Abstract
Trypanosomatids possess two homologues of Nopp140: a canonical Nopp140 and a Nopp140-like protein (TbNoLP) in which a GAR domain replaces the C-terminal SRP40 domain. Both are phosphorylated and coimmunoprecipitate with RNA polymerase I. Each paralogue has a distinct subnuclear localization, and depletion of TbNoLP produces an enlarged nucleolus in which TbNopp140-containing regions disperse. The restricted occurrence pattern of NoLP proteins reflects an intriguing convergence in evolution, suggestive of a function in nucleoplasmic small nucleolar ribonucleoprotein shuttling.
Nopp140 family proteins are characterized by a conserved C-terminal SRP40 domain (8). The precise function of these nucleolar proteins is unclear; however, there is evidence that they play a role in the biosynthesis of ribosomes and nucleocytoplasmic transport (5, 9). During ribosome biogenesis, Nopp140 is proposed to assist in the assembly of small nucleolar ribonucleoprotein (snoRNP) complexes (8, 17), which covalently modify precursor rRNA by 2′-O methylation or pseudouridylation. Different snoRNP complexes mediate these modifications (2, 12). 2′-O methylation complexes are guided by box C/D snoRNAs and contain the proteins fibrillarin and Snu13p and the two paralogues, Nop56p and Nop58p. Pseudouridylation complexes are guided by box H/ACA snoRNAs and contain Cbf5p, Gar1p, Nhp2p, and Nop10p. Intriguingly, Nopp140 interacts with both of these independent snoRNP complexes, yet it is not essential for the formation or catalytic activities of either complex in vitro or in vivo (16).
Trypanosoma brucei is unusual among eukaryotes as it harbors two separate RNA polymerase I (Pol I) compartments within its nucleus during certain stages of its life cycle. rRNAs are transcribed in the primary Pol I-containing organelle, the nucleolus. However, in bloodstream-form cells, genes encoding the variant surface glycoprotein are transcribed by Pol I in an extranucleolar expression site body (10). This presents trypanosomes with the unique challenge of linking the RNA polymerase II mRNA processing machinery to Pol I. Here, we describe two homologues of Nopp140 in trypanosomatids that are potentially involved in RNA processing and demonstrate an intriguing convergence in evolution.
Trypanosomatids encode two Nopp140-like proteins.
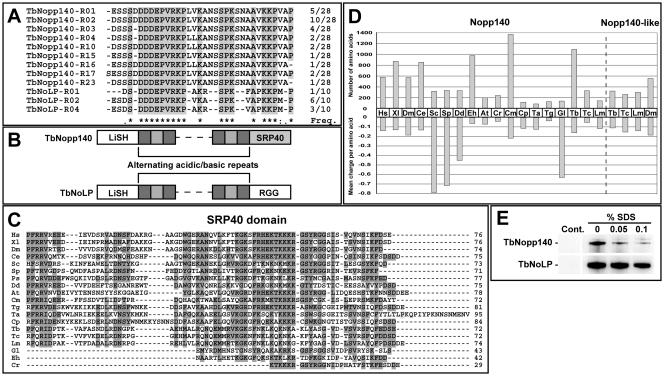
We generated a mouse monoclonal antibody (NUMAG) that detects the nucleolus and recognizes two proteins of ∼180 kDa and ∼80 kDa by Western blotting in T. brucei (11). Screening a T. brucei cDNA expression library identified clones encoding a repeat protein with a predicated mass of 128 kDa (accession no. XP_825186). We identified this protein as the T. brucei orthologue of Nopp140 (TbNopp140) due to the presence of the SRP40 C-terminal domain. Searching the completed T. brucei genome sequence revealed a related predicted protein of 54 kDa (accession no. XP_827326) This predicted protein, herein named TbNoLP (T. brucei Nopp140-like protein), is 19.7% identical across the N-terminal region and 51.4% identical across the repetitive central region to TbNopp140 but shares no detectable homology in the C-terminal domain (Fig. 1A and B). NUMAG binds to both proteins by shared epitopes in the central repetitive region (data not shown).
FIG. 1.
Properties of Nopp140 proteins. (A) Alignment of all central domain repeats from TbNopp140 and TbNoLP. (B) Domain diagram of TbNopp140 and TbNoLP. (C) Alignment of SRP40 domain across a broad range of eukaryotes: Hs, Homo sapiens; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Ps, Phytophthora sojae; Dd, Dictyostelium discoideum; At, Arabidopsis thaliana; Cm, Cyanidioschyzon merolae; Tg, Toxoplasma gondii; Ta, Theileria annulata; Cp, Cryptosporidium parvum; Tb, Trypanosoma brucei; Tc, Trypanosoma cruzi; Lm, Leishmania major; Gl, Giardia lamblia; Eh, Entamoeba histolytica; and Cr, Chlamydomonas reinhardtii. (D) Comparison between central domain size and mean charge per amino for Nopp140 and Nopp140-like proteins in the same set of organisms. (E) Immunoprecipitates from 108 cells extracted in RIPA buffer with the addition of sodium dodecyl sulfate (SDS; as indicated), and detected with the NUMAG monoclonal antibody. A Pol I-specific monoclonal was used (lanes 2 to 4) along with an unrelated control (Cont.; lane 1).
TbNopp140 and TbNoLP differ at their C terminus.
We used an iterative profile-based searching method to identify a large, diverse family of Nopp140 homologues in eukaryotes. This family is highly varied with respect to size: ranging from 25.9 kDa in the apicomplexan parasite Theileria to 155.2 kDa in the red alga Cyanidioschyzon (Fig. 1D). Characteristically, the C terminus of all Nopp140 proteins contains the SRP40 domain, which we redefine here as an ∼75-amino-acid region at the extreme C terminus (Fig. 1C). We generated a novel hidden Markov model for this domain and used it to identify several highly divergent homologues of this eukaryote-specific domain, including those of Entamoeba, Giardia, and Chlamydomonas, which have been truncated by up to ∼60% (Fig. 1C).
The central domains of Nopp140 homologues are poorly alignable. They do, however, share some traits. They are rich in amino acids A, D, E, K, P, and S (∼73% of all residues), which are arranged in repeats of alternating charge. Assuming all the available serines are phosphorylated (as has been shown for mammalian Nopp140) (8), the mean charge per residue for the repeat region is also well conserved (Fig. 1D). The central domains of four organisms from our analysis, Saccharomyces, Schizosaccharomyces, Dictyostelium, and Giardia, do not fit this pattern as their central domains are composed mainly of serine (61%, 50%, 32%, and 61%, respectively). The central domain of Saccharomyces Srp40p is phosphorylated to a much lesser extent than that of vertebrate Nopp140 (8), and this is likely to be true for other S-biased domains.
TbNoLP contains the N-terminal LisH and central repeats of a canonical Nopp140 protein. However it differs by replacement of the SRP40 domain with an RGG repeat containing a GAR domain similar to those in nucleolar proteins such as fibrillarin, nucleolin, and the kinetoplastid-specific Nopp44/46 (6). Intriguingly, a protein with an identical domain structure to TbNoLP is produced as a splice variant of DmNopp140 (15). Moreover, we have identified a hypothetical NoLP protein in Anopheles gambiae, suggestive of a more widespread distribution within insects. Additional Nopp140 variants can be found in other organisms, for example, the mammalian Treacher Collins syndrome protein, treacle (13). Like Nopp140, treacle contains an N-terminal LisH domain and a charged central domain, but lacks either an SRP40 domain or GAR domain at its C terminus.
TbNopp140 and TbNoLP have different subnuclear localizations.
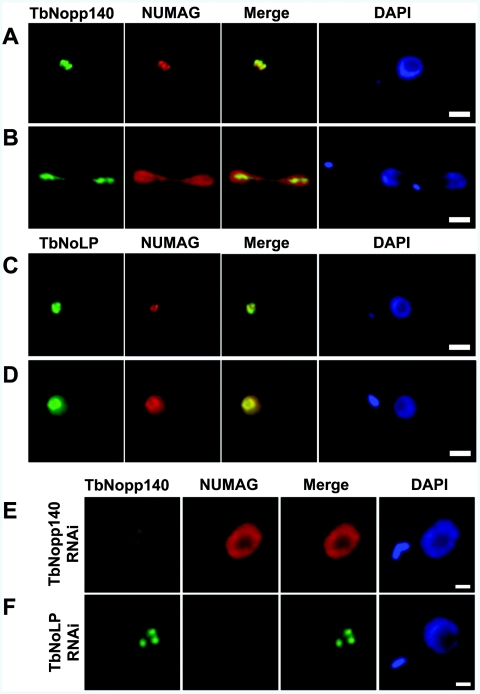
To determine the cellular localization of both TbNopp140 and TbNoLP, we used the NUMAG monoclonal antibody, which recognizes both proteins by Western blotting, and specific anti-TbNopp140 and anti-TbNoLP polyclonal antibodies. These antibodies behave differently following different fixation procedures. Following methanol fixation, NUMAG stains a subcompartment of the nucleolus (Fig. 2A) (11). Consistent with this, the affinity-purified anti-Nopp140 antiserum stains the same subcompartment (Fig. 2A and B). Anti-TbNoLP antiserum also stains the nucleolus after methanol fixation (Fig. 2C). The localization of TbNopp140 does not change following paraformaldehyde fixation (Fig. 2B). However, in paraformaldehyde-fixed cells, NUMAG and the anti-TbNoLP antiserum recognize both the nucleolus and the nucleoplasm (Fig. 2D). These patterns are maintained throughout the cell cycle, during which time the nucleolus remains intact and is segregated along the mitotic spindle (Fig. 2B). These contrasting fixation procedures reveal the dual localization of TbNoLP both within the nucleolus and the nucleoplasm.
FIG. 2.
Subnuclear localization of TbNopp140 and TbNoLP. (A and C) Cells fixed in −20°C methanol and labeled with NUMAG and either anti-TbNopp140 or anti-TbNoLP antibodies. (B and D) Cells fixed in paraformaldehyde and labeled with NUMAG and either anti-TbNopp140 or anti-TbNoLP antibodies. Scale bars, 2 μm. (E) RNAi-mediated knockdown of TbNoLP affects nucleolar morphology and localization of TbNopp140. Scale bars, 1 μm.
We could not detect TbNopp140 outside the nucleolus under any fixation condition used. As Nopp140 is a marker for Cajal bodies, we propose that functionally similar bodies are not present in trypanosomatids. Supporting this, we find no other proteins characteristic of Cajal bodies, such as Coilin, encoded within the trypanosome genomes. Additionally, we detected neither TbNopp140 nor an enrichment of TbNoLP in the bloodstream-form-specific extranucleolar Pol I expression site body (data not shown). Hence these proteins are unlikely to have a direct role in antigenic variation or Pol I-mediated variant surface glycoprotein gene transcription.
The behavior of the two Nopp140 homologues in immunofluorescence experiments is mirrored by their differential solubilization. The majority of TbNoLP, but not TbNopp140, is readily released from cells by treatment with nonionic detergent (see data in the supplemental material). Nucleolar TbNopp140 and a small pool of TbNoLP remain insoluble and are not released by treatment with RNase A or DNase I, suggesting that both are being held in the nucleolus by protein-protein interactions.
Both TbNopp140 and TbNoLP are phosphorylated and interact with Pol I.
Mammalian Nopp140 proteins are phosphorylated (9) and interact with Pol I (1). We asked whether this was true for both trypanosomatid Nopp140 homologues. In vivo labeling with [32P]phosphate followed by immunoprecipitation with the NUMAG antibody revealed that both TbNopp140 and TbNoLP are phosphorylated (see data in the supplemental material). We also performed coimmunoprecipitations using an anti-T. brucei Pol I monoclonal antibody (10) demonstrating an interaction with Pol I (Fig. 1E).
RNA interference (RNAi)-induced knockdown of TbNopp140 and TbNoLP.
To address the function of TbNopp140 and TbNoLP, we used an inducible RNAi system to specifically downregulate expression of each, both independently and simultaneously. Inducible knockdown of either TbNopp140 or TbNoLP resulted in minor slow-growth phenotypes detectable 4 days postinduction with simultaneous knockdown of both producing a more pronounced, earlier onset, phenotype (see data in the supplemental material). These growth defects phenocopy the yeast SRP40 knockout (18). Depletion of TbNopp140 resulted in no observable defects in nuclear or nucleolar morphology and had no impact on localization of TbNoLP. However, depletion of TbNoLP caused nucleolar enlargement, with Nopp140 foci becoming more dispersed (Fig. 2). Given the almost ubiquitous distribution of Nopp140 throughout Eukarya, it is surprising that neither deletion of the yeast protein, nor RNAi-mediated depletion of the trypanosomatid protein, produce more severe phenotypes.
NoLP proteins may function in snoRNP shuttling.
Trypanosomatids are unusual among eukaryotes as pseudouridylation is not limited to rRNA or sn(o)RNAs but is also found on every mRNA. This modification arises through trans-splicing of a pseudouridylated spliced leader sequence onto every pre-mRNA (7, 14). Drosophila also has an unusual capacity for RNA modification, exhibiting a large diversity of box H/ACA snoRNAs and the highest degree of pseudouridylation found in eukaryotes (3, 4). It is very suggestive that two organisms with such a dependence on RNA modification should have evolved NoLP proteins with a similar domain structure.
In trypanosomatids, pseudouridylation of spliced leader RNA is mediated by a canonical snoRNA-guided eukaryotic pathway (7), yet spliced leader RNA cannot be detected in the nucleolus where the modifying enzymes reside. We hypothesize that TbNoLP may recruit snoRNPs from the nucleolus so they can modify spliced leader RNA in the nucleoplasm. The enlargement of the nucleolus caused by TbNoLP knockdown may thus be indicative of accumulation of material within the nucleolus due to a breakdown in a TbNoLP-mediated snoRNP export process.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust and the E. P. Abraham Trust.
We acknowledge the technical support of Val Tilston and Lemy Tsikna.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Chen, H.-K., C.-Y. Pai, J.-Y. Huang, and N.-H. Yeh. 1999. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol. Cell. Biol. 19:8536-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henras, A. K., C. Dez, and Y. Henry. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14:335-343. [DOI] [PubMed] [Google Scholar]
- 3.Huang, Z. P., H. Zhou, H. L. He, C. L. Chen, D. Liang, and L. H. Qu. 2005. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA 11:1303-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang, Z. P., H. Zhou, D. Liang, and L. H. Qu. 2004. Different expression strategy: multiple intronic gene clusters of box H/ACA snoRNA in Drosophila melanogaster. J. Mol. Biol. 341:669-683. [DOI] [PubMed] [Google Scholar]
- 5.Ikonomova, R., T. Sommer, and F. Kepes. 1997. The Srp40 protein plays a dose-sensitive role in preribosome assembly or transport and depends on its carboxy-terminal domain for proper localization to the yeast nucleoskeleton. DNA Cell Biol. 16:1161-1173. [DOI] [PubMed] [Google Scholar]
- 6.Jensen, B. C., D. L. Brekken, A. C. Randall, C. T. Kifer, and M. Parsons. 2005. Species specificity in ribosome biogenesis: a nonconserved phosphoprotein is required for formation of the large ribosomal subunit in Trypanosoma brucei. Eukaryot. Cell 4:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8:237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier, U. T. 1996. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 271:19376-19384. [PubMed] [Google Scholar]
- 9.Meier, U. T., and G. Blobel. 1992. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 70:127-138. [DOI] [PubMed] [Google Scholar]
- 10.Navarro, M., and K. Gull. 2001. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759-763. [DOI] [PubMed] [Google Scholar]
- 11.Ogbadoyi, E., K. Ersfeld, D. Robinson, T. Sherwin, and K. Gull. 2000. Architecture of the Trypanosoma brucei nucleus during interphase and mitosis. Chromosoma 108:501-513. [DOI] [PubMed] [Google Scholar]
- 12.Tran, E., J. Brown, and E. S. Maxwell. 2004. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 29:343-350. [DOI] [PubMed] [Google Scholar]
- 13.Treacher Collins Syndrome Collaborative Group. 1996. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat. Genet. 12:130-136. [DOI] [PubMed] [Google Scholar]
- 14.Uliel, S., X. H. Liang, R. Unger, and S. Michaeli. 2004. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int. J. Parasitol. 34:445-454. [DOI] [PubMed] [Google Scholar]
- 15.Waggener, J. M., and P. J. DiMario. 2002. Two splice variants of Nopp140 in Drosophila melanogaster. Mol. Biol. Cell 13:362-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, C., C. C. Query, and U. T. Meier. 2002. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol. Cell. Biol. 22:8457-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Y., C. Isaac, C. Wang, F. Dragon, V. Pogacic, and U. T. Meier. 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol. Biol. Cell 11:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, Y., and U. T. Meier. 2003. Genetic interaction between a chaperone of small nucleolar ribonucleoprotein particles and cytosolic serine hydroxymethyltransferase. J. Biol. Chem. 278:23553-23560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.