Abstract
Infections of plants by the oomycete Phytophthora infestans typically result from zoospores, which develop from sporangia at cold temperatures. To help understand the relevant cold-induced signaling pathway, factors regulating the transcription of the zoosporogenesis-specific NIF (nuclear LIM-interactor-interacting factor) gene family were examined. Sequences required for inducing PinifC3 were identified by analyzing truncated and mutated promoters using the β-glucuronidase reporter in stable transformants. A 7-nucleotide (nt) sequence located 139 bases upstream of the major transcription start point (GGACGAG) proved essential for the induction of PinifC3 when sporangia were shifted from ambient to cold temperatures. The motif, named the cold box, also conferred cold inducibility to a promoter normally activated only during sexual development. An identical motif was detected in the two other zoosporogenesis-specific NIF genes from P. infestans and three Phytophthora sojae orthologues, and a closely related sequence was found in Phytophthora ramorum orthologues. The 7-nt motif was also found in the promoters of other zoosporogenesis-induced genes. The presence of a cold box-interacting protein in nuclear extracts of P. infestans sporangia was demonstrated using electrophoretic mobility shift assays. Furthermore, zoospore release and cold box-regulated transcription were stimulated by the membrane rigidizer dimethyl sulfoxide and inhibited by the membrane fluidizer benzyl alcohol. The data therefore delineate a pathway in which sporangia perceive cold temperatures through membrane rigidity, which activates signals that drive both zoosporogenesis and cold-box-mediated transcription.
Temperature represents an important environmental factor that affects all organisms. Cellular responses to temperature can be both immediate and long term. The former includes the rapid and transient heat or cold shock responses that are well described in both prokaryotes and eukaryotes (12, 16, 43, 48). Long-term mechanisms are exemplified by the cold acclimation phenomena of plants and fish, which involve altering lipid, osmolyte, protein, or other cellular components (48, 49). Many of these responses aim to enhance survival during stress, but specific temperature regimens may also be integral to normal development. For example, specific temperature treatments stimulate flowering in plants (37), pathogenic development in several fungi and bacteria that colonize animals (22, 30, 34), and appressorium formation in a phytopathogenic fungus (41).
Temperature also influences the germination of spores, particularly in the fungus-like eukaryotic microbes known as oomycetes. Asexual sporangia of the potato late blight pathogen Phytophthora infestans, for example, exhibit dual modes of germination when placed in liquid (23). Above 15°C, germ tubes typically emerge directly from sporangia. However, at lower temperatures the cytoplasm of sporangia is conditioned to cleave into six or more uninucleate biflagellated zoospores, which swim in search of a host (13, 42). Germination through zoospores, termed indirect germination, can occur in less than an hour and is not blocked by actinomycin or cycloheximide (10). The contribution of zoospores to disease is believed to be higher than that of directly germinating sporangia except at elevated temperatures, when the wall-lacking zoospores are more prone to desiccation. Therefore, Phytophthora sporangia appear to be designed to sense the environment and choose the optimal mode of germination.
Knowledge of the mechanisms regulating responses to cold in oomycetes or any organism is limited. Genes induced by cold temperatures have been described for many species (20, 35, 49) and some of the cognate transcription factors have been identified (6, 8). However, relatively little is known of the upstream components of such temperature perception pathways. An initial sensor of temperature in non-warm-blooded (poikilothermic) species, including plants, microbes, and some animals, has been proposed to be the plasma membrane (32). Its increased rigidity at lower temperatures may influence membrane-associated proteins to transmit signals to downstream targets, such as transcription factors for cold-regulated genes (9). Such membrane-associated proteins may include calcium channels (36), phospholipases (39), histidine kinases (2), and proteins that interact with the cytoskeleton (40).
In previous work we identified 70 genes induced during zoosporogenesis in P. infestans (46), a process which is induced conveniently in the laboratory by shifting sporangial suspensions from ambient to cold temperatures. Most such genes are up-regulated within minutes of cold treatment, before cytoplasmic reorganization is apparent within sporangia. Three of the zoosporogenesis-specific genes (PinifC1, PinifC2, and PinifC3) encode protein phosphatases known as nuclear LIM interactor-interacting proteins (NIFs). These are believed to control transcription by altering phosphorylation of the C-terminal domain of RNA polymerase, or by interacting with other regulators (18, 51). The PinifC genes appear to be controlled by phosphoinositols since their transcription is arrested by the phospholipase C inhibitor U-73122 and 2-aminoethoxydiphenylborate, which blocks calcium channels gated by inositol trisphosphate (45, 46). Therefore, the PinifC genes are promising targets for unraveling the mechanisms of cold perception in P. infestans.
This report demonstrates that a reduction in membrane rigidity stimulates both zoosporogenesis and expression of the PinifC genes. By analyzing truncated, chimeric, and mutated promoters, a 7-nucleotide (nt) motif sufficient for driving zoosporogenesis-induced transcription was identified, and a protein activity that binds the motif was documented.
MATERIALS AND METHODS
Growth of P. infestans.
Cultures of P. infestans isolate 1306 were maintained at 18°C, and developmental stages were obtained as described previously (25). Briefly, sporangia were purified by rubbing hyphal mats with a glass rod in water, followed by passage through 50-μm mesh to remove hyphal fragments. Cleavage (zoosporogenesis) was induced by placing sporangia in 10°C water for the times indicated in Results; in general, by 60 min cytoplasmic cleavage was visible in most sporangia, although zoospores had emerged from fewer than 10%.
Analysis of promoters in transgenic P. infestans.
Stable transformants were obtained using a protoplast method and stained histochemically for β-glucuronidase (GUS) activity (24). This involved derivatives of pOGUS (14) into which wild-type or mutagenized portions of the PinifC1, PinifC3, and M82 promoters were inserted into the ApaI and ClaI sites upstream of GUS. pOGUS also contains an npt gene for G418 selection.
Promoter fragments were obtained by PCR using the oligonucleotides listed in Table 1. The structures of the amplicons were verified by DNA sequencing. To generate the −254, −151, and −65 PinifC3 fragments, PCR employed primer NLIC3RC with C3F-255, C3-151, and C3-65, respectively. The M82 promoter was amplified using primers M82F and M82R. The 60-nt and 7-nt regions of the PinifC3 promoters destined for insertion upstream of M82 were amplified using C3-92Rp and C3-151 M, and M82C3-7F and M82R, respectively.
TABLE 1.
Oligonucleotides used for PCR
| Name | Sequence (5′ to 3′) |
|---|---|
| C3101MF | GATTGTCAAGGTTAGTGAGACAAGGCCCTTCCATG |
| C3101MR | AAGGGCCTTGTCTCACTAACCTTGACAATCAAAAC |
| C3111MF | GTAAGGTTTTTCGGTGACCTTGGCTGTCTCCAAG |
| C3111MR | AGACAGCCAAGGTCACCGAAAAACCTTACCACCC |
| C3121MF | CTCGAGGGTGTGCCTTGGGGGATTGTCAAGTGGC |
| C3121MR | TTGACAATCCCCCAAGGCACACCCTCGAGTCTCG |
| C3131MF | GCGGACGAGAAGATCTTTGTGTAAGGTTTTGATTG |
| C3131MR | AAAACCTTACACAAAGATCTTCTCGTCCGCTCTCG |
| C3141MF | CGAGGTCAGATATTCATCTCCTCGAGGGTGGTAA |
| C3141MR | CACCCTCGAGGAGATGAATATCTGACCTCGCCGTT |
| C3151MF | GGAGGAACGGATCTTGACTCGCGGACGAGACTCGA |
| C3151MR | TCTCGTCCGCGAGTCAAGATCCGTTCCTCCCTTAT |
| C392Rp | TTGGGCCCGAGACAGCCACTTGACAATC |
| C3F151 | AAGGGCCCCGAGGTCAGAGCGGACGAGAC |
| C3F254 | AAGGGCCCACCTGCCTCAGCGTAATTGTA |
| C3F65 | AAGGGCCCAAACTCGCTTTCAACGCACTG |
| M82C37F | AAGGGCCCGGACGAGTCTTCGGCAGCATTACCAC |
| M82F | AAGGGCCCTCTTCGGCAGCATTACCA |
| M82R | CCATCGATGGTTAAGGGTGGTCTTGGAA |
| NIFC1SF | ATGATGACGATGCCGACCAC |
| NIFC1SR | GCTTTGCTGTTATCTTCGGC |
| NIFC2SF | CAAGTCCAGTGGTCTATCGC |
| NIFC2SR | GCTCAATAGTGGTGCCCAAA |
| NIFC3SF | CTGAATATCACCTACCCACTG |
| NIFC3SR | GGAAACTGGCGGCAAGAGACT |
| NLIC12RA | AAGGGCCCGTCCGAAAGGGGTCTTGG |
| NLIC360F | CCCAAGCTTTATGGATAAGGGAGGAACGG |
| NLIC360R | GCTCTAGAAAGGGCCTTGGAGACAGC |
| NLIC3RC | CCATCGATTCTTTACTGAGATTGTCTCGTTA |
| GUSF | TAGAAACCCCAACCCGTGAA |
| GUSR | GCAATAACATACGGCGTGAC |
For site-directed mutagenesis of PiNifC3, a two-stage PCR strategy was used. The first involved separate reactions in which fragments extending leftward and rightward of the mutagenized sites were amplified, which were joined in the second stage using oligonucleotides NLIC1-2RA and NLIC3RC. The first/second-stage primer sets used to generate p151M, p141M, p131M, p121M, p111M, and p101M were NLIC12RA/C3151MR and NLIC3RC/C3151MF; NLIC12RA/C3141MR and NLIC3RC/C3141MF; NLIC12RA/C3131MR and NLIC3RC/C3131MF; NLIC12RA/C3121MR and NLIC3RC/C3121MF; NLIC12RA/C3111MR and NLIC3RC/C3111MF; and NLIC12RA/C3101MR and NLIC3RC/C3101MF, respectively.
RNA analysis.
RNA was extracted and blotted as described previously (25). Radiolabeled probes were generated from full-length cDNA clones or gene-specific fragments were amplified using primer pair NIFC1SF-NIFC1SR, NIFC2SF-NIFC2SR, or NIFC2SF-NIFC2SR. Signals were detected by phosphorimager analysis. Random amplification of cDNA end (RACE) products were generated with a kit (Invitrogen, Carlsbad, CA) using reverse transcription at 42°C and 50°C, cloned into pGEMT-EZ (Promega, Madison, WI), and sequenced. Reverse transcription-PCR was performed with primers GUSF and GUSR against DNase-treated RNA using a one-step kit (Invitrogen) and 35 cycles of PCR; in such assays, no bands were observed in controls lacking reverse transcriptase.
Sequence analyses.
Sequences were assembled using Seqman for Macintosh (DNASTAR, Madison, WI). Promoter alignments were performed using ClustalW. Phytophthora ramorum and Phytophthora sojae sequences were from assembly 1.0 of their draft genome sequences, which are available online from the Joint Genome Institute of the U.S. Department of Energy (Walnut Creek, CA).
Electrophoretic mobility shift assays.
Nuclear extracts were prepared using sporangia from 15 150-mm petri plates which were flooded with cold water, incubated at 10°C for 40 min to induce cleavage, homogenized in liquid nitrogen, suspended in buffer containing 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-KOH, pH 7.0, 10 mM MgCl2, 10 mM β-mercaptoethanol, 25% glycerol, and 0.5% Triton X-100, and processed as described but without Percoll centrifugation (33). DNA probes were amplified from PinifC3 using primers NLIC360F and NLIC360R. After end-labeling with 32P, 10,000 cpm was mixed with 10 μg of nuclear protein in binding buffer (125 mM HEPES, pH 7.9, 100 mM KCl, 25 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, 50% glycerol) for 15 min at 4°C, and electrophoresed in 4% acrylamide. For competition assays, unlabeled DNA was added 15 min before the labeled probe. Competitors were from either the native PinifC3 promoter, the mutated version present in plasmid p141M, or pBluescript II SK+ (Invitrogen).
RESULTS
Mapping transcription start points.
As described earlier (45), PinifC1, PinifC2, and PinifC3 are clustered within P. infestans and appear to be transcribed exclusively during zoosporogenesis. The PinifC1 and PinifC3 open reading frames are in opposite orientations, separated by a 1.4-kb intergenic region. PinifC2 resides 1.2 kb downstream of PinifC1. As a first step towards characterizing their promoters, transcription start sites were mapped by sequencing cDNAs primed by oligo(dT) or oligonucleotides binding near their translational start codons (5′ RACE).
The 5′ termini of the reaction products mapped to two regions, named TSP1 and TSP2. In PinifC1, PinifC2, and PinifC3, TSP1 is 116, 85, and 123 nt upstream of the ATG codon, respectively, while TSP2 is at 74, 42, and 75 nt, respectively. These data may reflect in vivo heterogeneity in initiation sites as is common in many species (21, 29), or artifacts during reverse transcription due to a high cytosine content between TSP1 and TSP2. Since nearly all 5′ RACE products ended at TSP2, including those assays performed at elevated temperatures, for simplicity TSP2 will be referred to as +1 in this article.
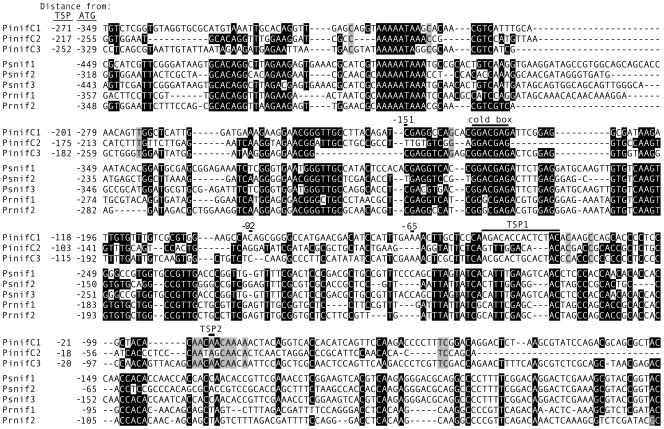
An alignment of sequences upstream of the ATG codons in the three P. infestans genes is shown in Fig. 1, along with the sites of TSP1 and TSP2. The figure also aligns promoters from putative P. ramorum and P. sojae homologues, which were extracted from draft genome data. Although the sequences from P. ramorum and P. sojae were not available during the experimental portion of our study, the significance of interspecies conservation at some sites will be discussed in more detail later. However, it is notable that only partial conservation exists near TSP1 and that the TSP2 region is well conserved within P. infestans (gray shading in Fig. 1) but not the other species. Neither region contains a consensus sequence present at the transcription start sites of some, but not all, genes from P. infestans (31).
FIG. 1.
Promoters of Phytophthora NIF genes. Shown (top to bottom) are alignments of portions of the promoters of the PinifC1, PinifC2, and PinifC3 genes of P. infestans, three putative homologues from P. sojae, and two from P. ramorum. The last five are extracted from sequences upstream of gene models estExt_fgenesh1_pg.C_260137, estExt_fgenesh1_pg.C_260134, estExt_fgenesh1_pg.C_260135, fgenesh1_pg.C_scaffold_35000061, and fgenesh1_pg.C_scaffold_35000062 from assembly 1.0 of their respective genome projects. Numbers in the left margin indicate distances from transcriptional and/or translational start points; only the latter are predicted for the P. sojae and P. ramorum genes. Black shading represents >70% identity among the eight genes, and gray shading indicates identity in P. infestans only. Indicated at the top of each panel for the P. infestans genes are approximate areas of predicted transcription start sites (TSP1 and TSP2), deletion endpoints used in functional studies of PinifC3, and the 7-nt cold box required for gene induction during zoosporogenesis. For PinifC1 and PinifC2, a majority of 5′ transcript termini appeared to be within TSP1, while for PinifC3 most were at TSP2.
Deletion analysis of the PinifC1/PinifC3 promoter region.
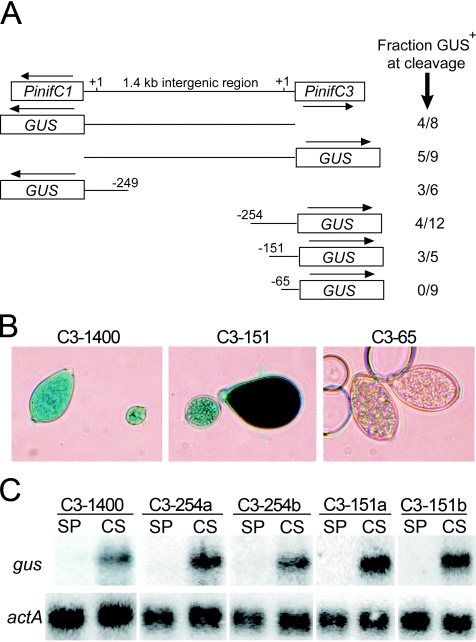
Truncated fragments of the PinifC1/PinifC3 intergenic region were used to determine whether that interval contained shared motifs required to express the two genes or two independent promoters. Fragments of approximately 350 nt were isolated from each end of the region, fused to the GUS reporter, and stably transformed into P. infestans. GUS was then scored histochemically in sporangia that had been placed at 10°C for 40 min to activate zoosporogenesis. In such “cleaving sporangia,” GUS expression was enabled by both the right and left fragments of the intergenic region in three of six and 4 of 12 transformants, respectively (Fig. 2A and B).
FIG. 2.
Deletion analyses of PinifC promoters. (A) Arrangement of PinifC1 and PinifC3 in the P. infestans genome, and GUS fusions made with fragments of their intergenic region. Indicated to the right for each fusion are the fraction of total transformants expressing GUS in cleaving sporangia; transformants in the negative class failed to express GUS at any life stage. Constructs represent, top to bottom, those with the entire 1.4-kb intergenic region, C1-1400::GUS or C3-1400::GUS; one with the 249-nt 5′ region of PinifC1, C1-249::GUS; and those with the 254-, 151-, and 65-nt regions 5′ to PinifC3, named C1-254::GUS, C1-151::GUS, and C1-65::GUS, respectively. (B) Representative transformants obtained using C3-1400::GUS and C3-151::GUS, which produce GUS in cleaving sporangia, and C3-65::GUS, which fails to express the reporter. (C) Blot analysis of RNA from freshly harvested undifferentiated sporangia (SP) and sporangia treated at 10°C for 40 min to induce cleavage (CS), hybridized with a probe for the GUS gene (gus) and actin (actA). The strains shown are a transformant expressing GUS from C3-1400::GUS, two strains with C3-254::GUS, and two strains with C3-151::GUS.
The failure of some transformants in these and later experiments to express GUS presumably reflects the general nature of transformation in P. infestans and not any feature specific to the particular PinifC3 promoter variant being tested. Even when the complete 1.4-kb intergenic region between PinifC1 and PinifC3 was used as a promoter, for example, only half of the transformants expressed GUS. A similar result was obtained using constitutive promoters such as ham34 and hsp70. Previous studies indicated that the failure to express GUS from constitutive promoters was usually due not to rearrangement or truncation of the transgene, but instead to position effects (27).
Other deletions narrowed the region required for PinifC3 transcription to an 86-nt interval. Transformants employing promoter fragments with 5′ endpoints 254 and 151 nt upstream of the transcription start site, but not one with the promoter fragment truncated to −65, produced GUS enzyme in cleaving sporangia (Fig. 2A and B). These transformants were also examined using RNA blotting, since enzyme levels may understate transcript accumulation during the rapid process of cleavage (45). In transformants utilizing either the −254 or −151 promoter deletions, strong mRNA induction was observed, similar to that obtained using the full 1.4-kb region (Fig. 2C). No GUS mRNA was observed using the −65 promoter (not shown).
Conserved blocks within these regions were revealed by promoter alignments (Fig. 1). Several are within the −65 to −151 interval of PinifC3, which, as described above, is sufficient to confer stage-specific transcription. These represent potential binding sites for a cold-activated transcription factor(s). They probably bind between −151 and −92 since little conservation exists downstream of the latter. Conserved blocks also reside within the −151 to −254 region, such as the AAAAATA at −206. The functions of such blocks are unclear, as their elimination did not block induction during cleavage. In theory, they may contribute quantitatively to transcript abundance. However, no trends in expression levels were noted in comparisons of transformants utilizing the full 1.4-kb intergenic region as a promoter versus the −254 or −151 fragment, although such analyses are difficult since in P. infestans neither the copy number nor the integration site of transgenes can be controlled. Integration-independent approaches for measuring promoters such as transient assays (26, 31), although more amenable to quantitative analysis, are not suited to studies of developmentally regulated promoters such as those of the PinifC genes.
Site-directed mutagenesis of PinifC3.
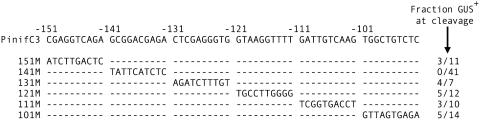
Based on the deletion and alignment studies, it seemed that bases required for activating transcription during cleavage might reside between −151 and −92. Therefore, a mutagenesis scheme was undertaken in which blocks of 10 bases spanning that interval were sequentially altered (Fig. 3). To maximize the nature of the mutations, adenines were replaced with cytosines, and guanines were replaced with thymines. Only changes in the −141 to −131 region yielded transformants that failed to transcribe GUS in cleaving sporangia, based on both histochemical staining and reverse transcription-PCR. Notably, this region contains a GGACGAG motif that is absolutely conserved in P. infestans and P. sojae and conserved at six of seven bases in P. ramorum. Alterations in other sites did not result in phenotypes appreciably different from that with the full 1.4-kb promoter, such as constitutive expression and drastic quantitative variation.
FIG. 3.
Site-directed mutagenesis of 60-nt region of PinifC3. Illustrated are the −92 to −151 portions of the wild-type gene and six mutant versions; dashes represent no change from the wild-type sequence. Indicated to the right are the fraction of total transformants expressing GUS in cleaving sporangia.
Using promoter chimeras to define the region needed for induction.
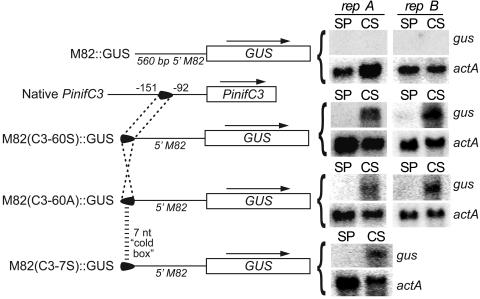
To better define the bases required for activating transcription during cleavage, and to test whether they act independently of other conserved portions of the PinifC3 promoter (such as those upstream of −151 or surrounding TSP2), a chimeric promoter approach was employed. This involved mating-induced gene M82 (17). Native M82 exhibits low expression in hyphae, strong expression in mating cultures, and very low expression in asexual sporangia and cleaving sporangia.
Initially, the 60-nt block between −151 and −92 was inserted at site −560 of the M82 promoter in its native 5′-to-3′ orientation. All GUS-expressing transformants (four of five total transformants) accumulated GUS mRNA in cleaving but not undifferentiated sporangia; this is shown in Fig. 4 for two strains containing plasmid M82(C3-60S)::GUS. A similar result was observed when the 60-nt region was in the opposite orientation, using plasmid M82(C3-60A)::GUS. It is therefore concluded that sequences within the 60-nt region determine cold inducibility, act in a position- and orientation-independent manner, and act through a mechanism dominant to the normal mode of M82 regulation.
FIG. 4.
Analysis of chimeric promoters. Shown on the left are, top to bottom, schematics of the M82 promoter fused to GUS; PinifC3, including the 60-nt region; a fusion between GUS, the M82 promoter, and the 60-nt region in the sense orientation; a fusion between GUS, the M82 promoter, and the 60-nt region in the antisense orientation; and a fusion between GUS, the M82 promoter, and the 7-nt cold box. On the right are blots of RNA from representative strains transformed with each plasmid (repA and repB), using the same tissues and probes as in Fig. 2.
Next, the conserved GGACGAG within the 10-nt region shown by site-directed mutagenesis to be critical for induction was tested in an M82 chimera. The relevant plasmid, M82(C3-7S)::GUS, resulted in cleavage-specific induction of the reporter in two of three transformants (Fig. 4). The 7-nt motif is therefore the presumed binding site for the transcription factor that activates PinifC3 when sporangia are shifted from ambient to cold temperatures. It was consequently named the cold box.
Presence of the cold box in other cleavage-associated genes.
Whether P. infestans genes coinduced with the PinifC family also contained the 7-nt motif was assessed. This involved obtaining 350 nt of sequences upstream of nine P. infestans genes (pic2, pic7, pic9, pic11, pic15, pic19, pic21, pic23, and pic27) (46), which were searched for the motif in the sense and antisense directions. The latter was appropriate since both orientations functioned in the chimeric promoter studies (Fig. 4). The 7-nt motif was detected four times, in four of the nine genes. This is significantly higher (P < 0.03) than would be expected for random DNA of the same G+C content (50.5% G+C), and higher (P < 0.05) than expected based on the frequency at which GGACGAG was found in either orientation in 20 Mb of random P. infestans DNA (once every 5,200 nt). The genome resources for P. infestans are currently insufficient to determine the frequency of the heptamer in its promoters; however, analyses of 75 kb of putative promoter sequences extracted from the draft genome sequence of P. sojae revealed that the heptamer is actually underrepresented in such regions, being present once every 11,800 nt.
To validate the importance of finding the 7-nt motif in the four zoosporogenesis-induced P. infestans genes, an attempt was made to check whether their P. ramorum or P. sojae homologues also contained the sequence in the same position relative to the ATG codon. This was difficult to calculate in most cases since homologues, based on BLASTN searches, were absent from both P. ramorum and P. sojae. However, one exception was pic23, in which GGACGAG was detected in the putative promoter regions of each of the P. infestans, P. ramorum (gene model fgenesh1_pg.C_ scaffold_29000029), and P. sojae (gene model estExt_fgenesh1_pg.C_130067) orthologues. Moreover, the 7-nt sequence was at approximately the same position in each of the three genes (−119, −135, and −125, respectively).
These results suggest that comparative promoter analysis may be useful for Phytophthora spp., but the findings should be considered preliminary. This is because the number of genes sampled was small, the transcription start sites of the P. ramorum and P. sojae genes are unmapped, and whether the genes from the latter two species are induced during cold treatment or zoosporogenesis is not known.
Nuclear factor that binds the cold box.
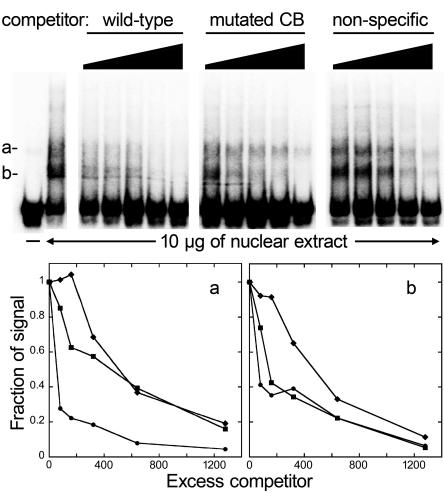
Electrophoretic mobility shift assays detected protein-binding activities that potentially regulate the PinifC loci. This involved nuclear extracts from cleaving sporangia and radiolabeled DNA corresponding to bases −151 to −92 of PinifC3. Two prominent shifted bands were detected (Fig. 5). Both appear to define specific DNA-protein interactions since they were eliminated effectively by unlabeled PinifC3 fragments but not by a nonspecific competitor of similar G+C content (54%) amplified from pBluescript II SK(+). The shift band of higher mobility (band b) was eliminated nearly equally by 60-nt DNAs representing the wild-type sequence and one with a mutated cold box. In contrast, the species of lower mobility (band a) was eliminated at least 20 times more effectively by the wild-type sequence than one with a mutated cold box. Therefore, band a likely results from association with the cold-box-binding transcription factor.
FIG. 5.
Electrophoretic mobility shift assay using the PinifC3 promoter. Labeled double-stranded DNA from the −151 to −92 region of PinifC3 was mixed with 10 μg of nuclear protein from cleaving sporangia and increasing amounts of unlabeled competitor DNA representing an approximately 80- to 1,280-fold excess. The competitors are, left to right, the normal −151 to −92 region (wild type); the −151 to −92 region containing an altered 7-nt cold box (mutated CB); and the pBluescript II SK(+) multiple cloning site (nonspecific). The graphs below the gel show quantitations of bands a and b (left and right panels, respectively), using increasing excess wild-type DNA competitor (circles), cold-box-mutated competitor (squares), and nonspecific competitor (diamonds).
Mechanisms for temperature perception in sporangia.
Temperature determines whether germination occurs by release of zoospores (which predominates below 15°C), or directly by elaboration of germ tubes through the sporangial wall. In other biological systems, membrane fluidity is proposed to be a primary sensor of temperature, acting by modifying the activity of membrane-associated proteins. The latter might regulate the cold-box-binding protein.
To investigate whether the mode of germination in P. infestans correlates with membrane fluidity, the effects of dimethyl sulfoxide (DMSO) and benzyl alcohol were measured. These compounds fluidize and rigidify plasma membranes, respectively, and have been used to study temperature-regulated processes in several organisms (38, 40).
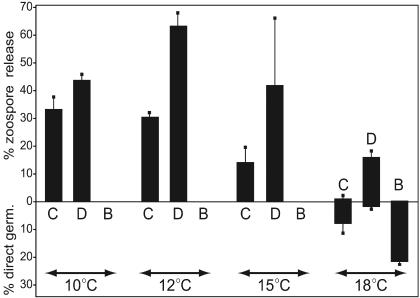
At every temperature tested, 35 mM DMSO (0.25%) stimulated zoospore release compared to that of the controls (Fig. 6). The effect was most striking at 18°C, when direct germination normally predominates, but DMSO caused 86% of germination events to involve zoospores. A similar trend was noted at higher and lower concentrations of DMSO; however, only the 35 mM data are reported here since it gave a noticeable effect and at that level forms only a minor component (1.4 mM) of the plasma membrane based on a log P (octanol-water partition coefficient) for DMSO of −1.35. In contrast to the effect of DMSO, 10 mM benzyl alcohol inhibited zoospore release at every temperature and stimulated direct germination at 18°C. However, benzyl alcohol did not enable direct germination to occur below 15°C.
FIG. 6.
Influence of modifiers of membrane rigidity on P. infestans germination. Sporangia (104/ml in water) were incubated for 30 min at the indicated temperatures in water as a control (C), 35 mM DMSO (D), or 10 mM benzyl alcohol (B). The average percentage of sporangia germinating directly or through zoospores was recorded; error bars represent standard deviations based on four independent replicates.
Effects of DMSO and benzyl alcohol on gene expression.
The influence of these chemicals on PinifC expression was consistent with a model in which transcription during zoosporogenesis is induced by the effect of cold on membrane fluidity. DMSO enhanced the level of mRNA from the three PinifC genes in sporangia kept at 12°C for 30 min, while benzyl alcohol suppressed expression (Fig. 7A). As illustrated in Fig. 7B, the same result was observed for GUS driven by a chimeric M82-PinifC promoter in plasmid M82(C3-60S)::GUS. It follows that membrane rigidification induces transcription by acting through the cold box.
FIG. 7.
Influence of modifiers of membrane rigidity on transcription. (A) Blot of RNA from undifferentiated sporangia (SP) and cleaving sporangia (CS) treated with 35 mM DMSO (D), 10 mM benzyl alcohol (B), or no chemical (−). Blots were hybridized with specific probes for PinifC1, PinifC2, PinifC3, or actA. (B) Same as panel A except that the RNA was from a transformant expressing M82(C3-60S)::GUS, and a probe for GUS (gus) was employed.
DISCUSSION
For more than a century, it has been recognized that temperature is a critical factor in the epidemiology of potato late blight (reference 4 and references therein). Early researchers noted that a low temperature (or at least the avoidance of warmer temperatures) was important for causing sporangia to release zoospores (13). Computer programs used to forecast late blight often incorporate measurements of the time that potato fields are at low temperature and give a higher score if conditions are optimal for zoospore emergence (47, 50). Since such programs are used by growers to plan fungicide applications, understanding how cool temperatures affect sporangia is of practical as well as fundamental importance.
This paper presents the first data suggesting that an increase in membrane rigidity is responsible for triggering the cellular and molecular events that characterize zoosporogenesis. Since zoospores can be produced in the absence of de novo transcription or translation (10), zoosporogenesis-induced genes such as the PinifC loci are probably required not for zoosporogenesis but instead for later stages, such as those between cyst germination and appressorium development. Nevertheless, zoosporogenesis and PinifC loci undoubtedly are regulated by common signals. Since previous research suggested that phospholipid messengers were required for both zoosporogenesis and PinifC3 expression (46), the signaling pathway may involve a membrane-bound phospholipase. The nature of that enzyme remains obscure since we have been unable to convincingly show that inositol trisphosphate accumulates in chilled sporangia (unpublished). Nevertheless, oscillations in the activity of a phospholipase, kinase, or other enzyme may be induced by conformational changes resulting from a transition of the plasma membrane from a liquid crystalline to a more rigid state, or by altered access to activators due to reduced phospholipid “flip-flop” between inner and outer leaflets (5).
The best-understood examples of the effect of membrane fluidity on signaling proteins involve phospholipases C and D in eukaryotes and histidine kinases in bacteria (39, 44). In Phytophthora spp., such enzymes or other membrane-associated proteins such as Ca2+ channels may participate in detecting cold, and indeed there is evidence that multiple mechanisms for transducing the temperature signal exist. Our previous study of the effect of inhibitors on zoosporogenesis-specific transcription supported the presence of four signaling mechanisms; these involved an inositol trisphosphate-regulated Ca2+ channel, an inositol trisphosphate-independent Ca2+ channel, a diacylglycerol-dependent pathway, and a Ca2+-plus-phospholipid-independent pathway (46). Multiple membrane-associated effectors may therefore exist, and participants independent of the membrane such as cold-induced microtubule disassembly cannot be discounted (1). A logical next step in identifying such effectors could involve determining how the transcription factor that binds the GGACGAG cold box becomes activated.
The cold box was identified based on functional tests performed prior to the availability of genome sequence data for P. ramorum and P. sojae. Nevertheless, a retrospective look at the feasibility of employing phylogenetic footprinting to identify binding sites for transcription factors in the genus is useful. In other systems, regulatory modules within promoters have been predicted by searching for regions that stand out from a nonconserved background (19). The best results are obtained using species of moderate overall genome similarity (60 to 70%), as in comparisons between humans and rodents or between Saccharomyces species (11, 28). Predictions are more difficult between close species, such as primates, since distinguishing functional from passive conservation is challenging (3). Our analyses suggest that the three Phytophthora species are suitably distant to perform phylogenetic footprinting, since the blocks of conservation evident in the alignment in Fig. 1 encompassed 51% of the promoter region. This compares with average identities in overall coding sequences between P. infestans and P. ramorum and between P. infestans and P. sojae of about 82% (unpublished data).
However, phylogenetic footprinting in Phytophthora spp. will benefit from more-accurate transcript mapping and expression data for all three species, and comprehensive genome data from P. infestans (fortunately, public funding for an eightfold draft sequence of P. infestans was recently obtained). For example, while the 7-nt cold box was 100% conserved between P. infestans and P. sojae, there was one mismatch in P. ramorum. This could indicate divergence in the specificity of the cold-box-binding protein. Alternatively, the NIF genes from P. ramorum may be expressed differently than the P. infestans genes; unfortunately, our ability to test this is impaired by quarantine restrictions concerning this invasive pathogen in California (15). Another limitation on comparative studies is a dearth of functional data on oomycete promoters, especially binding sites for general transcription factors. The PinifC genes contain the first stage-specific promoters to be dissected; however, even for constitutive promoters, only a few functional studies have been performed (7, 31).
Acknowledgments
This work was supported by awards to H.J. from the U.S. Department of Agriculture National Research Initiative.
We thank W. Prakob for sharing data from preliminary analyses of the M82 promoter.
REFERENCES
- 1.Abdrakhamanova, A., Q. Y. Wang, L. Khokhlova, and P. Nick. 2003. Is microtubule disassembly a trigger for cold acclimation? Plant Cell Physiol. 44:676-686. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar, P. S., A. M. Hernandez-Arriaga, L. E. Cybulski, A. C. Erazo, and D. de Mendoza. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boffelli, D., J. McAuliffe, D. Ovcharenko, K. D. Lewis, I. Ovcharenko, L. Pachter, and E. M. Rubin. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299:1391-1394. [DOI] [PubMed] [Google Scholar]
- 4.Bourke, P. M. A. 1964. Emergence of potato blight, 1843-46. Nature 203:805-808. [Google Scholar]
- 5.Bratton, D. L., V. A. Fadok, D. A. Richter, J. M. Kailey, S. C. Frasch, T. Nakamura, and P. M. Henson. 1999. Polyamine regulation of plasma membrane phospholipid flip-flop during apoptosis. J. Biol. Chem. 274:28113-28120. [DOI] [PubMed] [Google Scholar]
- 6.Brigulla, M., T. Hoffmann, A. Krisp, A. Volker, E. Bremer, and U. Volker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., and R. Roxby. 1997. Identification of a functional CT-element in the Phytophthora infestans piypt1 gene promoter. Gene 198:159-164. [DOI] [PubMed] [Google Scholar]
- 8.Chinnusamy, V., M. Ohta, S. Kanrar, B. H. Lee, X. Hong, M. Agarwal, and J. K. Zhu. 2003. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17:1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinnusamy, V., K. Schumaker, and J.-K. Zhu. 2004. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 5:225-236. [DOI] [PubMed] [Google Scholar]
- 10.Clark, M. C., D. L. Melanson, and O. T. Page. 1978. Purine metabolism and differential inhibition of spore germination in Phytophthora infestans. Can. J. Microbiol. 24:1032-1038. [DOI] [PubMed] [Google Scholar]
- 11.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 12.Craig, E. A. 1985. The heat shock response. CRC Crit. Rev. Biochem. 18:239-280. [DOI] [PubMed] [Google Scholar]
- 13.Crosier, W. 1934. Studies in the biology of Phytophthora infestans (Mont.) de Bary, memoir 155. Cornell University Agricultural Experiment Station, Ithaca, N.Y.
- 14.Cvitanich, C., and H. S. Judelson. 2003. A gene expressed during sexual and asexual sporulation in Phytophthora infestans is a member of the Puf family of translational regulators. Eukaryot. Cell 2:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson, J. M., S. Werres, M. Garbelotto, E. M. Hansen, and D. M. Rizzo. 2003. Sudden Oak Death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. 1:1-21. [Google Scholar]
- 16.Ermolenko, D. N., and G. I. Makhatadze. 2002. Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59:1902-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabritius, A.-L., C. Cvitanich, and H. S. Judelson. 2002. Stage-specific gene expression during sexual development in Phytophthora infestans. Mol. Microbiol. 45:1057-1066. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes, A. O., C. W. Campagnoni, K. Kampf, J. M. Feng, V. W. Handle, V. Schonmann, E. R. Bongarzone, S. Reyes, and A. T. Campagnoni. 2004. Identification of a protein that interacts with the golli-myelin basic protein and with nuclear LIM interactor in the nervous system. J. Neurosci. Res. 75:461-471. [DOI] [PubMed] [Google Scholar]
- 19.Fickett, J. W., and W. W. Wasserman. 2000. Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol. 11:19-24. [DOI] [PubMed] [Google Scholar]
- 20.Graumann, P., and M. A. Marahiel. 1996. Some like it cold: response of microorganisms to cold shock. Arch. Microbiol. 166:293-300. [DOI] [PubMed] [Google Scholar]
- 21.Gudas, J. M., J. L. Fridovich-Keil, M. W. Datta, J. Bryan, and A. B. Pardee. 1992. Characterization of the murine thymidine kinase-encoding gene and analysis of transcription start point heterogeneity. Gene 118:205-216. [DOI] [PubMed] [Google Scholar]
- 22.Ignatov, A., and E. J. Keath. 2002. Molecular cell biology and molecular genetics of Histoplasma capsulatum. Int. J. Med. Microbiol. 292:349-361. [DOI] [PubMed] [Google Scholar]
- 23.Judelson, H. S., and F. A. Blanco. 2005. The spores of Phytophthora: weapons of the plant destroyer. Nat. Microbiol. Rev. 3:47-58. [DOI] [PubMed] [Google Scholar]
- 24.Judelson, H. S., R. Dudler, C. M. J. Pieterse, S. E. Unkles, and R. W. Michelmore. 1993. Expression and antisense inhibition of transgenes in Phytophthora infestans is modulated by choice of promoter and position effects. Gene 133:63-69. [DOI] [PubMed] [Google Scholar]
- 25.Judelson, H. S., and S. Roberts. 2002. Novel protein kinase induced during sporangial cleavage in the oomycete Phytophthora infestans. Eukaryot. Cell 1:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judelson, H. S., B. M. Tyler, and R. W. Michelmore. 1992. Regulatory sequences for expressing genes in oomycete fungi. Mol. Gen. Genet. 234:138-146. [DOI] [PubMed] [Google Scholar]
- 27.Judelson, H. S., and S. L. Whittaker. 1995. Inactivation of transgenes in Phytophthora infestans is not associated with their deletion, methylation, or mutation. Curr. Genet. 28:571-579. [DOI] [PubMed] [Google Scholar]
- 28.Kolbe, D., J. Taylor, L. Elnitski, P. Eswara, J. Li, W. Miller, R. Hardison, and F. Chiaromonte. 2004. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 14:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leubner-Metzger, G., B. A. Horwitz, O. C. Yoder, and B. G. Turgeon. 1997. Transcripts at the mating type locus of Cochliobolus heterostrophus. Mol. Gen. Genet. 256:661-673. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H. 2002. Coregulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292:299-311. [DOI] [PubMed] [Google Scholar]
- 31.McLeod, A., C. D. Smart, and W. E. Fry. 2004. Core promoter structure in the oomycete Phytophthora infestans. Eukaryot. Cell 3:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murata, N., and D. A. Los. 1997. Membrane fluidity and temperature perception. Plant Physiol. 115:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagata, O., T. Takashima, M. Tanaka, and N. Tsukagoshi. 1993. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible taka-amylase A gene. Mol. Gen. Genet. 237:251-260. [DOI] [PubMed] [Google Scholar]
- 34.Nally, J. E., J. F. Timoney, and B. Stevenson. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phadtare, S., and M. Inouye. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp deletion strains of Escherichia coli. J. Bacteriol. 186:7007-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plieth, C. 1999. Temperature sensing by plants: calcium-permeable channels as primary sensors: a model. J. Membr. Biol. 172:121-127. [DOI] [PubMed] [Google Scholar]
- 37.Reeves, P. H., and G. Coupland. 2000. Response of plant development to environment: control of flowering by daylength and temperature. Curr. Opin. Plant Biol. 3:37-42. [DOI] [PubMed] [Google Scholar]
- 38.Regev, R., Y. G. Assaraf, and G. D. Eytan. 1999. Membrane fluidization by ether, other anesthetics, and certain agents abolishes P-glycoprotein ATPase activity and modulates efflux from multidrug-resistant cells. Eur. J. Biochem. 259:18-24. [DOI] [PubMed] [Google Scholar]
- 39.Ruelland, E., C. Cantrel, M. Gawer, J.-C. Kader, and A. Zachowski. 2002. Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sangwan, V., B. L. Orvar, J. Beyerly, H. Hirt, and R. S. Dhindsa. 2002. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31:629-638. [DOI] [PubMed] [Google Scholar]
- 41.Staples, R. C., H. C. Hoch, P. Freve, and T. M. Bourett. 1989. Heat shock-induced development of infection structures by bean rust uredospore germlings. Exp. Mycol. 13:149-157. [Google Scholar]
- 42.Sujkowski, L. S. 1987. The influence of temperature on Phytophthora infestans Mont. de Bary. J. Phytopathol. 120:271-275. [Google Scholar]
- 43.Sun, W., M. Van Montagu, and N. Verbruggen. 2002. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta 1577:1-9. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, I., D. A. Los, Y. Kanesaki, K. Mikami, and N. Murata. 2000. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tani, S., K. S. Kim, and H. S. Judelson. 2004. A cluster of NIF transcriptional regulators with divergent patterns of spore-specific expression in Phytophthora infestans. Fungal Genet. Biol. 42:42-50. [DOI] [PubMed] [Google Scholar]
- 46.Tani, S., E. Yatzkan, and H. S. Judelson. 2004. Multiple pathways regulate the induction of genes during zoosporogenesis in Phytophthora infestans. Mol. Plant-Microbe Interact. 17:330-337. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, M. C., N. V. Hardwick, N. J. Bradshaw, and A. M. Hall. 2003. Relative performance of five forecasting schemes for potato late blight (Phytophthora infestans). I. Accuracy of infection warnings and reduction of unnecessary, theoretical, fungicide applications. Crop Prot. 22:275-283. [Google Scholar]
- 48.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 49.Thomashow, M. F. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:571-599. [DOI] [PubMed] [Google Scholar]
- 50.Ullrich, J., and H. Schrodter. 1966. Das Problem der Vorhersage des Aufretens der Kartoffelkrautfaule (Phytophthorainfestans) und die Moglichkeit seiner Losung durch eine Negativprognose. Nachrbl. Dtsch. Pflanzenschutzd. 18:33-40. [Google Scholar]
- 51.Yeo, M., P. S. Lin, M. E. Dahmus, and G. N. Gill. 2003. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 278:26078-26085. [DOI] [PubMed] [Google Scholar]