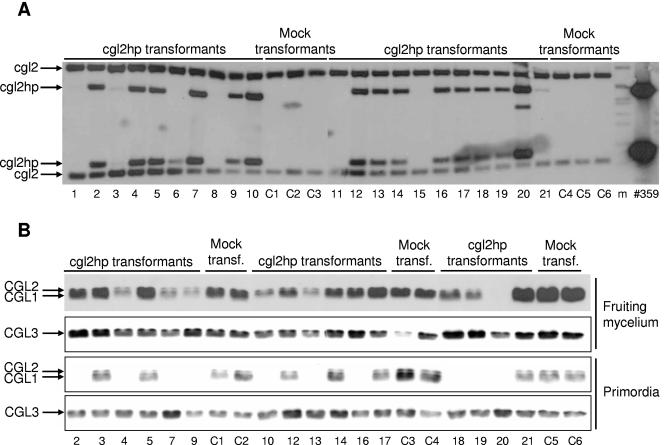
Southern analysis of oidial DNA revealed that 15 of the 21 analyzed cgl2hp-transformants carried the silencing cassette (Fig. 4A). Fourteen of these positive transformants, two negative transformants lacking the silencing cassette (transformants 3 and 21), and all six control transformants were cultivated under fruiting conditions and analyzed at the protein level by immunoblotting of WCEs prepared from fruiting mycelium and immature fruiting bodies (primordia) using a specific antiserum directed against CGL1 and CGL2 (see Materials and Methods for details). As a control an antiserum to a novel lectin, CGL3, which is also induced during fruiting body formation but whose coding region reveals much lower sequence similarity to cgl2 (53% identity; M. Wälti and A. Grünler, unpublished results), was used. Figure 4B shows that galectin genes cgl1 and cgl2 were slightly silenced in some (transformants 4, 7, 9, 10, 13, and 19) and efficiently in one (transformant 20) of the transformants at the stage of fruiting mycelium. At the stage of the primordia, however, 10 of the 14 analyzed transformants harboring a cgl2 silencing cassette revealed efficient silencing of both cgl1 and cgl2. Southern analysis of primordial DNA showed that the failure of silencing in transformants 5, 12, 14, and 17 was not due to a loss of the hairpin construct (data not shown). No significant change in CGL1/CGL2 protein levels was observed in any of the transformants lacking a silencing cassette (transformants 3, 21, and C1 to -6). CGL3 was not affected by expression of the cgl2 hairpin RNA since its levels were the same in all transformants. In conclusion, we demonstrated efficient, specific, and simultaneous silencing of two endogenous and developmentally regulated isogenes by coexpression of a hairpin RNA harboring sequences from only one of the two genes.
FIG. 4.
Molecular analysis of cgl2 silencing in C. cinerea strain AmutBmut. Individual transformants with cgl2-silencing construct 359 (lanes 1 to 21) were analyzed and compared to mock transformants carrying the vector control 336 (lanes C1 to C6). (A) Analysis of DNA integration by Southern blotting. Genomic DNA of the individual transformants was analyzed by using the restriction enzyme HindIII and a cgl2 hybridization probe (see Materials and Methods for details). The positions of the endogenous cgl2 and the introduced cgl2hp fragments are indicated. A HindIII digest of plasmid 359 served as a positive control. DNA-Molecular-Weight-Marker VII (Roche) was used as size standard (m). (B) Analysis of CGL1/2 and CGL3 protein levels in WCEs prepared from fruiting mycelium and primordia of the individual transformants (see Materials and Methods for details). A total of 5 μg of WCE from the indicated sample of the indicated transformants (numbering refers to panel A) was analyzed by immunoblotting with specific antisera against CGL1/2 and related protein CGL3 as described in Materials and Methods. In case of the anti-CGL1/2 blots, the HRP-coupled secondary antibody was detected either by using ECL (fruiting mycelium) or by using less sensitive chloronaphthol development (primordia). The positions of the individual proteins are indicated.
In addition to the expression studies described above, we examined the set of transformants phenotypically during the complete developmental pathway of fruiting body formation. None of the silenced transformants revealed any significant temporal or morphological difference compared to the control transformants (data not shown). The number and viability of basidiospores produced by the analyzed silenced fruiting bodies appeared normal (data not shown), suggesting that the galectins CGL1 and CGL2 are not necessary, at least for late stages of fruiting body development in C. cinerea. It is still possible that the galectins are involved in early steps of fruiting body formation as only one of the analyzed transformants (transformant 20) appeared to efficiently silence the cgl1 and cgl2 genes at this stage.
Hairpin RNA-induced cytosine methylation of transcribed and nontranscribed DNA at target and silencing loci.
The observed decrease in mRNA level upon expression of homologous hairpin RNAs (Fig. 3) could be due to interference at the transcriptional and/or posttranscriptional level. RNA silencing at the transcriptional level is often accompanied by cytosine and/or histone methylation of the involved chromatin (reviewed in reference 40). In order to determine whether the expression of hairpin RNAs in C. cinerea leads to any change in the cytosine methylation of the homologous DNA, we analyzed both target and silencing loci by Southern analysis using pairs of isoschizomers with different sensitivities to this DNA modification (see Fig. 1 for the location of the restriction sites and the hybridization probes used). The pairs of isoschizomers used, HpaII-MspI and Sau3AI-MboI, allow probing of different types of DNA methylation. Cleavage of the recognition sequence CCGG by HpaII is blocked by methylation of the second cytosine (CpG methylation), whereas MspI-mediated cleavage of the same sequence is insensitive to such a modification. On the other hand, both enzymes are sensitive to a methylation of the first cytosine (CpNpG methylation). Cleavage of the recognition sequence GATC by Sau3AI is blocked by cytosine methylation at overlapping CpG and CpNpG sites, whereas cleavage of the same sequence by MboI is not affected by this modification. On the other hand, MboI but not Sau3AI is blocked by adenine methylation by prokaryotic dam-methylase. In the analysis shown in Fig. 5A, we included three silenced GFPhp transformants (Fig. 2, transformants 1, 7, and 8), two mock transformants (Fig. 2, transformants 11 and 13), and the original GFP transformant and strain KK7 (Fig. 2, strains 15 and 16, respectively). In the first experiment, we applied both pairs of isoschizomers and a GFP hybridization probe (Fig. 5A, upper two panels). The appearance of high-molecular-weight bands in the HpaII-digested but not in the MspI-digested DNA suggests the presence of abundant CpG methylation in DNA regions containing GFP sequences. This methylation was only observed in the silenced GFPhp transformants and not in the control strains, suggesting that it is induced by expression of the homologous hairpin RNAs. Only a few minor high-molecular-weight bands in the MspI-digested DNA of silenced transformants compared to control strains were detected, suggesting the occurrence but low abundance of CpNpG methylation. Sau3AI digests did not reveal any difference compared to analogous MboI digests, suggesting the absence of adenine methylation and cytosine methylation at the few CpG or CpNpG sites overlapping with the GATC sequences in the GFP target and silencing loci. In order to test the locus specificity of the GFPhp-induced CpG methylation, we hybridized the same Southern blots with probes for the two marker genes, pab1 and trp1, and the cgl3 gene (Fig. 5A, lower three panels). No methylation was observed in the cgl3 locus in the silenced transformants, demonstrating the sequence specificity of the epigenetic changes. On the other hand, the appearance of HpaII-specific high-molecular-weight bands in the case of both marker genes demonstrates that CpG methylation occurred both at the target and silencing locus and was not restricted to the DNA regions containing GFP sequences but rather spread into the adjacent DNA regions, albeit to a lesser extent. Interestingly, CpNpG methylation was slightly more pronounced in these cases than in the case of the GFP-containing sequences. CpG methylation of both target and silencing loci was confirmed by isolating the HindIII fragments containing the respective loci and subjecting the isolated DNA fragments to the methylation analysis described above using the isoschizomer pair HpaII-MspI and the GFP hybridization probe (Fig. 5B; see Materials and Methods for details). The results suggest that methylation of the silencing locus was more pronounced than that of the target locus.
FIG. 5.
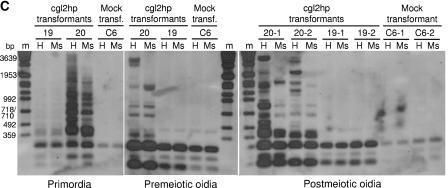
DNA methylation analysis in GFP and cgl2 silencing. Individual transformants of C. cinerea strains KK7 silenced for GFP (A and B) and AmutBmut silenced for cgl2 (C) were analyzed for methylation of their genomic DNA in comparison to corresponding mock transformants and original strains. DNA-Molecular-Weight-Marker VII (Roche) was used as a size standard (m). (A) DNA methylation analysis in GFP silencing. Oidial DNA of the indicated KK7 transformants and original strains (numbering refers to Fig. 2) was analyzed by Southern blotting with the indicated pairs of isoschizomeric restriction endonucleases, HpaII/MspI (H/Ms) or Sau3AI/MboI (S/Mb), and hybridization probes for the indicated genes (see Materials and Methods for details). (B) Cytosine methylation of GFP target and silencing locus. GFP target and silencing locus of indicated KK7 transformants (numbering refers to Fig. 2) were isolated as HindIII fragments as described in Materials and Methods and analyzed for cytosine methylation by Southern blotting using restriction endonucleases HpaII (H) and MspI (Ms) and a GFP hybridization probe. (C) Cytosine methylation in cgl2 silencing during fruiting body formation. Genomic DNA from different tissues (primordia, premeiotic oidia of mycelium before fruiting, and postmeiotic oidia of mycelium derived from two independent basidiospores [-1, -2] of respective fruiting bodies) of individual transformants (numbering refers to Fig. 4) was analyzed for cytosine methylation by Southern blotting using the restriction endonucleases HpaII (H) and MspI (Ms) and a cgl2 hybridization probe.
Cytosine methylation in the course of cgl2 silencing was examined analogously by analyzing genomic DNA of 2 silenced (transformants 19 and 20) and one mock transformant (C6) isolated from premeiotic oidia, primordia and postmeiotic oidia (Fig. 5C). The latter were collected from mycelia that had been derived from two independent basidiospores (indicated by the extension behind the numbers of the respective transformants). Methylation occurred in no developmental stage of the silenced transformant 19 but in all three analyzed developmental stages of transformant 20. The latter transformant carries multiple copies of the complete and also truncated versions of the cgl2 silencing cassette and revealed cgl2 silencing already at the mycelial stage (see Fig. 4A). Based on the intensity of the high-molecular-weight bands, methylation appeared to be least pronounced in premeiotic oidia and most pronounced in primordia. The difference in the pattern of high-molecular-weight bands between HpaII- and MspI-digested DNA demonstrates the occurrence of CpG methylation. However, the abundance of high-molecular-weight bands in the MspI lanes of transformant 20 versus transformant 19 suggests that CpNpG methylation was more pronounced in this case than in the GFP silencing experiment (see Fig. 5A).
In summary, these results suggest that silencing by constitutive high-level expression of hairpin RNAs in C. cinerea is accompanied by cytosine methylation of homologous and adjacent DNA regions, whereas the transient expression of hairpin RNAs does not lead to such epigenetic changes.
DISCUSSION
The number of reports about successful applications of RNA-mediated gene silencing (RNA silencing) as a reverse genetics tool in fungi is rising (see the introduction for references). In most studies RNA silencing was achieved by the transformation of heritable constructs expressing additional copies of the target gene (cosuppression), antisense RNA, or hairpin RNA, where the sequence homologous to the target gene forms the double-stranded stem structure. Systematic comparisons between the three different approaches in fungi and plants suggest that hairpin RNA is the most efficient trigger of RNA silencing in these organisms (19, 29, 57). This approach was also shown to be successful in Drosophila melanogaster (31), Caenorhabditis elegans (61), Trypanosoma brucei (55), and mammalian cells albeit in the latter case the stem regions of the hairpin loop have to be kept short in order not to elicit an interferon response (48). In plants and flies, the silencing efficiency could be further enhanced by introducing an intron in the hairpin loop (31, 57).
Here we demonstrate efficient silencing of both exogenous and endogenous genes by expression of homologous hairpin RNAs in the homobasidiomycete C. cinerea. The regions of homology forming the stem of the hairpin comprised in both cases the whole coding region of the target genes (450 and 711 bp, respectively) but may be shortened in future applications. In contrast to the studies in plants and flies, we did not observe any significant influence of the size of the loop or the presence of an intron in the loop. However, this conclusion is hampered by the fact that experimental attempts to demonstrate the splicing of the loop intron failed (data not shown) and that all silencing constructs used in the present study contained additional introns as a prerequisite for their expression. The lack of silencing in some of the transformants despite the presence of a complete silencing cassette may be due to a lack of expression caused by the surrounding DNA of the ectopic insertion point. Whereas in the GFP silencing experiment both target and silencing construct were constitutively expressed, cgl2 silencing was achieved by developmentally regulated (transient) coexpression of silencing construct and target gene(s).
The simultaneous silencing of cgl1 and cgl2 using the cgl2 hairpin construct is, apart from a report on Cladosporium fulvum (54), the only demonstration of this feature of RNA-mediated gene silencing in a fungus. The effect is possibly restricted to highly homologous genes (the coding regions of cgl1 and cgl2 are 87% identical) since the less homologous cgl3 gene (53% overall identity to cgl2 in its coding region) is not affected. However, it should be noted that such target specificity always depends on the actual sequence alignment since short regions of high identity can also lead to unwanted off-target effects.
With regard to the possible mechanisms of RNA-induced gene silencing in C. cinerea, translational inhibition can be excluded based on the observed reduction in mRNA levels. The observed cytosine methylation induced by high-level expression of the hairpin RNA but not by transient expression suggests that RNA silencing in C. cinerea occurs via posttranscriptional degradation of mRNA but can be reinforced at the transcriptional level by epigenetic changes. Interestingly, this RNA-induced DNA methylation in C. cinerea is not found in the two model fungi for RNA silencing: Schizosaccharomyces pombe and Neurospora crassa. In S. pombe, RNA-induced histone methylation leads to transcriptional silencing without detectable DNA methylation (62). In N. crassa, DNA methylation depends on histone methylation, but these processes are apparently independent of RNAi (9, 17, 60). On the other hand, the occurrence of de novo cytosine methylation in C. cinerea with preference for CpG sites and inheritance through meiosis is in agreement with previous reports (16, 65). In addition, transgene-induced cytosine methylation was reported for another homobasidiomycete, S. commune (53). The spreading of the RNA-induced DNA methylation in C. cinerea from homologous to adjacent nonhomologous sequences over a distance of several hundred base pairs suggests the involvement of RNAi-directed heterochromatin (including histone modification) may be in conjunction with RNA-directed DNA methylation (reviewed in reference 40). In this respect, the situation in C. cinerea and possibly in homobasidiomycetes in general may be more related to the one in animals and plants, where both pathways for RNA-induced epigenetic changes exist (reviewed in reference 40).
Gene targeting by RNA silencing is attractive even for fungi where classical gene knockout procedures are feasible. First of all, the hypermorphic mechanism of RNA silencing implies that the technique is also applicable to polyploid and polykaryotic organisms. The approach also offers solutions to the frequent lack of multiple marker genes in fungi. Simultaneous silencing of several unrelated genes by introducing a single chimeric hairpin construct has been demonstrated (15, 37, 43). In addition, as also shown in the present study, a single hairpin construct is able to simultaneously silence genes with high homology to the target gene. The latter application might be useful in C. cinerea since this fungus contains large families of highly homologous genes (25). The approach might also be useful for studying essential genes since the incomplete shutdown of expression may still support growth but be sufficient to cause a phenotype that can be indicative for the function of the gene; if the residual growth is too low under these conditions, the hairpin construct can be put under the control of an inducible promoter which allows cultivation of the organism under permissive conditions before studying its behavior under restrictive conditions. Finally, the use of expression cassettes minimizes cloning efforts since several hundred base pairs of the transcribed region appear to be sufficient for efficient silencing. Of course, there are also disadvantages of the RNA silencing approach. The most severe problem, in our opinion, is the fact that, due to its hypermorphic nature, the silencing phenotype cannot be complemented. This makes it impossible to perform more subtle studies of gene function, e.g., by introducing point mutations. On the other hand, the limited specificity of silencing might complicate the interpretation of phenotypes. A possible remedy of this problem, other than the careful selection of the homology region of the hairpin RNA, may be the overexpression of the intended target as a means of complementing the defect. Finally, a 90% reduction of gene expression may not be sufficient to cause a phenotype in all cases.
In accordance with the observed silencing, the C. cinerea genome sequence harbors at least two potential orthologs of each dicer, argonaute, and RdRP. On the other hand, in the Ustilago maydis genome sequence no obvious orthologs of these proteins are found and, indeed, Keon et al. (33) recently reported no evidence for antisense suppression in this basidiomycete despite a considerable expression level of the antisense RNA. Thus, predictions on the applicability of RNA silencing in a specific fungus based on its genome sequence are tempting but probably not very reliable. For example, Cryptococcus neoformans, another basidiomycete lacking both an obvious dicer and an RdRP ortholog, was reported to mediate gene silencing by the expression of hairpin and antisense RNAs (20, 37). Even more surprising, efficient RNA silencing by a hairpin construct was recently reported for the yeast S. cerevisiae, which is widely believed to be silencing deficient due to the absence of any orthologs of proteins described above (8). In conclusion, the presence of orthologs in the genome of an organism is a strong hint for the presence of RNA silencing, but their absence is not necessarily linked to a lack of this process. Therefore, experimental evidence for such a mechanism is required before considering its application as a reverse genetics tool.
Finally, it is remarkable that the lack of two proteins that accumulate to such a high level in the fruiting body is without obvious effect on the build up of this organ. Of course, it is still possible that there are subtle differences escaping our analysis thus far. Also, we cannot completely exclude that the galectins play a role during the initiation of fruiting body formation since, with the exception of a single transformant, expression of the cgl2 hairpin RNA under the control of the cgl2 promoter allowed residual expression of cgl1 and cgl2 at the stage of the fruiting mycelium. We will construct a constitutively expressed cgl2 silencing cassette to test this possibility. However, the chances for a phenotype are small since fruiting of this single transformant, in which cgl2hp is probably already constitutively expressed, also appeared normal.
During the revision of this manuscript, Namekawa et al. (45) described the silencing of a C. cinerea gene involved in meiosis using an analogous approach. The silencing construct used consisted of cDNA sequences without any introns, arguing against a crucial role of introns in such constructs. In addition to this study, these authors show that the silencing acts in trans in a heterokaryotic situation.
Acknowledgments
We thank W. Nickel (Heidelberg University Biochemistry Center, Heidelberg, Germany) for providing the anti-GFP antiserum and M. Bednar for excellent technical assistance.
REFERENCES
- 1.Almeida, R., and R. C. Allshire. 2005. RNA silencing and genome regulation. Trends Cell Biol. 15:251-258. [DOI] [PubMed] [Google Scholar]
- 2.Bertossa, R. C., U. Kües, M. Aebi, and M. Künzler. 2004. Promoter analysis of cgl2, a galectin encoding gene transcribed during fruiting body formation in Coprinopsis cinerea (Coprinus cinereus). Fungal Genet. Biol. 41:1120-1131. [DOI] [PubMed] [Google Scholar]
- 3.Binninger, D. M., L. Le Chevanton, C. Skrzynia, C. D. Shubkin, and P. J. Pukkila. 1991. Targeted transformation in Coprinus cinereus. Mol. Gen. Genet. 227:245-251. [DOI] [PubMed] [Google Scholar]
- 4.Binninger, D. M., C. Skrzynia, P. J. Pukkila, and L. A. Casselton. 1987. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 6:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulianne, R. P., Y. Liu, M. Aebi, B. C. Lu, and U. Kües. 2000. Fruiting body development in Coprinus cinereus: regulated expression of two galectins secreted by a non-classical pathway. Microbiology 146(Pt. 8):1841-1853. [DOI] [PubMed] [Google Scholar]
- 6.Burns, C., K. E. Gregory, M. Kirby, M. K. Cheung, M. Riquelme, T. J. Elliott, M. P. Challen, A. Bailey, and G. D. Foster. 2005. Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet. Biol. 42:191-199. [DOI] [PubMed] [Google Scholar]
- 7.Cerutti, H. 2003. RNA interference: traveling in the cell and gaining functions? Trends Genet. 19:39-46. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Q., Q. Ding, J. Thorpe, R. J. Dohmen, and J. N. Keller. 2005. RNA interference toward UMP1 induces proteasome inhibition in Saccharomyces cerevisiae: evidence for protein oxidation and autophagic cell death. Free Radic. Biol. Med. 38:226-234. [DOI] [PubMed] [Google Scholar]
- 9.Chicas, A., C. Cogoni, and G. Macino. 2004. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 32:4237-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogoni, C. 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55:381-406. [DOI] [PubMed] [Google Scholar]
- 11.Cottrell, T. R., and T. L. Doering. 2003. Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol. 11:37-43. [DOI] [PubMed] [Google Scholar]
- 12.Day, P. R., and G. E. Anderson. 1961. Two linkage groups in Coprinus lagopus. Genet. Res. 2:414-423. [Google Scholar]
- 13.De Backer, M. D., M. Raponi, and G. M. Arndt. 2002. RNA-mediated gene silencing in non-pathogenic and pathogenic fungi. Curr. Opin. Microbiol. 5:323-329. [DOI] [PubMed] [Google Scholar]
- 14.Del Sal, G., G. Manfioletti, and C. Schneider. 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 16:9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald, A., J. A. Van Kan, and K. M. Plummer. 2004. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 41:963-971. [DOI] [PubMed] [Google Scholar]
- 16.Freedman, T., and P. J. Pukkila. 1993. De novo methylation of repeated sequences in Coprinus cinereus. Genetics 135:357-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag, M., D. W. Lee, G. O. Kothe, R. J. Pratt, R. Aramayo, and E. U. Selker. 2004. DNA methylation is independent of RNA interference in Neurospora. Science 304:1939. [DOI] [PubMed] [Google Scholar]
- 18.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415.11313466 [Google Scholar]
- 19.Goldoni, M., G. Azzalin, G. Macino, and C. Cogoni. 2004. Efficient gene silencing by expression of double-stranded RNA in Neurospora crassa. Fungal Genet. Biol. 41:1016-1024. [DOI] [PubMed] [Google Scholar]
- 20.Gorlach, J. M., H. C. McDade, J. R. Perfect, and G. M. Cox. 2002. Antisense repression in Cryptococcus neoformans as a laboratory tool and potential antifungal strategy. Microbiology 148:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Granado, J. D., K. Kertesz-Chaloupková, M. Aebi, and U. Kües. 1997. Restriction enzyme-mediated DNA integration in Coprinus cinereus. Mol. Gen. Genet. 256:28-36. [DOI] [PubMed] [Google Scholar]
- 22.Hamada, W., and P. D. Spanu. 1998. Co-suppression of the hydrophobin gene HCf-1 is correlated with antisense RNA biosynthesis in Cladosporium fulvum. Mol. Gen. Genet. 259:630-638. [DOI] [PubMed] [Google Scholar]
- 23.Hammond, T. M., and N. P. Keller. 2005. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169:607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Hoegger, P. J., M. Navarro-Gonzalez, S. Kilaru, M. Hoffmann, E. D. Westbrook, and U. Kües. 2004. The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45:9-18. [DOI] [PubMed] [Google Scholar]
- 26.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 27.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James, T. Y., R. P. Boulianne, A. P. Bottoli, J. D. Granado, M. Aebi, and U. Kües. 2002. The pab1 gene of Coprinus cinereus encodes a bifunctional protein for para-aminobenzoic acid (PABA) synthesis: implications for the evolution of fused PABA synthases. J. Basic Microbiol. 42:91-103. [DOI] [PubMed] [Google Scholar]
- 29.Kadotani, N., H. Nakayashiki, Y. Tosa, and S. Mayama. 2003. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact. 16:769-776. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Kalidas, S., and D. P. Smith. 2002. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron 33:177-184. [DOI] [PubMed] [Google Scholar]
- 32.Kamada, T. 2002. Molecular genetics of sexual development in the mushroom Coprinus cinereus. Bioessays 24:449-459. [DOI] [PubMed] [Google Scholar]
- 33.Keon, J. P., J. W. Owen, and J. A. Hargreaves. 1999. Lack of evidence for antisense suppression in the fungal plant pathogen Ustilago maydis. Antisense Nucleic Acid Drug Dev. 9:101-104. [DOI] [PubMed] [Google Scholar]
- 34.Kertesz-Chaloupkova, K., P. J. Walser, J. D. Granado, M. Aebi, and U. Kües. 1998. Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet. Biol. 23:95-109. [DOI] [PubMed] [Google Scholar]
- 35.Kües, U. 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64:316-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Liu, H., T. R. Cottrell, L. M. Pierini, W. E. Goldman, and T. L. Doering. 2002. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lugones, L. G., K. Scholtmeijer, R. Klootwijk, and J. G. Wessels. 1999. Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol. Microbiol. 32:681-689. [DOI] [PubMed] [Google Scholar]
- 39.Ma, B., M. B. Mayfield, and M. H. Gold. 2001. The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 67:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matzke, M. A., and J. A. Birchler. 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6:24-35. [DOI] [PubMed] [Google Scholar]
- 41.May, G., L. Le Chevanton, and P. J. Pukkila. 1991. Molecular analysis of the Coprinus cinereus mating type A factor demonstrates an unexpectedly complex structure. Genetics 128:529-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meister, G., and T. Tuschl. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343-349. [DOI] [PubMed] [Google Scholar]
- 43.Mouyna, I., C. Henry, T. L. Doering, and J. P. Latge. 2004. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 237:317-324. [DOI] [PubMed] [Google Scholar]
- 44.Nakayashiki, H., S. Hanada, B. Q. Nguyen, N. Kadotani, Y. Tosa, and S. Mayama. 2005. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42:275-283. [DOI] [PubMed] [Google Scholar]
- 45.Namekawa, S. H., K. Iwabata, H. Sugawara, F. N. Hamada, A. Koshiyama, H. Chiku, T. Kamada, and K. Sakaguchi. 2005. Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology 151:3669-3678. [DOI] [PubMed] [Google Scholar]
- 46.Nicolas, F. E., S. Torres-Martinez, and R. M. Ruiz-Vazquez. 2003. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 22:3983-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101:12248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raponi, M., and G. M. Arndt. 2003. Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 31:4481-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robzyk, K., and Y. Kassir. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schuurs, T. A., E. A. Schaeffer, and J. G. Wessels. 1997. Homology-dependent silencing of the SC3 gene in Schizophyllum commune. Genetics 147:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segers, G. C., W. Hamada, R. P. Oliver, and P. D. Spanu. 1999. Isolation and characterisation of five different hydrophobin-encoding cDNAs from the fungal tomato pathogen Cladosporium fulvum. Mol. Gen. Genet. 261:644-652. [DOI] [PubMed] [Google Scholar]
- 55.Shi, H., A. Djikeng, T. Mark, E. Wirtz, C. Tschudi, and E. Ullu. 2000. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA 6:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, N. A., S. P. Singh, M. B. Wang, P. A. Stoutjesdijk, A. G. Green, and P. M. Waterhouse. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407:319-320. [DOI] [PubMed] [Google Scholar]
- 58.Spiering, M. J., C. D. Moon, H. H. Wilkinson, and C. L. Schardl. 2005. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swamy, S., I. Uno, and T. Ishikawa. 1984. Morphogenic effects of mutations at the A and B incompatibility factors in Coprinus cinereus. J. Gen. Microbiol. 130:3219-3224. [Google Scholar]
- 60.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 61.Tavernarakis, N., S. L. Wang, M. Dorovkov, A. Ryazanov, and M. Driscoll. 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24:180-183. [DOI] [PubMed] [Google Scholar]
- 62.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 63.Walser, P. J., U. Kües, and M. Aebi. 2002. A quick method to isolate pure DNA from asexual spores of Coprinus cinereus for screening approaches. Fungal Genet. Newsl. 49:15-16. [Google Scholar]
- 64.Zadra, I., B. Abt, W. Parson, and H. Haas. 2000. xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Appl. Environ. Microbiol. 66:4810-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zolan, M. E., and P. J. Pukkila. 1986. Inheritance of DNA methylation in Coprinus cinereus. Mol. Cell. Biol. 6:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]